Introduction
Cardiovascular disease (CVD) is the leading cause of
human mortality worldwide. The prevalence and incidence of coronary
heart disease (CHD) is increasing in numerous countries, including
China (1). CHD is a complex
disease that involves a variety of genetic and environmental
factors. Increased concentrations of low-density lipoprotein
cholesterol (LDL-C) in the blood is a well-established risk factor
for CHD (2) and the primary target
for lipid-lowering therapy in the prevention and treatment of CVD
(3).
Apolipoprotein B (apoB) is the main apolipoprotein
component of LDL-C and is important in the transport and metabolism
of LDL-C. APOB gene variants (including rs562338 and
rs503662) have been shown to be associated with an increased
concentration of LDL-C in European and American populations
(2,4,5).
High levels of plasma apoB and LDL-C were also shown to increase
the risk of CVD (6). A number of
studies recommend the use of apoB instead of LDL-C as a predictor
of CVD (7–9).
Cleaved fragments of lipoprotein(a) (LPA) protein
are capable of attaching to atherosclerotic lesions and thus
promote thrombogenesis (10,11).
Elevated levels of plasma LPA are associated with atherosclerosis
(12). LPA gene variants
may contribute to the risk of CHD by regulating the level of lipids
(13). SNP rs7767084 of the
LPA gene was observed to be associated with levels of LDL-C
(14) and CHD (15). However, rs7767084 of the LPA
gene was not associated with CHD risk in the Hispanic population
(16).
Lysosomal lipase A (LIPA) is able to catalyze the
hydrolysis of cholesteryl esters and triglycerides. SNP rs2246942
of the LIPA gene was demonstrated to be significantly
associated with the risk of CHD in European and South Asian
populations (17). A recent study
observed a significant association between rs1412444 of the
LIPA gene and risk of CHD in South Asian and European
populations (18).
The present study examined four gene variants
involved in lipid metabolism: rs562338 and rs503662 of the
APOB gene; rs7767084 of the LPA gene; and rs2246942
of the LIPA gene. We performed a case-control study to
investigate their contribution to the risk of CHD in the Han
Chinese population. A meta-analysis of three case-control studies
among Han Chinese individuals was also performed to establish the
role of LPA rs7767084 in CHD.
Materials and methods
Sample collection
A total of 483 unrelated inpatients were enrolled
from Lihuili Hospital (Ningbo, Zhejiang, China). In addition, 330
healthy individuals (including 86 males and 244 females; mean age,
63.44±9.21 years) who originated from Ningbo City (China) were
recruited as healthy controls. Patients were diagnosed by
standardized coronary angiography according to Seldinger’s method
(19), and assessed by at least
two independent cardiologists. Patients with CHD (n=290; 209 males
and 81 females; mean age, 61.98±9.49 years) demonstrated at least
one of the following criteria: i) ≥50% coronary artery occlusion of
one or more major coronary arteries (20); ii) a history of prior angioplasty;
or iii) a history of coronary artery bypass surgery. Non-CHD
controls (n=193; 98 males and 95 females; mean age, 58.65±9.36
years) with a <50% occlusion in the major coronary artery and no
atherosclerotic vascular disease were selected from the inpatient
population. All samples were obtained from individuals of Han
Chinese ethnicity originating from Ningbo, China. Subjects with
congenital heart disease, cardiomyopathy and liver or renal
diseases were excluded from the study. Blood samples were collected
in 3.2% citrate sodium-treated tubes and then stored at −80°C. The
protocol of our study was approved by the ethical committee of
Lihuili Hospital (Ningbo, Zhejiang, China). Written informed
consent was obtained from all patients.
SNP genotyping
Human genomic DNA was prepared from peripheral blood
samples using the nucleic acid extraction automatic analyzer
(Lab-Aid 820, Xiamen City, China) and was quantified using the
PicoGreen® double strand (dsDNA) DNA Quantification kit
(Molecular Probes Inc., Eugene, USA). Amplification was performed
on the ABI Geneamp® PCR System 9700, Dual 384-Well
Sample Block Module (Applied Biosystems, Foster City, CA) for the
polymerase chain reaction (PCR). PCR conditions included an initial
denaturation stage at 94°C for 15 sec, followed by 45 amplification
cycles (including 94°C for 20 sec, 56°C for 30 sec and primer
extension at 72°C for 1 min) and a final extension stage for 3 min
at 72°C. Primer extension for genotyping was performed on the
Sequenom® Mass-ARRAY iPLEX® (Sequenom, San
Diego, CA, USA) platform according to the manufacturer’s
instructions (21). The primer
sequences of the four SNPs used for the PCR assays are shown in
Table I.
 | Table IPrimer sequences for the four
SNPs. |
Table I
Primer sequences for the four
SNPs.
| SNP | Primer type | Primer
sequence |
|---|
| rs562338 | 1st PCR primer |
ACGTTGGATGCAGCCTAAATGTTCATTGTC |
| 2nd PCR primer |
ACGTTGGATGCCATGGTTTGCATACATCAC |
| rs503662 | 1st PCR primer |
ACGTTGGATGGATAGTATGTGTGGCAGAAG |
| 2nd PCR primer |
ACGTTGGATGACCCTGAATCTAACACAATC |
| rs7767084 | 1st PCR primer |
ACGTTGGATGTTGGGCTGGTCACTTTTGTC |
| 2nd PCR primer |
ACGTTGGATGGTGACTCCAGAATGAAGCTC |
| rs2246942 | 1st PCR primer |
ACGTTGGATGGGAAAGATCTCCAAGATAT |
| 2nd PCR primer |
ACGTTGGATGCTTATTTTTCCCTTGCCTCC |
Meta-analysis
We systematically searched databases, including
PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and the China
National Knowledge Infrastructure (CNKI; http://www.cnki.net/) for all available case-control
studies relating to rs7767084 of the LPA gene and CHD. We
selected studies based on the following criteria: i) The study was
an original study with an abstract in citation; ii) the study used
a case-control or a prospective design; iii) the study contained
complete data with genotype and allele frequencies; and iv) the
genotype frequencies of controls were reported in Hardy-Weinberg
equilibrium (HWE). Statistical heterogeneity between studies was
estimated using the Q-test. An I2 value >50%
indicated a significant heterogeneity among the studies included in
the meta-analysis (22). A
random-effects model based on the inverse-variance method was used
for the studies with high heterogeneity. Publication bias was
estimated using funnel plots (23).
Statistical analysis
HWE was analyzed using the Arlequin program (version
3.5) (24). Genotype and allele
frequencies were compared between CHD cases and each of the two
controls using the PASW Statistics 18.0 software (SPSS, Inc.,
Somers, NY, USA). The odds ratio (OR) with 95% confidence interval
(95% CI) were calculated using an online tool (http://faculty.vassar.edu/lowry/odds2x2.html). Power
and Sample Size Calculation software (v3.0.43) was used to
determine the power of the study (25). Correlation between genotype and the
extent of CHD disease was also performed using the PASW Statistics
18.0 software. Meta-analyses were performed using RevMan software
(version 5.1, Copenhagen: The Nordic Cochrane Centre, The Cochrane
Collaboration, 2011). P<0.05 was considered to indicate a
statistically significant result.
Results
Genetic tests
No departure from HWE was observed for the four
lipid metabolism gene variants. SNPs rs503662 and rs562338 of the
APOB gene were extremely rare in our samples (minor allele
frequencies <1%), therefore they were not included in the
further analysis. Genotype and allele frequencies of rs7767084 and
rs2246942 are shown in Table II.
No significant association with CHD was observed for the two SNPs.
Further genetic tests under the recessive and dominant inheritance
models were performed for rs7767084 and rs2246942, and the results
of these tests are shown in Table
III and Table IV,
respectively. In the recessive model, a significant association
between the rs2246942-GG genotype and risk of CHD was detected (CHD
cases versus healthy controls: P=0.04; OR=1.63; 95%
CI=1.02–2.60).
 | Table IIDistribution of genotype and allele
frequencies between CHD cases and the two control groups. |
Table II
Distribution of genotype and allele
frequencies between CHD cases and the two control groups.
| A, SNP
rs7767084 |
|---|
|
|---|
| Genotype
frequencies (%) | | | | Allele frequencies
(%) | | | |
|---|
|
| | | |
| | | |
|---|
| Group | TT | TC | CC | χ2 | P-value | HWE | C | T | χ2 | P-value | OR (95% CI) |
|---|
| CHD cases | 149 (51.4) | 121 (41.7) | 20 (6.9) | | | 0.49 | 161 (27.8) | 419 (72.2) | | | |
| Control 1 | 105 (54.4) | 74 (38.3) | 14 (7.3) | 0.55 | 0.76 | 0.85 | 102 (26.4) | 284 (73.6) | 0.21 | 0.65 | 1.07 (0.80,
1.43) |
| Control 2 | 171 (51.8) | 127 (38.5) | 32 (9.7) | 1.85 | 0.40 | 0.24 | 191 (28.9) | 469 (71.1) | 0.21 | 0.65 | 0.94 (0.74,
1.21) |
|
| B, SNP
rs2246942 |
|
| Genotype
frequencies (%) | | | | Allele frequencies
(%) | | | |
|
| | | |
| | | |
| Group | AA | AG | GG | χ2 | P-value | HWE | G | A | χ2 | P-value | OR (95% CI) |
|
| CHD cases | 115 (39.7) | 128 (44.1) | 47 (16.2) | | | 0.26 | 222 (38.3) | 358 (61.7) | | | |
| Control 1 | 69 (35.8) | 96 (49.7) | 28 (14.5) | 1.46 | 0.48 | 0.56 | 152 (39.4) | 234 (60.6) | 0.12 | 0.73 | 0.96 (0.73,
1.24) |
| Control 2 | 136 (41.3) | 158 (48.0) | 35 (10.7) | 4.22 | 0.12 | 0.27 | 228 (34.7) | 430 (65.3) | 1.75 | 0.19 | 1.17 (0.93,
1.48) |
 | Table IIIGenetic analysis of the two gene
variants under the recessive model. |
Table III
Genetic analysis of the two gene
variants under the recessive model.
| A, SNP
rs7767084 |
|---|
|
|---|
| Genotype
frequencies | | |
|---|
|
| | |
|---|
| Group | CC | TT+TC | P-value | OR (95% CI) |
|---|
| Total |
| CHD cases | 20 | 270 | | |
| Control 1 | 14 | 179 | 0.88 | 0.95 (0.47,
1.92) |
| Control 2 | 32 | 298 | 0.21 | 0.69 (0.39,
1.24) |
| Male |
| CHD cases | 18 | 191 | | |
| Control 1 | 4 | 94 | 0.15 | 2.21 (0.73,
6.73) |
| Control 2 | 6 | 80 | 0.64 | 1.26 (0.48,
3.28) |
| Female |
| CHD cases | 2 | 79 | | |
| Control 1 | 10 | 85 | 0.04 | 0.21 (0.05,
1.01) |
| Control 2 | 26 | 218 | 0.02 | 0.21 (0.05,
0.91) |
|
| B, SNP
rs2246942 |
|
| Genotype
frequencies | | |
|
| | |
| Group | GG | AA+AG | P-value | OR (95% CI) |
|
| Total |
| CHD cases | 47 | 243 | | |
| Control 1 | 28 | 165 | 0.61 | 1.14 (0.69,
1.89) |
| Control 2 | 35 | 294 | 0.04 | 1.63 (1.02,
2.60) |
| Male |
| CHD cases | 35 | 174 | | |
| Control 1 | 14 | 84 | 0.58 | 1.21 (0.62,
2.36) |
| Control 2 | 7 | 79 | 0.06 | 2.27 (0.97,
5.33) |
| Female |
| CHD cases | 12 | 69 | | |
| Control 1 | 14 | 81 | 0.99 | 1.01 (0.44,
2.32) |
| Control 2 | 28 | 215 | 0.43 | 1.33 (0.64,
2.77) |
 | Table IVGenetic analysis of the two gene
variants under the dominant model. |
Table IV
Genetic analysis of the two gene
variants under the dominant model.
| A, SNP
rs7767084 |
|---|
|
|---|
| Genotype
frequencies | | |
|---|
|
| | |
|---|
| Group | TC+CC | TT | P-value | OR (95% CI) |
|---|
| Total |
| CHD cases | 141 | 149 | | |
| Control 1 | 88 | 105 | 0.51 | 1.13 (0.78,
1.63) |
| Control 2 | 159 | 171 | 0.91 | 1.02 (0.74,
1.40) |
| Male |
| CHD cases | 98 | 111 | | |
| Control 1 | 46 | 52 | 0.99 | 1.00 (0.62,
1.61) |
| Control 2 | 38 | 48 | 0.67 | 1.11 (0.67,
1.85) |
| Female |
| CHD cases | 43 | 38 | | |
| Control 1 | 42 | 53 | 0.24 | 1.43 (0.79,
2.59) |
| Control 2 | 121 | 123 | 0.59 | 1.15 (0.69,
1.90) |
|
| B, SNP
rs2246942 |
|
| Genotype
frequencies | | |
|
| | |
| Group | AG+GG | AA | P-value | OR (95% CI) |
|
| Total |
| CHD cases | 175 | 115 | | |
| Control 1 | 124 | 69 | 0.39 | 0.85 (0.58,
1.23) |
| Control 2 | 193 | 136 | 0.67 | 1.07 (0.78,
1.48) |
| Male |
| CHD cases | 118 | 91 | | |
| Control 1 | 64 | 34 | 0.14 | 0.69 (0.42,
1.13) |
| Control 2 | 49 | 37 | 0.93 | 0.98 (0.59,
1.63) |
| Female |
| CHD cases | 57 | 24 | | |
| Control 1 | 60 | 35 | 0.31 | 1.38 (0.73,
2.61) |
| Control 2 | 144 | 99 | 0.07 | 1.63 (0.95,
2.81) |
CHD
CHD is the leading cause of human mortality
worldwide. However, higher rates of CHD are observed in males
compared with females across all age groups. In addition, coronary
disease occurs up to 10 years later in females (26). Due to the genetic and habitual
differences between genders, we performed a breakdown association
test by gender to examine whether gender as a factor may influence
the contribution of SNPs to CHD risk. Subsequently, we identified a
significant association at the genotype level (Table V). Further tests under the dominant
(Table IV) and recessive
(Table III) models were also
performed. In the recessive model, we observed a significant
protective effect of rs7767084-CC against CHD in females (CHD cases
versus non-CHD controls: P=0.04, OR=0.21, 95% CI=0.05–1.01; CHD
cases versus healthy controls: P=0.02, OR=0.21, 95% CI=0.05–0.91).
In addition, there was a correlation towards a significant
association of rs2246942-GG with CHD in males under the recessive
model (CHD cases versus healthy controls: P=0.06, OR=2.27, 95%
CI=0.97–5.33). No significant association was identified in the
dominant model.
 | Table VGenetic testing of the two gene
variants stratified by gender. |
Table V
Genetic testing of the two gene
variants stratified by gender.
| A, SNP
rs7767084 |
|---|
|
|---|
| Genotype
frequencies (%) | | | | Allele frequencies
(%) | | | |
|---|
|
| | | |
| | | |
|---|
| Group | TT | TC | CC | χ2 | P-value | HWE | C | T | χ 2 | P-value | OR (95% CI) |
|---|
| Male |
| CHD cases | 111 | 80 | 18 | | | 0.51 | 116 | 302 | | | |
| Control 1 | 52 | 42 | 4 | 2.26 | 0.32 | 0.21 | 50 | 146 | 0.34 | 0.56 | 1.12 (0.76,
1.65) |
| Control 2 | 48 | 32 | 6 | 0.30 | 0.86 | 0.83 | 44 | 128 | 0.29 | 0.59 | 1.12 (0.75,
1.67) |
| Female | | | | | | | | | | | |
| CHD cases | 38 | 41 | 2 | | | 0.02 | 45 | 117 | | | |
| Control 1 | 53 | 32 | 10 | 7.85 | 0.02 | 0.14 | 52 | 138 | 0.01 | 0.93 | 1.02 (0.64,
1.63) |
| Control 2 | 123 | 95 | 26 | 6.86 | 0.03 | 0.24 | 147 | 341 | 0.32 | 0.57 | 0.89 (0.60,
1.32) |
|
| B, SNP
rs2246942 |
|
| Genotype
frequencies (%) | | | | Allele frequencies
(%) | | | |
|
| | | |
| | | |
| Group | AA | AG | GG | χ 2 | P-value | HWE | G | A | χ 2 | P-value | OR (95% CI) |
|
| Male |
| CHD cases | 91 | 83 | 35 | | | 0.04 | 153 | 265 | | | |
| Control 1 | 34 | 50 | 14 | 3.51 | 0.17 | 0.52 | 78 | 118 | 0.58 | 0.45 | 0.87 (0.62,
1.24) |
| Control 2 | 37 | 42 | 7 | 4.37 | 0.11 | 0.30 | 56 | 116 | 0.87 | 0.35 | 1.20 (0.82,
1.74) |
| Female |
| CHD cases | 24 | 45 | 12 | | | 0.22 | 69 | 93 | | | |
| Control 1 | 35 | 46 | 14 | 1.11 | 0.57 | 0.86 | 74 | 116 | 0.48 | 0.49 | 1.16 (0.76,
1.78) |
| Control 2 | 99 | 116 | 28 | 3.26 | 0.20 | 0.49 | 172 | 314 | 2.70 | 0.10 | 1.35 (0.94,
1.95) |
In the CHD group, a correlation test was performed
between the two gene variants and the number of coronary arteries
with occlusion. No correlation between either of the two gene
variants and CHD severity was observed (Table VI). For rs7767084 of the
LPA gene, our meta-analysis included three case-control
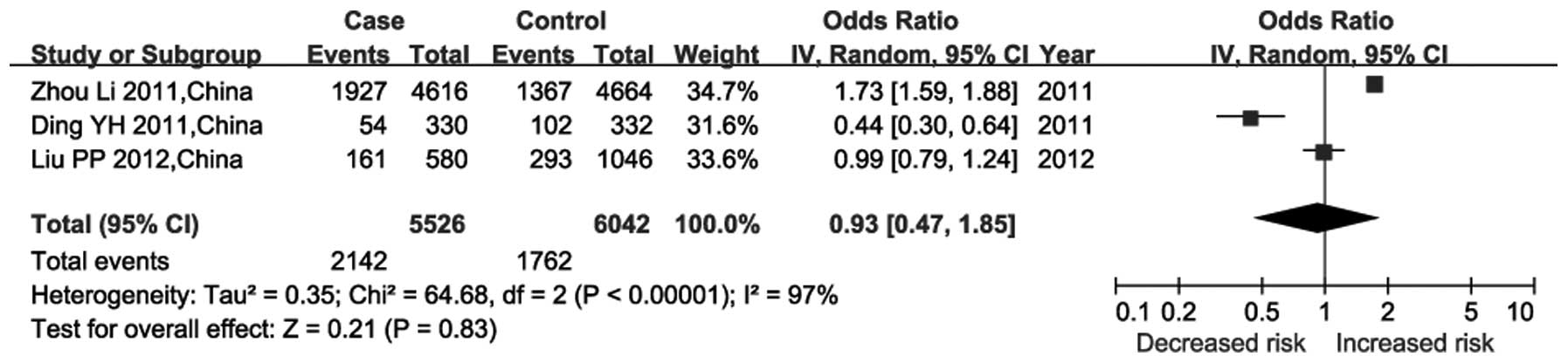
studies among the Han Chinese population (Fig. 1). The random-effects model was used
since significant heterogeneity was observed among these studies
(P<10−5; I2=97%). The results of the
meta-analysis indicated that rs7767084 was not associated with risk
of CHD (P=0.83; df=2; Z=0.21; combined OR=0.93; 95% CI=0.47–1.85).
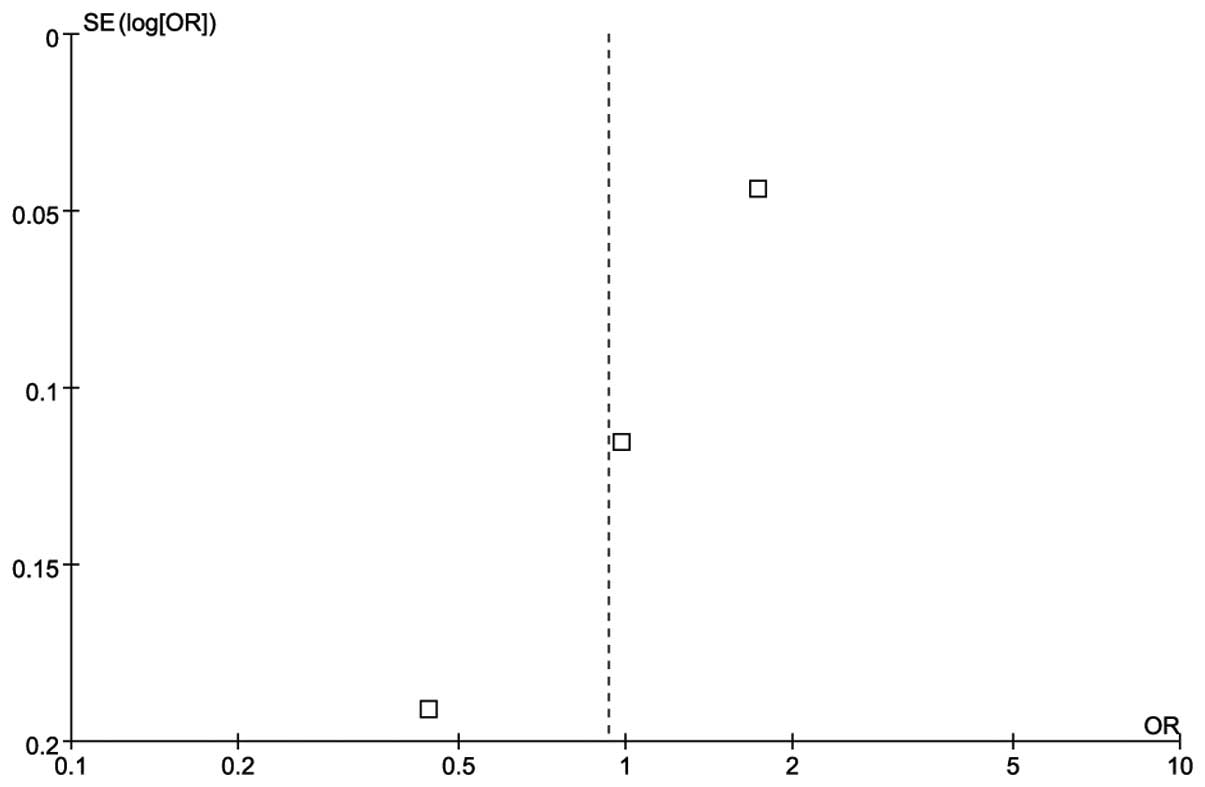
There was no publication bias according to the funnel plot
(Fig. 2).
 | Table VICorrelation between two gene variants
and the number of stenoses in CHD cases under the dominant and
recessive models. |
Table VI
Correlation between two gene variants
and the number of stenoses in CHD cases under the dominant and
recessive models.
| | Number of
stenoses | |
|---|
| |
| |
|---|
| Model | N | 1 | 2 | ≥3 | P-value |
|---|
| Dominant |
| rs7767084 |
| TT | 129 | 43 | 32 | 54 | 0.23 |
| TC+CC | 121 | 51 | 25 | 45 | |
| rs2246942 |
| AA | 98 | 34 | 24 | 40 | 0.56 |
| AG+GG | 152 | 60 | 33 | 59 | |
| Recessive |
| rs7767084 |
| CC | 15 | 5 | 1 | 9 | 0.26 |
| TC+TT | 235 | 89 | 56 | 90 | |
| rs2246942 |
| GG | 41 | 18 | 13 | 10 | 0.08 |
| AG+AA | 209 | 76 | 44 | 89 | |
Discussion
Two SNPs of the APOB gene (rs562338 and
rs503662) were detected at extremely low levels in our samples.
According to the information in the online HapMap dataset, the
minor allele frequencies in the HapMap-HCB (Han Chinese in Beijing)
are 1.1% for rs562338 and 1.1% for rs503662, in contrast with 22.5%
and 31.7% in the HapMap-CEU (CEPH; Utah residents with ancestry
from northern and western Europe), respectively (http://hapmap.ncbi.nlm.nih.gov/). These findings
support our data and implicate a significant ethnic difference for
the two SNPs. We were unable to observe a significant association
between rs7767084 and the risk of CHD in the case-control study and
the subsequent meta-analysis. This negative result in the Chinese
population agrees with the previous results of a large-scale
case-control study in the Hispanic population (16). Notably, a further breakdown test by
gender demonstrated that the rs7767084-CC genotype acts as a
protective factor against CHD in females under the recessive model
(Table II). This gender-dependent
result is novel and a further study on a larger scale is warranted.
In the present study, genotype rs2246942-GG of the LIPA gene
was shown to increase the risk of CHD by 63% in the recessive model
(CHD cases versus healthy controls: P=0.04). In addition, a
correlation between genotype rs2246942-GG with an increased risk of
CHD was observed in males under the recessive model (CHD cases
versus healthy controls: P=0.06, OR=2.27). Another SNP of the
LIPA gene (rs2246833) is located only 968 bp away from
rs2246942, and was previously implicated with an increased risk of
CHD in European and South Asian populations (17). Our results suggest that rs2246942
of the LIPA gene is likely to contribute to the risk of CHD
in the male Chinese population under the recessive model.
ApoB regulates the concentration of plasma LDL-C and
is directly associated with CHD (27). Recent studies have shown that
APOB polymorphisms (XbaI, MspI and 3′VNTR) are associated
with the risk of CHD in the Chinese population (28,29).
LPA may contribute to CVD through complex mechanisms that involve
proatherogenic and prothrombotic pathways (30,31).
LPA accumulates in the arterial wall of patients with CHD (32) and contributes to cholesterol
deposition (33). Previous studies
have reported that SNPs (rs10455872 and rs3798220) in the
LPA region are associated with a higher risk of CHD
(34–37). The LIPA gene encodes
lysosomal acid lipase (LAL) (38,39),
which hydrolyzes cholesteryl esters and triglycerides delivered to
the lysosome. A loss of LAL function results in the accumulation of
triglycerides and cholesteryl esters in the cell, and eventually
causes the formation of atherosclerotic plaques (40). LIPA gene mutations may cause
the cholesteryl ester storage disease and Wolman’s disease
(41), which often accompany
premature CVD.
CHD is a complex disease involving numerous genes.
Although a total of 813 Han Chinese individuals were included in
this study, it is not well powered for analyses, demonstrating that
the power of the test under the recessive model is 53.5% for
rs2246942-GG and 69% for rs7767084-CC in females. Meanwhile, the
ratio of males and females enrolled in our sample requires
adjustment to ensure a more balanced case-control study. All
P-values provided in this study were not corrected by the number of
tests, thus there is a chance that this study may include false
positive results.
In conclusion, a gender-dependent association
between rs7767084 of the LPA gene and CHD was observed in
the female Chinese population under the recessive model. In
addition, a possible explanation for the contribution of
rs2246942-GG of the LIPA gene to the risk of CHD in the male
Chinese population under the recessive model was identified.
Acknowledgements
The study was supported by grants from the National
Natural Science Foundation of China (nos. 31100919 and 30772155),
K.C. Wong Magna Fund in Ningbo University, Key Program of Education
Commission of Zhejiang Province (no. Z201017918), Zhejiang
Provincial Program for the Cultivation of High level Innovative
Health Talents, Natural Science Foundation of Zhejiang Province
(no. Y206608) and Youth and Doctor Foundation of Ningbo (no.
2005A610016).
References
|
1
|
Yu J, Huang J, Liang Y, et al: Lack of
association between apolipoprotein C3 gene polymorphisms and risk
of coronary heart disease in a Han population in East China. Lipids
Health Dis. 10:2002011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sandhu MS, Waterworth DM, Debenham SL, et
al: LDL-cholesterol concentrations: a genome-wide association
study. Lancet. 371:483–491. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baigent C, Keech A, Kearney PM, et al:
Efficacy and safety of cholesterol-lowering treatment: prospective
meta-analysis of data from 90,056 participants in 14 randomised
trials of statins. Lancet. 366:1267–1278. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Willer CJ, Sanna S, Jackson AU, et al:
Newly identified loci that influence lipid concentrations and risk
of coronary artery disease. Nat Genet. 40:161–169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lettre G, Palmer CD, Young T, et al:
Genome-wide association study of coronary heart disease and its
risk factors in 8,090 African Americans: the NHLBI CARe Project.
PLoS Genet. 7:e10013002011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
St-Pierre AC, Cantin B, Dagenais GR, et
al: Apolipoprotein-B, low-density lipoprotein cholesterol, and the
long-term risk of coronary heart disease in men. Am J Cardiol.
97:997–1001. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moss AJ, Goldstein RE, Marder VJ, et al:
Thrombogenic factors and recurrent coronary events. Circulation.
99:2517–2522. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Talmud PJ, Hawe E, Miller GJ and Humphries
SE: Nonfasting apolipoprotein B and triglyceride levels as a useful
predictor of coronary heart disease risk in middle-aged UK men.
Arterioscler Thromb Vasc Biol. 22:1918–1923. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Walldius G and Jungner I: The apoB/apoA-I
ratio: a strong, new risk factor for cardiovascular disease and a
target for lipid-lowering therapy - a review of the evidence. J
Intern Med. 259:493–519. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kamstrup PR, Tybjærg-Hansen A and
Nordestgaard BG: Genetic evidence that lipoprotein(a) associates
with atherosclerotic stenosis rather than venous thrombosis.
Arterioscler Thromb Vasc Biol. 32:1732–1741. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Szilágyi S, Péter A, Magyar MT, et al:
Recurrent arterial thrombosis associated with the antithrombin
basel variant and elevated lipoprotein(a) plasma level in an
adolescent patient. J Pediatr Hematol Oncol. 34:276–279.
2012.PubMed/NCBI
|
|
12
|
Terres W, Tatsis E, Pfalzer B, et al:
Rapid angiographic progression of coronary artery disease in
patients with elevated lipoprotein(a). Circulation. 91:948–950.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deshmukh HA, Colhoun HM, Johnson T, et al:
Genome-wide association study of genetic determinants of LDL-c
response to atorvastatin therapy: importance of Lp(a). J Lipid Res.
53:1000–1011. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Teslovich TM, Musunuru K, Smith AV, et al:
Biological, clinical and population relevance of 95 loci for blood
lipids. Nature. 466:707–713. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Trégouët DA, König IR, Erdmann J, et al:
Genome-wide haplotype association study identifies the
SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery
disease. Nat Genet. 41:283–285. 2009.PubMed/NCBI
|
|
16
|
Qi L, Ma J, Qi Q, et al: Genetic risk
score and risk of myocardial infarction in Hispanics. Circulation.
123:374–380. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Butterworth AS, Braund PS, Farrall M, et
al; IBC 50K CAD Consortium. Large-scale gene-centric analysis
identifies novel variants for coronary artery disease. PLoS Genet.
7:e10022602011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peden JF, Hopewell JC, Saleheen D, et al:
Coronary Artery Disease (C4D) Genetics Consortium: A genome-wide
association study in Europeans and South Asians identifies five new
loci for coronary artery disease. Nat Genet. 43:339–344. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Higgs ZC, Macafee DA, Braithwaite BD and
Maxwell-Armstrong CA: The Seldinger technique: 50 years on. Lancet.
366:1407–1409. 2005.PubMed/NCBI
|
|
20
|
Reilly MP, Li M, He J, et al:
Identification of ADAMTS7 as a novel locus for coronary
atherosclerosis and association of ABO with myocardial infarction
in the presence of coronary atherosclerosis: two genome-wide
association studies. Lancet. 377:383–392. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gabriel S, Ziaugra L and Tabbaa D: SNP
genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc
Hum Genet. Chapter 2(Unit 2): 122009.PubMed/NCBI
|
|
22
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Excoffier L and Lischer HE: Arlequin suite
ver 3.5: a new series of programs to perform population genetics
analyses under Linux and Windows. Mol Ecol Resour. 10:564–567.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dupont WD and Plummer WD Jr: Power and
sample size calculations. A review and computer program. Control
Clin Trials. 11:116–128. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Emslie C: Women, men and coronary heart
disease: a review of the qualitative literature. J Adv Nurs.
51:382–395. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Robinson JG, Wang S and Jacobson TA:
Meta-analysis of comparison of effectiveness of lowering
apolipoprotein B versus low-density lipoprotein cholesterol and
nonhigh-density lipoprotein cholesterol for cardiovascular risk
reduction in randomized trials. Am J Cardiol. 110:1468–1476. 2012.
View Article : Google Scholar
|
|
28
|
Li S, Lei ZW, Chen Z, et al: Relationship
between apolipoprotein E and apolipoprotein B polymorphisms in
youths with coronary heart disease. Zhonghua Yi Xue Yi Chuan Xue Za
Zhi. 20:241–243. 2003.(In Chinese).
|
|
29
|
Huang G, Zhong H, Re HM, et al: Coalition
of DNA polymorphisms of ApoB and ApoAI genes is related with
coronary artery disease in Kazaks. J Geriatr Cardiol. 9:33–37.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Boffa MB, Marcovina SM and Koschinsky ML:
Lipoprotein(a) as a risk factor for atherosclerosis and thrombosis:
mechanistic insights from animal models. Clin Biochem. 37:333–343.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kamstrup PR: Lipoprotein(a) and ischemic
heart disease - a causal association? A review. Atherosclerosis.
211:15–23. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rath M, Niendorf A, Reblin T, et al:
Detection and quantification of lipoprotein(a) in the arterial wall
of 107 coronary bypass patients. Arteriosclerosis. 9:579–592.
1989.PubMed/NCBI
|
|
33
|
Kiechl S and Willeit J: The mysteries of
lipoprotein(a) and cardiovascular disease revisited. J Am Coll
Cardiol. 55:2168–2170. 2010.PubMed/NCBI
|
|
34
|
Clarke R, Peden JF, Hopewell JC, et al;
PROCARDIS Consortium. Genetic variants associated with Lp(a)
lipoprotein level and coronary disease. N Engl J Med.
361:2518–2528. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chasman DI, Shiffman D, Zee RY, et al:
Polymorphism in the apolipoprotein(a) gene, plasma lipoprotein(a),
cardiovascular disease, and low-dose aspirin therapy.
Atherosclerosis. 203:371–376. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luke MM, Kane JP, Liu DM, et al: A
polymorphism in the protease-like domain of apolipoprotein(a) is
associated with severe coronary artery disease. Arterioscler Thromb
Vasc Biol. 27:2030–2036. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schunkert H, König IR, Kathiresan S, et
al: Large-scale association analysis identifies 13 new
susceptibility loci for coronary artery disease. Nat Genet.
43:333–338. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Anderson RA and Sando GN: Cloning and
expression of cDNA encoding human lysosomal acid lipase/cholesteryl
ester hydrolase. Similarities to gastric and lingual lipases. J
Biol Chem. 266:22479–22484. 1991.PubMed/NCBI
|
|
39
|
Anderson RA, Rao N, Byrum RS, et al: In
situ localization of the genetic locus encoding the lysosomal acid
lipase/cholesteryl esterase (LIPA) deficient in Wolman disease to
chromosome 10q23.2-q23.3. Genomics. 15:245–247. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zschenker O, Illies T and Ameis D:
Overexpression of lysosomal acid lipase and other proteins in
atherosclerosis. J Biochem. 140:23–38. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Klima H, Ullrich K, Aslanidis C, et al: A
splice junction mutation causes deletion of a 72-base exon from the
mRNA for lysosomal acid lipase in a patient with cholesteryl ester
storage disease. J Clin Invest. 92:2713–2718. 1993. View Article : Google Scholar : PubMed/NCBI
|