Introduction
Lung cancer is the most common cause of
cancer-related mortality worldwide, and a poor five-year survival
rate (15%) highlights the importance of gaining an improved
understanding of this malignancy to improve prevention, diagnosis
and treatment (1). A previous
study showed that tumor initiation and propagation is induced by a
specific population of self-renewing tumor cells, termed cancer
stem cells (CSCs) or tumor propagating cells (2). Previous studies have indicated that
similar to a number of solid tumors, human lung cancers,
particularly non-small cell lung carcinoma (NSCLC), harbor CSC
populations (3–5).
Certain molecules are being investigated as putative
markers of CSCs in malignancies, including lung cancer (6). CD133 (also termed prominin-1), a
cell-surface glycoprotein comprising five transmembrane domains and
two large glycosylated extracellular loops, has been previously
used as a biomarker to separate cancer stem cells from a variety of
solid human tumors, including brain (7), breast (8), liver (9), pancreas (10), colon (11) and lung (12,13)
tumors. Although CD133 has been used to enrich lung cancer CSCs in
several studies, the potential for the use of CD133 as a key marker
remains unclear (14–16). In addition, the use of CD133 as a
prognostic marker in lung cancer has not yet been confirmed due to
conflicting studies (17–22). To investigate these divergent
observations, the involvement of CD133 was analyzed using NSCLC
cells and clinical specimens to evaluate a possible correlation
between CD133 expression and clinical-pathological variables in
patients with NSCLC. To the best of our knowledge, this was the
first study to investigate NSCLC tumor and non-tumor tissue, with
respect to CSC-marker expression profiles and their relevance to
the clinicopathological parameters of lung cancer. The current
results demonstrated that elevated expression of CD133 was
associated with early clinical stages, larger tumor size and poor
differentiation of NSCLC.
Materials and methods
Cell lines and isolation of
CD133-positive cells
NSCLC cells, A549, H1299, SPC-A1, SK-MES-1 and a
human bronchial epithelial cell line (HBE), were cultured
separately in RPMI-1640 medium (Gibco, Invitrogen Life
Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine
serum (FBS), 100 IU/ml penicillin and 100 mg/ml streptomycin in a
5% CO2 humidified atmosphere at 37°C. For isolation of
CD133-positive cells, the parental H1299 cells were labeled with 1
ml CD133 per liter micromagnetic beads per million cells, using a
CD133 cell isolation kit (CD133 MicroBead kit; Miltenyi Biotec,
Auburn, CA, USA). The isolated CD133-positive cells were cultured
in medium consisting of serum-free Dulbecco’s modified Eagle’s
medium (DMEM)/F-12 medium (Gibco, Invitrogen Life Technologies)
supplemented with 20 ng/ml human epidermal growth factor (EGF)
(PeproTech, Rocky Hill, NJ, USA), 10 ng/ml human basic fibroblast
growth factor (bFGF; PeproTech) and 2% B-27 serum-free supplements
(Invitrogen Life Technologies). The medium was replaced or
supplemented with fresh growth factors twice weekly until floating
aggregates formed.
To determine the percentage of single cells capable
of regenerating novel spheres, cells were plated at a density of
1,000 cells/ml in ultralow-attachment 6-well plates (Corning Life
Sciences, Union City, CA, USA) to obtain the novel spheres. The
total number of tumor spheres was counted following 10 days of
culture. Efficiency of sphere formation was calculated by the
following equation: (Total number of spheres formed / total number
of living cells seeded) × 100.
Patients and tissue samples
NSCLC and matched adjacent normal biopsies were
obtained from 30 patients (see supplementary Table I) undergoing pulmonary resection at
the Zhoushan Hospital of Zhejiang province (Zhoushan, Zhejiang,
China) between January 2009 and May 2010. All samples were
collected with informed consent obtained at an internal review. The
ethics board committees of the Zhoushan Hospital of Zhejiang
province approved the current study. Patients recruited into the
study had not undergone chemotherapy or radiotherapy prior to
surgery. Surgical specimens of the resected tumors were collected
following the confirmation of diagnosis by pathological
examination. Tumor sections and matched adjacent noncancerous
tissues, were placed in separated cryovials and snap-frozen in
liquid nitrogen until analysis. All cases were reviewed by two
pathologists and diagnosis was confirmed according to criteria
previously established by the National Comprehensive Cancer Network
(23).
 | Table IPrimer sequences of CSC-associated
genes for qPCR. |
Table I
Primer sequences of CSC-associated
genes for qPCR.
| Gene (accession
no.) | Primers sequence
(5′-3′) | Product size
(bp) | Tm (°C) |
|---|
| OCT4A
(NM_002701) | F:
GTGGAGAGCAACTCCGATG | 86 | 60 |
| R:
TGCTCCAGCTTCTCCTTCTC | | |
| SOX2 (NM_003106) | F:
CGAGTGGAAACTTTTGTCGGA | 74 | 60 |
| R:
TGTGCAGCGCTCGCAG | | |
| NANOG
(NM_024865) | F:
ATTCAGGACAGCCCTGATTCTTC | 76 | 60 |
| R:
TTTTTGCGACACTCTTCTCTGC | | |
| MDR1 (NM_000927) | F:
TGGCAAAGAAATAAAGCGACTGA | 76 | 60 |
| R:
CAGGATGGGCTCCTGGG | | |
| ABCG2
(NM_004827) | F:
CCCCAGGCCTCTATAGCTCAGATCA | 164 | 60 |
| R:
TCCACGGCTGAAACACTGCTGA | | |
| CD133
(NM_006017) | F:
CACTACCAAGGACAAGGCGT | 134 | 60 |
| R:
TCCTTGATCGCTGTTGCCAT | | |
| GAPDH
(NM_002046) | F:
CATCATCCCTGCCTCTACTG | 180 | 60 |
| R:
GCCTGCTTCACCACCTTC | | |
Flow cytometry and fluorescence-activated
cell sorting (FACS) analyses
For flow cytometry analysis, cells
(106/100 μl) were dissociated using non-enzymatic
solution Cellstripper (Mediatech, Herndon, VA, USA) and incubated
with the appropriate dilution of control or specific Phycoerythrin
(PE)-conjugated anti-CD133/2 antibody (Miltenyi Biotec) at 4°C for
10 min. All samples were measured using a FACSCalibur flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and analyzed
with CellQuest software (BD Biosciences).
RNA extraction and quantitative
polymerase chain reaction (qPCR)
Total RNA from specimens or cells was extracted
using TRIzol (Invitrogen Life Technologies), according to the
manufacturer’s instructions and the concentration was determined
using a Bioanalyzer UV spectrophotometer Q3000 (Quawell Technology,
Inc., San Jose, CA, USA). Complementary DNA (cDNA) was reverse
transcribed by 3 μg total RNA in a 20 μl reaction using 0.5 μg
Oligo(dT)18 primer and 200 units of RevertAid™ M-MuLV
Reverse Transcriptase (Fermentas, Burlington, ON, Canada) and
treated with RNase-Free DNase (Qiagen, Valencia, CA, USA). The
primers of the housekeeping gene glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) and target genes CD133, octamer-binding
transcription factor 4 (OCT4A), sex determining region Y-box 2
(SOX2) and Nanog homeobox (NANOG), multidrug resistance protein 1
(MDR1) and ATP-binding cassette subfamily G member 2 (ABCG2) are
shown in Table I. The cDNA
amplification was performed in a final reaction volume of 20 μl
containing 0.5 μM concentrations of each primer, 8 μl 2.5X Real
Master mix, 1 μl 20X SYBR solution (Tiangen Biotech, Shanghai,
China) and 2 μl 1:10 diluted cDNA. PCR was prepared in triplicate
and heated to 95°C for 10 min, followed by 40 cycles of the
following sequences: Denaturation at 95°C for 15 sec, annealing at
60°C for 20 sec and extension at 68°C for 35 sec. The expression of
genes was detected by qPCR using Applied Biosystems 7500 Real-Time
RT-PCR system (Applied Biosystems, Carlsbad, CA, USA). The cycle
threshold (Ct) values were calculated with the SDS 2.0.1 software
(Applied Biosystems). All target gene Ct values were determined in
reference to GAPDH using the 2−ΔΔCt method (24).
Chemotherapy resistance studies
Parental H1299 cells and sorted cells
(5×103) were plated in 96-well plates. Cisplatin and
paclitaxel (Sigma-Aldrich, St. Louis, MO, USA) were added to the
culture medium at a final concentration of 25 μg/ml and 20 μM,
respectively, for 24 h. Subsequently, cell viability was analyzed
by an MTT assay.
In vivo analysis of tumor growth
Female nude mice (strain, BALB/c; age, 4 weeks) were
obtained from the Shanghai Laboratory Animal Center (Shanghai,
China). Specific numbers of parental and sorted cells
(103, 104 or 106 cells) were
subcutaneously injected into the flank of the nude mice. Tumor
growth was monitored and volume was measured weekly using a caliper
(volume = width × length2 × π/6) for a maximum of 8
weeks. Subsequently, mice were euthanized by cervical vertebra
dislocation and the tumors were collected, fixed in formalin and
embedded in paraffin for further use.
Statistical analysis
Statistical analyses were performed with the
GraphPad Prism 5.0 statistical software (GraphPad Software, Inc.,
La Jolla, CA, USA). The paired t-test, unpaired t-test and
Mann-Whitney U test were used to analyze the correlation between
mRNA expression levels and the clinicopathological features. The
paired sample t-test was used to compare the differences of mRNA
expression between lung tumors and the surrounding normal tissue.
Survival analysis was estimated by the Kaplan-Meier method and the
log-rank test was used to compare the survival between groups. The
Cox proportional hazard regression model was used to analyze the
risk factors for lung cancer. P<0.05 was considered to indicate
a statistically significant difference.
Results
Isolation and characterization of
CD133-positive cells from lung cancer cell lines
To investigate the involvement of CD133 in the link
between CSCs and lung cancer biology, CD133-positive cells were
isolated from lung cancer cell lines using the magnetic bead method
to determine the expression levels of CD133. CD133 was shown to be
highly expressed in, 95C, SPC-A1, A549 and H1299 NSCLC lines. The
highest expression levels were observed in the H1299 cells
(Fig. 1A). Thus, H1299 cells were
used for all subsequent experiments.
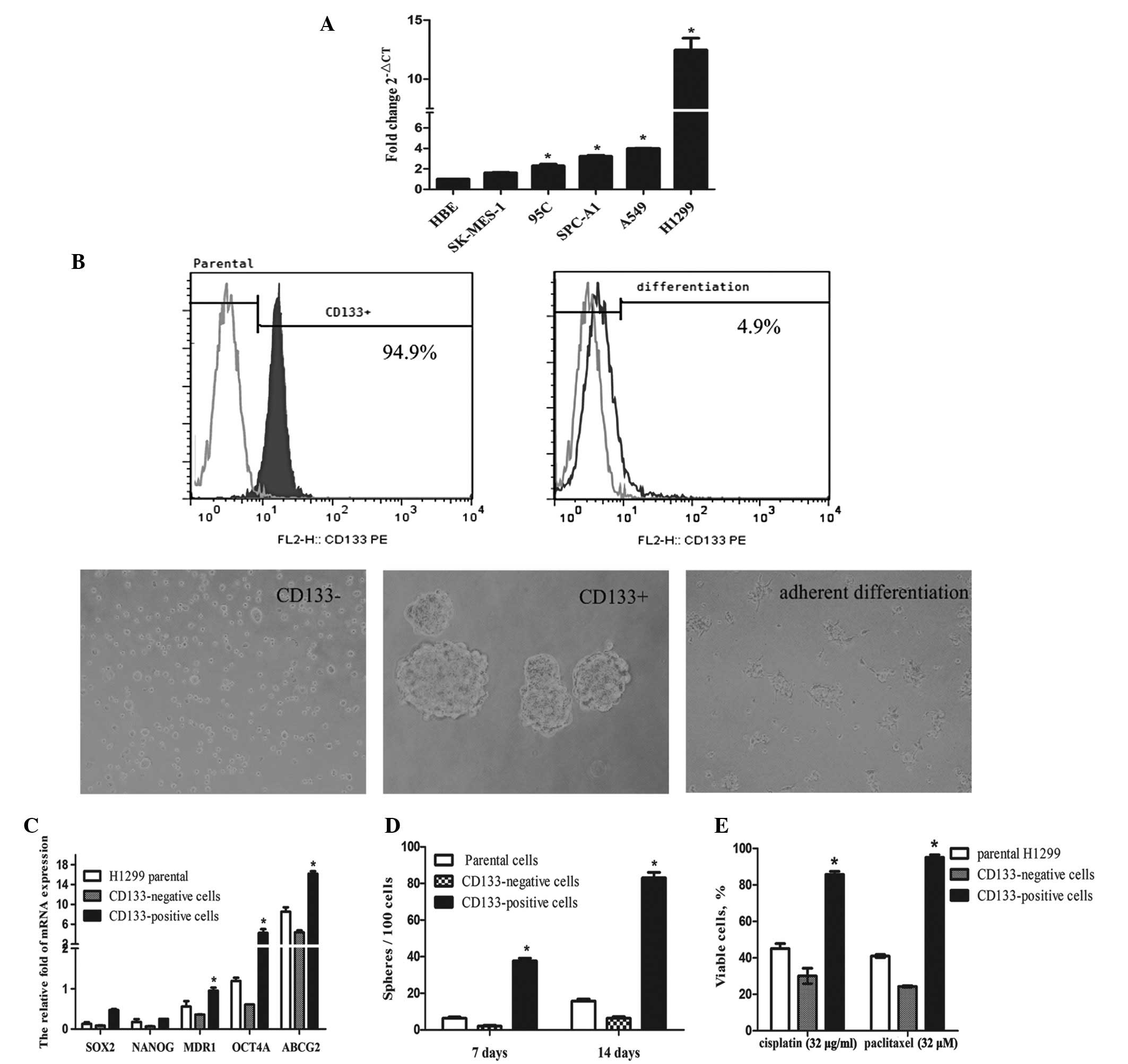 | Figure 1Isolation and characterization of
CD133-positive and CD133-negative cells from the H1299 cell line.
(A) Expression levels of CD133 were detected by qPCR in a normal
bronchial epithelial cell line and a number of non-small cell lung
carcinoma cell lines (HBE, SK-MES-1, 95C, SPC-A1, A549 and H1299).
The average Ct value from triplicate assessment in the candidate
mRNAs was calculated. *P<0.05, vs. HBE cells. (B)
Using a magnetic bead method, CD133-positive cells isolated from
H1299 cells were identified by flow cytometry. The image shows
spheroid-like bodies formed by CD133-positive and CD133-negative
cells in serum-free medium containing bFGF and EGF. The
differentiated cells, which were identified to be CD133-positive
cells, were grown in adherent conditions in differentiating culture
medium for 4 days. Scale bar, 100 μm. (C) mRNA expression levels of
cancer stem cell association genes: OCT4A, SOX2, NANOG, MDR1 and
ABCG2 in CD133-positive, CD133-negative and H1299 parental cells
were determined by qPCR. The fold changes in the target mRNAs were
normalized with that of GAPDH. *P<0.05, vs.
CD133-negative and H1299 parental cells. (D) An analysis of the
ability of different cell groups to form spheroid-like bodies in
the serum-free medium containing bFGF and EGF at the indicated
days. *P<0.05, vs. CD133-negative and H1299 parental
cells. (E) Effect of cisplatin and paclitaxel on the proliferation
of parental H1299, CD133-positive or CD133-negative cells. The
cells (5×103) were plated in a 96-well plate and treated
with the indicated concentrations of cisplatin and paclitaxel for
24 h. The survival rate was determined by a cell counting kit-8
assay. Data are presented as the mean ± SD of three independent
experiments. *P<0.05, vs. CD133-negative and H1299
parental cells. qPCR, quantitative polymerase chain reaction; Ct,
cycle threshold; bFGF, basic fibroblast growth factor; EGF.
epidermal growth factor; OCT4A, octamer-binding transcription
factor 4; SOX2, sex determining region Y-box 2; NANOG, Nanog
homeobox; MDR1, multidrug resistance protein 1; ABCG2, ATP-binding
cassette subfamily G member 2; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase. |
The isolated CD133-positive cells identified by flow
cytometry formed floating sphere-like bodies in DMEM/F-12
serum-free medium with bFGF and EGF. When CD133-positive cells were
grown under adherent conditions in culture medium supplemented with
10% FBS, the cells acquired the typical morphological features of
parental H1299 cells with the loss of CD133 expression (decrease in
expression from 94.9 to 4.9%; Fig.
1B). qPCR results demonstrated that the expression of stemness
genes (OCT4A, SOX2 and NANOG) and drug resistant genes (MDR1 and
ABCG2) in CD133-positive cells were higher compared with
those in the CD133-negative cells (Fig. 1C). The ability to form
spheroid-like bodies was significantly higher in the CD133-positive
cells compared with the parental or CD133-negative cells
(P<0.05; Fig. 1D). The
multidrug chemotherapy resistant abilities of the CD133-positive
cells and CD133-negative cells were then investigated. Compared
with the CD133-negative and parental cells, CD133-positive cells
sorted from H1299 cells were more resistant to cisplatin and
paclitaxel (P<0.05; Fig.
1E).
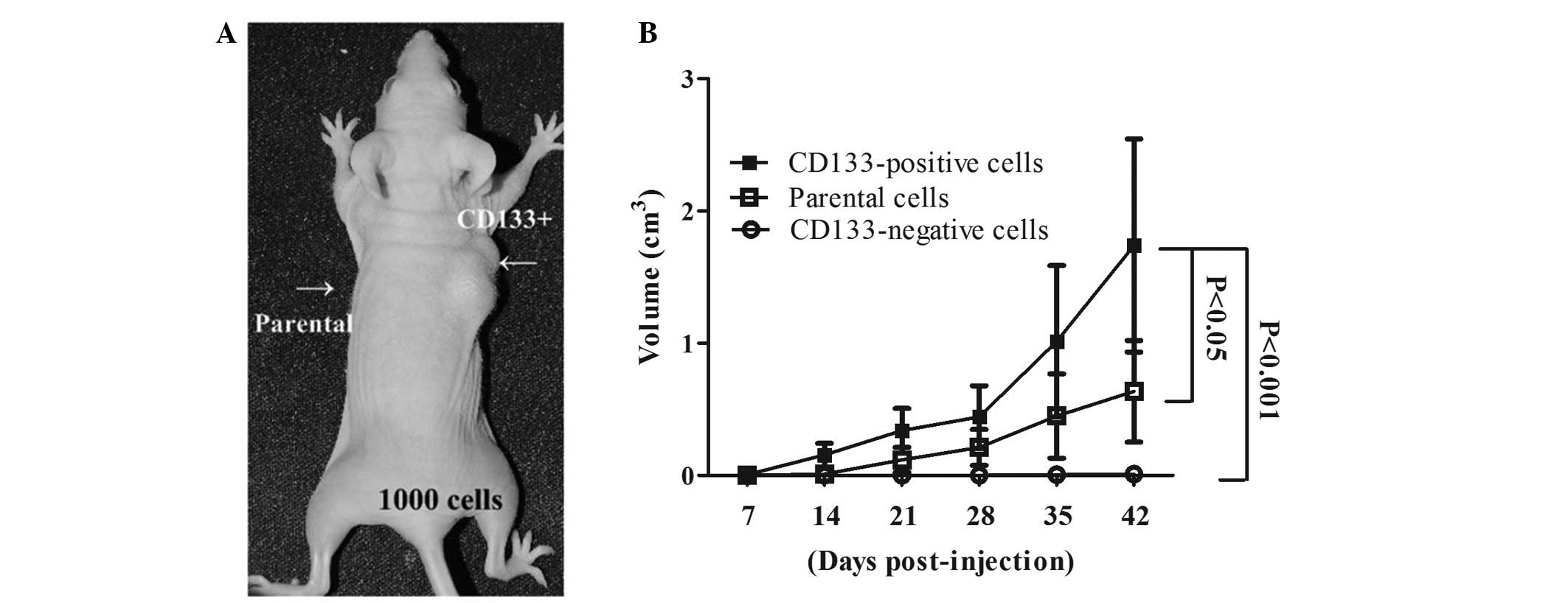
To analyze the tumorigenic potential of
CD133-positive cells grown under sphere-forming conditions, during
which this property of stem cells may be maintained, specific
numbers of H1299 CD133-positive, CD133-negative and parental cells
were subcutaneously implanted into the flanks of nude mice (BALB/c
strain). As shown in Table II,
tumor growth was observed in all groups in which mice were
inoculated with 103–106 CD133-positive cells,
whereas no tumor growth was observed following inoculation with
103 H1299 parental or CD133-negative cells (Fig. 2A). Tumor growth curves showed that
tumors that had developed from the CD133-positive cells grew at a
faster rate compared with those that had developed from the
parental or CD133-negative cells (Fig.
2B). These observations indicated that CD133-positive lung
cancer cells may be used to enrich tumor-initiating cells under
sphere-forming conditions.
 | Table IITumor formation derived from parental
and sorted H1299 cells. |
Table II
Tumor formation derived from parental
and sorted H1299 cells.
| Cells | 103
cells | 105
cells | 106
cells |
|---|
| Parental | 0/4 | 1/4 | 3/4 |
| CD133-positive | 2/4 | 4/4 | 4/4 |
| CD133-negative | 0/4 | 0/4 | 1/4 |
Patient information
Tumor specimens from 30 patients (19 paired samples
of lung adenocarcinomas and 11 paired samples of lung squamous-cell
carcinomas) with previously untreated NSCLC were recruited into the
current study. The patients consisted of 23 males and 7 females.
The median age was 61.1 years, with nine cases (30%) aged >65
years. Based on the World Health Organization criteria, 15 patients
(50%) presented with stage I disease, six patients (20%) with stage
II disease and 9 patients (30%) with stage III/IV disease. Of the
30 patients, 14 (46.67%) had tumors with nodal metastasis and 15
(30%) had a primary tumor >3 cm in diameter. The majority of the
patients (22; 73.33%) were smokers.
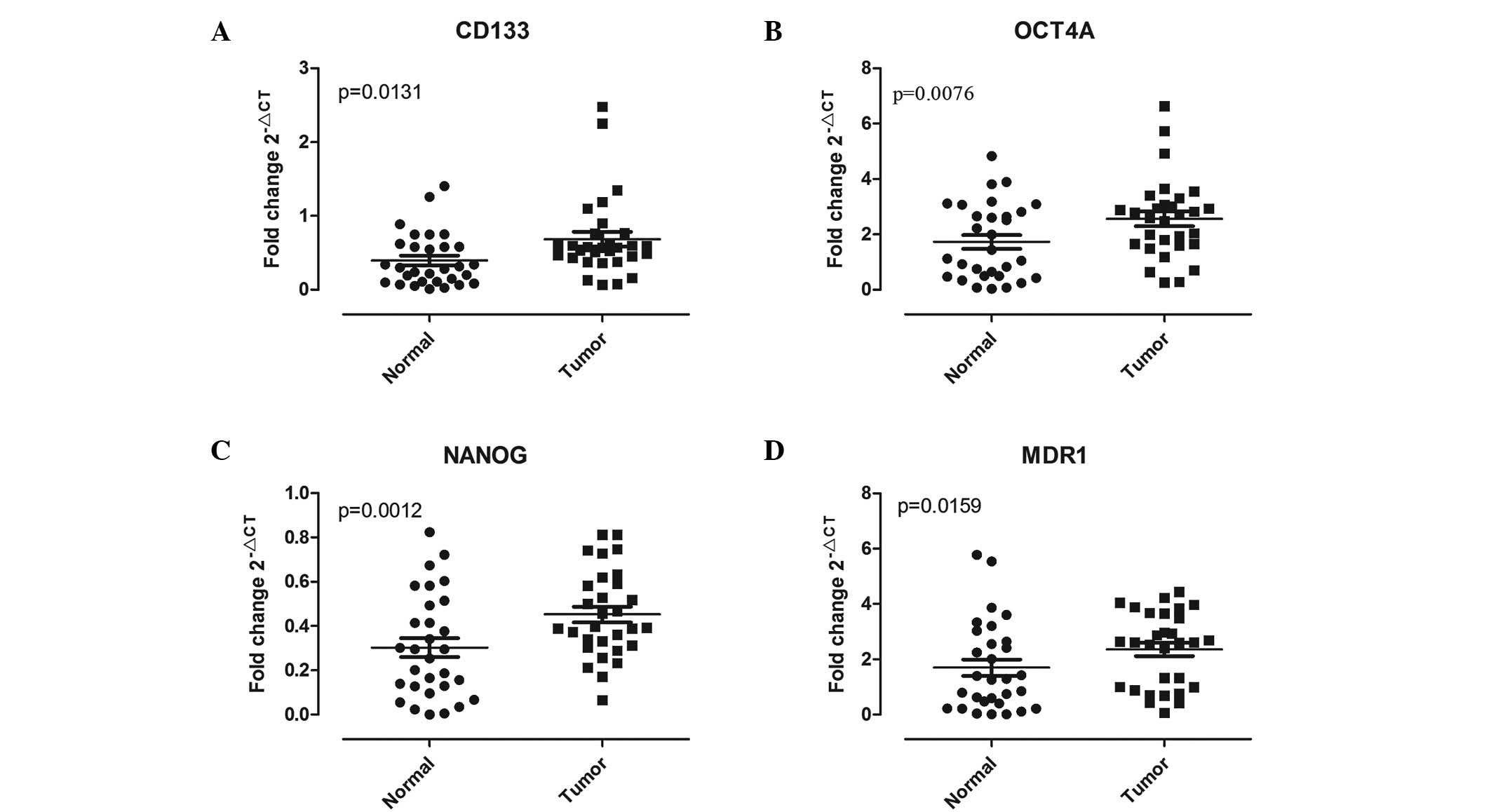
Cancer stem cell-associated gene
expression in lung tissue
A primary aim of the current study was to determine
whether there was a difference in the frequency of CSC-associated
gene expression between paired lung carcinoma and the corresponding
noncancerous lung tissue. The results from the qPCR analysis showed
that, compared with the adjacent normal lung tissues, the levels of
CSC-associated biomarkers, such as OCT4A, CD133, NANOG and MDR1,
were significantly increased in lung cancer tissues (P<0.05;
Fig. 3). The mRNA levels of CD133
(Fig. 3A) and MDR1 (Fig. 3D) were observed to be significantly
higher in the tumor tissue (CD133, P=0.0131 and MDR1, P=0.0159)
compared with that in adjacent normal lung tissue. In addition,
compared with the normal counterpart, higher NANOG and OCT4A
expression was observed in the malignant tissue (P=0.0012 and
P=0.0076) in 23 and 19 of 30 pairs (76.67 and 63.33%, respectively;
Fig. 3C and B).
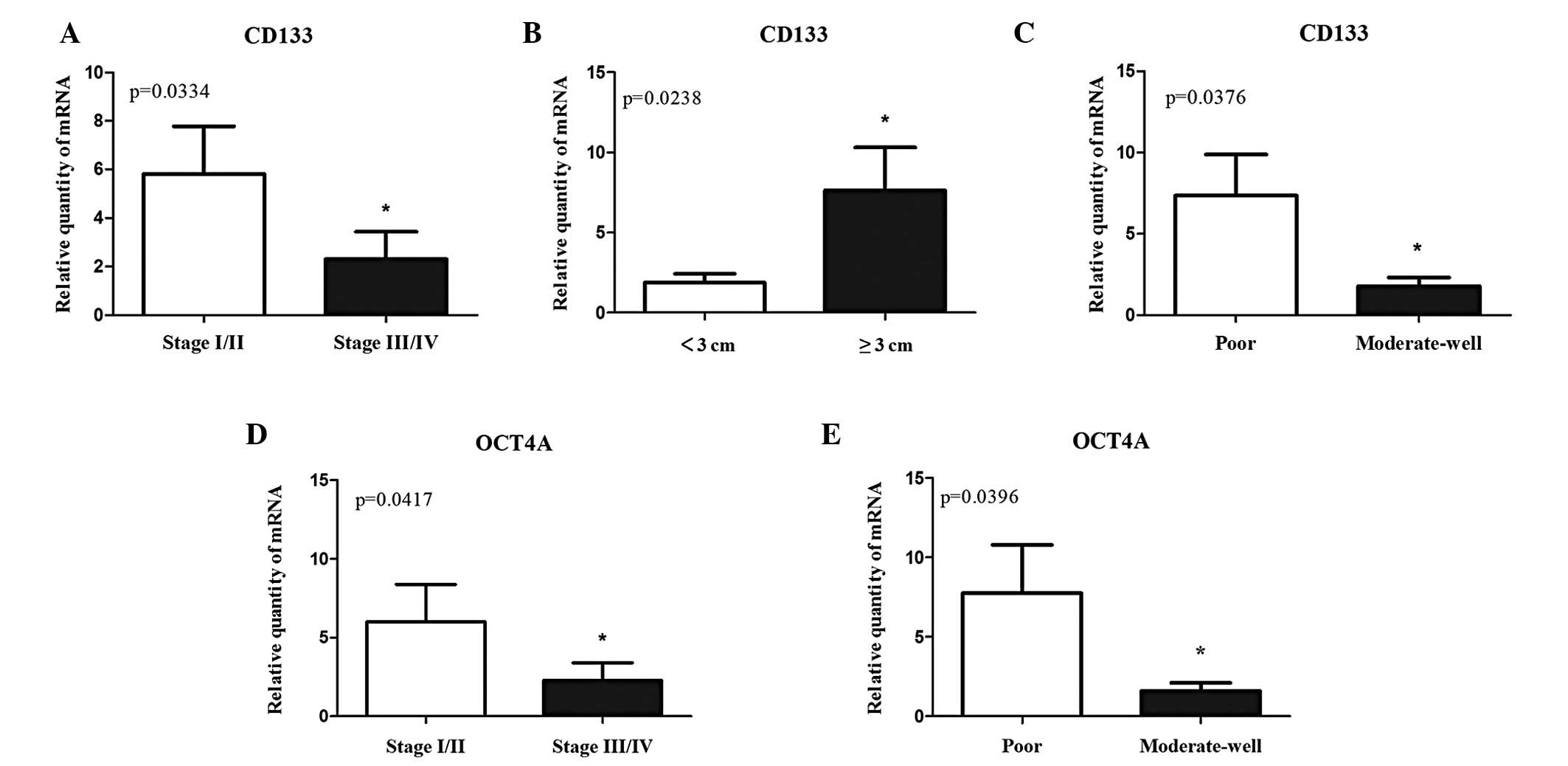
Correlation between the expression of
cancer stem cell-associated genes and clinical-pathological
features of NSCLC
The correlation between the expression of
CSC-associated genes and clinicopathological features of NSCLC was
investigated to gain an improved understanding of the potential
involvement of these genes in NSCLC development and progression
using the Mann-Whitney U test (Table
III). No substantial correlation between the expression of
NANOG or MDR1 and the tumor stage, nodal status, tumor size and
histological type was observed, with the exception of tumor
differentiation. However, overexpression of CD133 in lung tumors
was observed to be markedly associated with clinical stage, tumor
size and differentiation of NSCLC (P=0.0334, 0.0238 and 0.0376,
respectively; Fig. 4A–C). In
addition, patients with squamous-cell lung carcinoma or a tumor ≥3
cm in diameter exhibited higher expression levels of CD133.
Notably, overexpression of OCT4A in tumors was positively
associated with a tumor stage I or II and moderate-well
differentiated tumors (P=0.0417 and 0.0396, respectively; Fig. 4D and E). In addition, patients with
poor differentiation had higher levels of CD133 and OCT4A.
 | Table IIICorrelation between the expression of
CSC-associated genes and the clinicopathological factors in
patients with NSCLC. |
Table III
Correlation between the expression of
CSC-associated genes and the clinicopathological factors in
patients with NSCLC.
| | OCT4A | | NANOG | | MDR1 | | CD133 | |
|---|
| |
| |
| |
| |
| |
|---|
| Variables | n=30 | Mean ± SEM | P-value | Mean ± SEM | P-value | Mean ± SEM | P-value | Mean ± SEM | P-value |
|---|
| Gender | | | 0.1285 | | 0.0470a | | 0.0558 | | 0.1224 |
| Male | 23 | 6.049±2.184 | | 9.195±4.362 | | 11.75±8.49 | | 5.821±1.829 | |
| Female | 7 | 1.081±0.1406 | | 1.461±0.3930 | | 1.310±0.3004 | | 1.235±0.2130 | |
| Age (years) | | | 0.4385 | | 0.0388a | | 0.2452 | | 0.2917 |
| ≤60 | 11 | 5.485±4.112 | | 6.080±4.851 | | 19.26±17.88 | | 4.816±3.389 | |
| >60 | 19 | 4.546±1.412 | | 8.149±4.632 | | 3.550±0.6745 | | 4.713±1.262 | |
| Histological
type | | | 0.0034a | | 0.6498 | | 0.1138 | | 0.0024a |
| Squamous
carcinoma | 11 | 11.17±4.099 | | 9.460±4.631 | | 22.93±17.53 | | 10.19±3.336 | |
|
Adenocarcinoma | 19 | 1.256±0.2140 | | 6.192±4.690 | | 1.429±0.2178 | | 1.604±0.4545 | |
| Pathological
stage | | | 0.0417a | | 0.1539 | | 0.1132 | | 0.0334a |
| I, II | 21 | 6.009±2.272 | | 9.769±4.767 | | 12.47±9.301 | | 5.800±1.976 | |
| III, IV | 9 | 2.280±1.125 | | 1.841±0.4646 | | 1.944±0.608 | | 2.302±1.125 | |
| Smoking status | | | 0.0710 | | 0.0516 | | 0.0292a | | 0.0827 |
| Nonsmokers | 8 | 1.046±0.1265 | | 1.472±0.3405 | | 1.264±0.2641 | | 1.201±0.1875 | |
| Current
smokers | 22 | 6.288±2.272 | | 9.542±4.551 | | 12.24±8.870 | | 6.041±1.900 | |
| Tumor size
(cm) | | | 0.0680 | | 0.0970 | | 0.1776 | | 0.0238a |
| <3 | 15 | 1.838±0.5074 | | 2.435±0.8169 | | 2.090±0.554 | | 1.865±0.5416 | |
| ≥3 | 15 | 7.942±3.247 | | 12.35±6.576 | | 16.53±12.99 | | 7.636±2.669 | |
| Grade of
differentiation | | | 0.0396a | | 0.0142a | | 0.0248a | | 0.0376a |
| Moderate-well | 16 | 1.611±0.4888 | | 2.257±0.8798 | | 1.877±0.5741 | | 1.761±0.5459 | |
| Poor | 14 | 7.759±3.043 | | 11.88±6.166 | | 15.82±12.17 | | 7.366±2.516 | |
| Lymph node
status | | | 0.1904 | | 0.3940 | | 0.3237 | | 0.2530 |
| N0 | 16 | 5.645±2.796 | | 11.64±6.213 | | 15.14±12.21 | | 5.565±2.329 | |
| N+ | 14 | 4.028±1.892 | | 2.535±0.6427 | | 2.782±0.8021 | | 3.820±1.629 | |
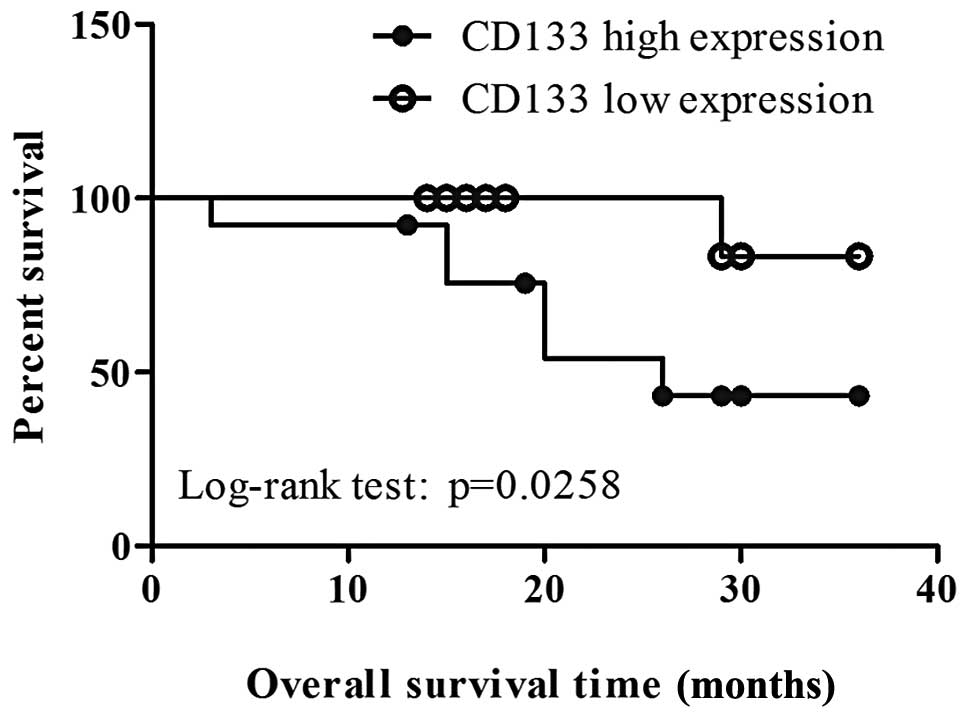
Correlation between cancer stem
cell-associated gene expression and overall survival of NSCLC
patients
The expression of CSC-associated genes and its
correlation with overall survival of NSCLC patients was
investigated (fold change >2 was considered as high expression).
The results showed that higher levels of expression of CD133 in
NSCLC was independently correlated with shorter overall survival
(log-rank test: P=0.0258, Fig. 5).
However, no significant correlation between the expression of OCT4A
and the overall survival was observed (log-rank test: P=0.1395). In
addition, tumor size >3 cm was observed to be associated with
decreased overall survival (log-rank test: P=0.015, data not
shown).
The Kaplan-Meier survival curve showed a trend for
an improved outcome in patients with lower CD133 mRNA signals;
however, the Cox hazard regression analysis indicated that CD133
was not considered to be an independent factor in predicting the
prognosis of patients with NSCLC (hazard ratio: 0.179; 95%
confidence interval: 0.010–3.205; P=0.243).
Discussion
Lung cancer is characterized by the difficulty
surrounding early diagnosis, high rates of metastasis, recurrence
and poor prognosis. Thus, investigating novel diagnostic strategies
and novel prognostic markers is urgently required to improve the
clinical outcome of this malignancy (1). Increased numbers of CD133-positive
cancer stem cells have been observed in NSCLC (13). In the current study, isolated
CD133-positive cells were shown to harbor stem cell-like
characteristics, as they readily form anchorage-independent
floating spheres, possess greater proliferative potential and
exhibit enhanced tumor regenerating capacity compared with their
CD133-negative counterparts. In addition, CD133-positive, but not
CD133-negative or parental NSCLC cells formed tumors in the nude
mice xenograft model. These results demonstrated the CSC status of
CD133-positive cells in NSCLC.
The correlation of CSC-associated gene expression
(CD133, OCT4A, NANOG and MDR1) in NSCLC samples, with patient
characteristics, tumor pathology and overall survival was
investigated. The current results indicated the presence of CSCs in
NSCLC tissue, as self-renewal and multipotency are critical
characteristics of CSCs (4). Based
on the present observations, the correlation between increased
CD133 expression and early stage tumors, larger tumor size and
poorly differentiated NSCLC tumors may be to be due to the
self-renewal properties of aberrant CSCs. This implicated the
importance of CD133-positive cells in the initiation of NSCLC. The
reduced levels of CD133 in the later stages of NSCLC may be
explained by the hypothesis that cells expressing CD133 undergo
asymmetric division, generating a diverse phenotype and therefore
resulting in reduced CD133 positivity. However, further studies are
required to confirm this.
Previous studies have shown that the expression of
CSC antigens was associated with a poor prognosis (20–22,25,26).
However, the current observations, which are consistent with the
results of other studies (17–19),
demonstrated that while the expression of CD133 in the surgically
resected specimens was involved in NSCLC carcinogenesis, CD133
alone may not be used as an independent biomarker in the prediction
of the prognosis of NSCLC. The origin of these discrepancies
remains unclear; however, specific NSCLC samples may be the reason
for variability between the results. Notably, the present specimens
were obtained from patients with all stages of NSCLC and contained
various histological types (adenocarcinoma and squamous
carcinoma).
Studies in human embryonic stem cells indicated that
OCT4A and the homeobox protein NANOG were two key transcription
factors that cooperatively maintained pluripotency (27,28).
The present results indicated that OCT4A was significantly
associated with the clinical stage and differentiation of NSCLC.
Consistent with this, Chen et al(29) observed that knockdown of the OCT4A
gene in CD133-positive stem-like lung cancer cells significantly
diminished the ability of tumor invasion and colony formation, and
increased apoptotic activities. The aforementioned observations
indicate that the expression of OCT4A was involved in maintaining
the stem cell-like properties of lung cancer cells. Whether the
combination of the two CSC markers, OCT4 and CD133, may enhance the
predictive validity of patient progression and survival remains to
be elucidated.
In conclusion, the results of the present study show
that CD133-positive NSCLC cells exhibit clonogenic, tumorigenic and
drug-resistant properties. The elevated expression of CD133 was
associated with early stage tumors, larger tumor size and poor
differentiation of NSCLC, indicating that highly expressed CD133 is
involved in the carcinogenesis of NSCLC. However, expression of
CD133 in CSCs is not considered to be an independent factor in the
prediction of the prognosis of patients with NSCLC. Further studies
are required to determine the association between CD133 expression
and overall survival in NSCLC patients, with or without other
CSC-associated genes.
Acknowledgements
This study was supported by grants from the Zhejiang
Provincial Natural Science Foundation of China (grant no. Y2101395)
and the Medical Bureau of Zhoushan (grant no. 2010G02).
References
|
1
|
Spira A and Ettinger DS: Multidisciplinary
management of lung cancer. N Engl J Med. 350:379–392. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Klaveren RJ, van’t Westeinde SC, de
Hoop BJ and Hoogsteden HC: Stem cells and the natural history of
lung cancer: implications for lung cancer screening. Clin Cancer
Res. 15:2215–2218. 2009.PubMed/NCBI
|
|
4
|
Sullivan JP, Minna JD and Shay JW:
Evidence for self-renewing lung cancer stem cells and their
implications in tumor initiation, progression, and targeted
therapy. Cancer Metastasis Rev. 29:61–72. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim CF, Jackson EL, Woolfenden AE,
Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT and Jacks T:
Identification of bronchioalveolar stem cells in normal lung and
lung cancer. Cell. 121:823–835. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rosen JM and Jordan CT: The increasing
complexity of the cancer stem cell paradigm. Science.
324:1670–1613. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao P, Lu Y, Jiang X and Li X:
Clinicopathological significance and prognostic value of CD133
expression in triple-negative breast carcinoma. Cancer Sci.
102:1107–1111. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yin S, Li J, Hu C, Chen X, Yao M, Yan M,
Jiang G, Ge C, Xie H, Wan D, Yang S, Zheng S and Gu J: CD133
positive hepatocellular carcinoma cells possess high capacity for
tumorigenicity. Int J Cancer. 120:1444–1450. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giangreco A, Groot KR and Janes SM: Lung
cancer and lung stem cells: strange bedfellows? Am J Respir Crit
Care Med. 175:547–553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eramo A, Lotti F, Sette G, Pilozzi E,
Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C and De
Maria R: Identification and expansion of the tumorigenic lung
cancer stem cell population. Cell Death Differ. 15:504–514. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meng X, Li M, Wang X, Wang Y and Ma D:
Both CD133+ and CD133− subpopulations of A549
and H446 cells contain cancer-initiating cells. Cancer Sci.
100:1040–1046. 2009.
|
|
15
|
Qiu X, Wang Z, Li Y, Miao Y, Ren Y and
Luan Y: Characterization of sphere-forming cells with stem-like
properties from the small cell lung cancer cell line H446. Cancer
Lett. 323:161–170. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Akunuru S, James Zhai Q and Zheng Y:
Non-small cell lung cancer stem/progenitor cells are enriched in
multiple distinct phenotypic subpopulations and exhibit plasticity.
Cell Death Dis. 3:e3522012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salnikov AV, Gladkich J, Moldenhauer G,
Volm M, Mattern J and Herr I: CD133 is indicative for a resistance
phenotype but does not represent a prognostic marker for survival
of non-small cell lung cancer patients. Int J Cancer. 126:950–958.
2010.PubMed/NCBI
|
|
18
|
Herpel E, Jensen K, Muley T, Warth A,
Schnabel PA, Meister M, Herth FJ, Dienemann H, Thomas M and
Gottschling S: The cancer stem cell antigens CD133, BCRP1/ABCG2 and
CD117/c-KIT are not associated with prognosis in resected
early-stage non-small cell lung cancer. Anticancer Res.
31:4491–4500. 2011.PubMed/NCBI
|
|
19
|
Shien K, Toyooka S, Ichimura K, Soh J,
Furukawa M, Maki Y, Muraoka T, Tanaka N, Ueno T, Asano H, et al:
Prognostic impact of cancer stem cell-related markers in non-small
cell lung cancer patients treated with induction chemoradiotherapy.
Lung Cancer. 77:162–167. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cortes-Dericks L, Galetta D, Spaggiari L,
Schmid RA and Karoubi G: High expression of octamer-binding
transcription factor 4A, prominin-1 and aldehyde dehydrogenase
strongly indicates involvement in the initiation of lung
adenocarcinoma resulting in shorter disease-free intervals. Eur J
Cardiothorac Surg. 41:e173–e181. 2012. View Article : Google Scholar
|
|
21
|
Woo T, Okudela K, Mitsui H, Yazawa T,
Ogawa N, Tajiri M, Yamamoto T, Rino Y, Kitamura H and Masuda M:
Prognostic value of CD133 expression in stage I lung
adenocarcinomas. Int J Clin Exp Pathol. 4:32–42. 2010.PubMed/NCBI
|
|
22
|
Li F, Zeng H and Ying K: The combination
of stem cell markers CD133 and ABCG2 predicts relapse in stage I
non-small cell lung carcinomas. Med Oncol. 28:1458–1462. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ettinger DS, Akerley W, Borghaei H, et al:
Non-Small Cell Lung Cancer, Version 2.2013. J Natl Compr Canc Netw.
11:645–653. 2013.PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
25
|
Zeppernick F, Ahmadi R, Campos B, Dictus
C, Helmke BM, Becker N, Lichter P, Unterberg A, Radlwimmer B and
Herold-Mende CC: Stem cell marker CD133 affects clinical outcome in
glioma patients. Clin Cancer Res. 14:123–129. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sullivan JP, Spinola M, Dodge M, Raso MG,
Behrens C, Gao B, Schuster K, Shao C, Larsen JE, Sullivan LA, et
al: Aldehyde dehydrogenase activity selects for lung adenocarcinoma
stem cells dependent on notch signaling. Cancer Res. 70:9937–9948.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nichols J, Zevnik B, Anastassiadis K, Niwa
H, Klewe-Nebenius D, Chambers I, Schöler H and Smith A: Formation
of pluripotent stem cells in the mammalian embryo depends on the
POU transcription factor Oct4. Cell. 95:379–391. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ,
Tsai TH, Chou SH, Chien CS, Ku HH and Lo JF: Positive correlations
of Oct-4 and Nanog in oral cancer stem-like cells and high-grade
oral squamous cell carcinoma. Clin Cancer Res. 14:4085–4095. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen YC, Hsu HS, Chen YW, Tsai TH, How CK,
Wang CY, Hung SC, Chang YL, Tsai ML, Lee YY, Ku HH and Chiou SH:
Oct-4 expression maintained cancer stem-like properties in lung
cancer-derived CD133-positive cells. PLoS One. 3:e26372008.
View Article : Google Scholar : PubMed/NCBI
|