Introduction
Tuberculosis is a chronic infectious disease caused
by the bacterium Mycobacterium tuberculosis, and it remains
a significant public health risk worldwide. Cosmopolitan
tuberculosis pestilence has quickly returned due to the misuse of
antituberculosis drugs and infection with HIV that has been
apparent since the 1980’s (1,2).
Since becoming the first elected antituberculosis drug, the
clinical therapeutic efficacy of RFP has severely degraded due to
the appearance of drug-resistant strains and the L-form mutations
of Mycobacterium tuberculosis. At present, drug resistance
is an issue preventing the elimination of tuberculosis (3). According to the statistics, mutations
in the rpoB gene in Mycobacterium tuberculosis account for
90% of the RFP-resistant strains, but there are no studies
regarding the resistance of the L-form (4,5). In
the present study, a DNA sequence analysis technique was applied
for investigating the mutational characteristics of rpoB in the
Mycobacterium tuberculosis L-form, in order to provide a
more complete understanding of its resistance.
Materials and methods
Experimental subjects
Subjects included male patients with pneumoconiosis
complicated by tuberculosis (n=114) who were aged between 41 and 70
years old, with a mean age of 53.35±9.28 years, and who were
treated in the Affiliated Hospital of Anhui University of Science
and Technology (Hefei, China) between July 2010 and July 2012. All
the subjects were instructed to expectorate phlegm originating from
the bottom of the trachea into sterile wide-mouthed bottles
subsequent to gargling several times. The H37Rv quality control
strain was provided by the Center of Biological Products of the
Department of Health (Beijing, China). Patient consent was obtained
from all the subjects. Approval was obtained from the Ethics
Committee of the School of Medicine, Anhui University of Science
and Technology (Huainan, China).
Experimental methods
DNA extraction
The sputum specimens of the patients were
inactivated by autoclave. Genomic DNA was extracted using a DNA
extraction kit (Takara Biotech Co., Ltd., Dalian, China). The
inactivated specimens were lysed with 200 μl DNA lysate (10 mmol/l
Tris-HCl, 100 mmol/l NaCl, 25 mmol/l EDTA, 1% SDS, 0.2 mg/ml
proteinase K) and incubated at 55°C for 1–3 h, then at 95°C for 5
min. The lysate was extracted twice with phenol:chloroform:isoamyl
alcohol (volume ratio 25:24:1). DNA was precipitated by adding
ammonium acetate. 2.5 volumes of pure ethanol were added, followed
by incubation at −20°C overnight. DNA was pelleted by
centrifugation at 12,000 × g for 15 min, washed with 70% ethanol,
air-dried and dissolved in the TE buffer [10 mmol/l Tris-HCl (pH
7.4), 1 mmol/l EDTA] (6).
Primer contrivance
The primer was synthesized by Sangon Biotect Co.,
Ltd., (Shanghai, China) based on the conserved Mycobacterium
tuberculosis genomic DNA sequence (a 69-bp mutational
duplication conserved sequence). The primer sequences of rpoB used
in this experiment were as follows: Forward: 5′-CGG ATG ACC ACC
CAGG AC-3′ and reverse: 5′-GGT TTA GAT CGG CAC AT-3′; product size,
258 bp.
PCR analysis and sequence analysis
The total PCR reaction volume was 50 μl and included
5.0 μl 10X buffer (15 mmol/l MgCl2), 4.0 μl dNTPs (200
μmol/l), 1.0 μl of each primer (25 pmol/μl), 0.5 μl Taq DNA
polymerase (5 U/μl) and 4 μl DNA template. Deionized water was
added for a total volume of 50 μl. The reaction conditions were:
95°C for 5 min of denaturation, then 94°C for 30 sec, 55°C for 90
sec and 72°C for 30 sec, for 33 cycles in total, followed by
maintenance at 72°C for 10 min. The amplified product (5 μl) was
determined by electrophoresis with a 2% agarose gel (containing 0.5
mg/ml ethidium bromide). Images were captured and analyzed with
Image Master TotalLab software (TotalLab, Ltd., Newcastle, UK).
This revealed whether the rpoB mutation was present, indicated by
the specific bands at 258 bp. The sequence determination of the DNA
sequences of the PCR product was detected by an ABI PRISM 7700
sequencer (Takara Biotech Co., Ltd.).
Statistical analysis
Statistical analyses were performed using SPSS 12.0
software (SPSS, Inc., Chicago, IL, USA).
Results
PCR analysis
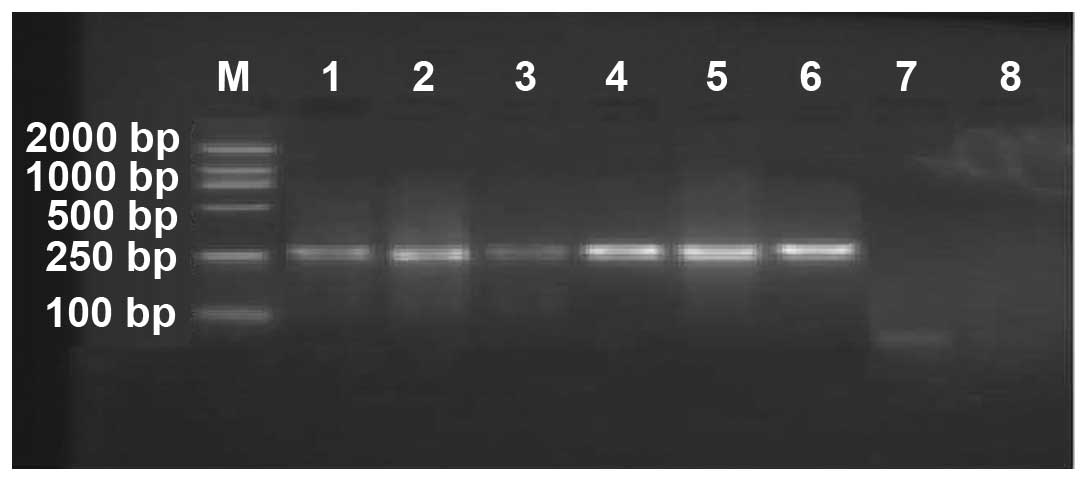
In the total 42 cases of Mycobacterium
tuberculosis L-form isolated from the sputum samples, 31
RFP-resistant strains and 11 RFP-sensitive strains were identified
(Fig. 1).
Sequence analysis of DNA of Mycobacterium
tuberculosis L-form
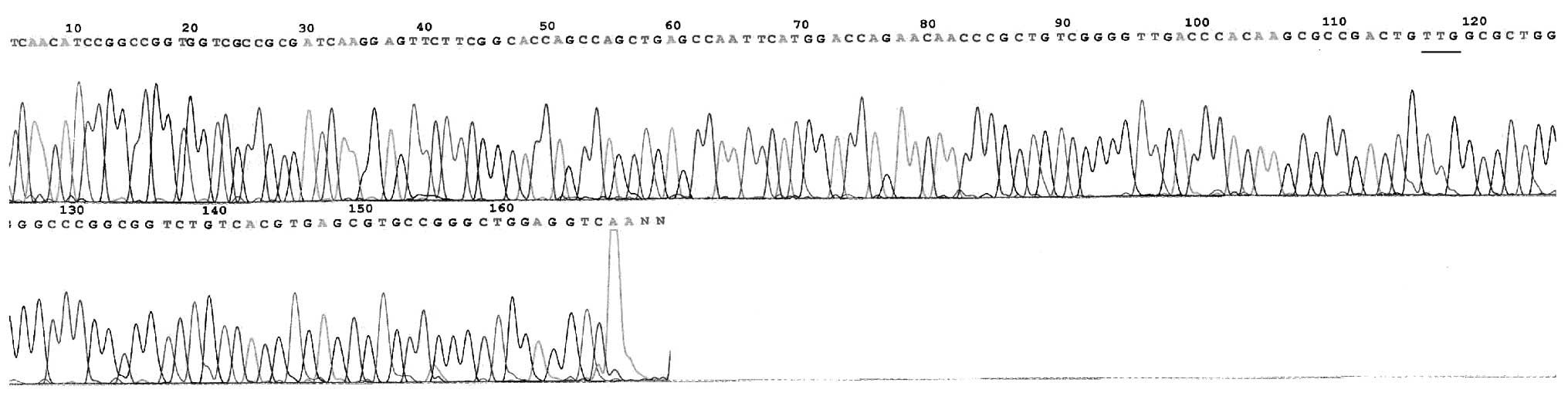
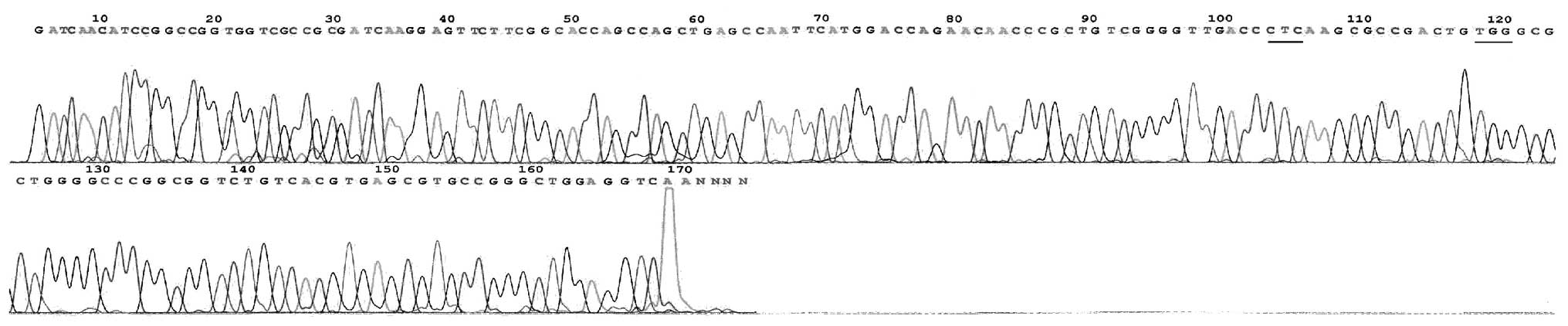
In total, there were 29 mutational strains of the
rpoB gene within the 31 RFP-resistant strains. The rate of mutation
was observed to be 93.55% (29/31; Table I), mainly concentrated in codon 531
(51.61%, 16/31) and 526 (32.26%, 10/31) and occurring by base
substitutions, including 27 unit point mutations and two two-site
point mutations. The mutation of codon 516 was a novel observation
(Fig. 2–11). No rpoB mutation was identified in
the 11 RFP-sensitive strains.
 | Table IMutation characteristics of the rpoB
gene in 31 clinical drug-resistant strains of Mycobacterium
tuberculosis L-forms. |
Table I
Mutation characteristics of the rpoB
gene in 31 clinical drug-resistant strains of Mycobacterium
tuberculosis L-forms.
| Strain | Location of amino
acids, codon | Change of codon | Change of amino
acids | Percentage, % |
|---|
| 10 | 531 | TCG→TTG | Ser→Leu | 32.26 |
| 6 | 531 | TCG→TGG | Ser→Trp | 19.35 |
| 5 | 526 | CAC→GAC | Gln→Asp | 16.13 |
| 2 | 526 | CAC→TAC | Gln→Tyr | 6.45 |
| 1 | 526 | CAC→CTC | Gln→Leu | 3.23 |
| 1 | 526 | CAC→CGC | Gln→Arg | 3.23 |
| 1 | 526 | CAC→GTC | Gln→Val | 3.23 |
| 1 | 516 | GAC→GGC | Asp→Gly | 3.23 |
| 1 | 511, 526 | CTG→CCG, CAC→CTC | Leu→Pro, Gln→Leu | 3.23 |
| 1 | 526, 531 | CAC→CTC, TCG→TGG | Gln→Leu, Ser→Trp | 3.23 |
| 2 | - | - | - | 6.45 |
Discussion
Pneumoconiosis is a significant occupational disease
in employees of coal mines, and once pneumoconiosis associates with
tuberculosis, it develops more rapidly and worsens the tuberculosis
of the patient. The prognoses of patients with the disease appear
to be poor, and the therapeutic effects on these cases are not
good, as may be expected. At present, the therapeutic regimen
includes treating the pneumoconiosis and the tuberculosis using
antituberculosis drugs, since there is a lack of radically curative
drugs for the treatment of pneumoconiosis.
The molecular anti-RFP mechanism of Mycobacterium
tuberculosis was correlated with the mutation of the rpoB gene,
which was coded by the β subunit of RNA polymerase. RFP usually
inhibits the transcription of RNA polymerase with the purpose of
killing infected cells (7,8). In total, 96% of drug-resistant
strains are caused by mutations in the rpoB gene, by an insertion
or deletion of basic groups, mainly concentrated around the
determining ~80-bp region of RFP. In 35 types of mutations, 43% are
missense mutations (9). Mutations
in site 513 have been shown to result in greater drug resistance
[minimal inhibition concentration (MIC)>32 μg/ml], while
mutations at sites 514, 521 and 533 lead to lower drug resistance
(MIC<12.5 μg/ml) (10–12). In theory, drug-resistant strains of
Mycobacterium tuberculosis may occur due to two main
reasons. Firstly, resistance may occur due to a mutation in the β
subunit of the RNA polymerase of the target molecule by drug
action. Secondly, the ingesting capability of the infected cells
may be decreased due to a change in the osmosis of the cell wall
(13).
The Mycobacterium tuberculosis L-form is also
known as the Mycobacterium cell wall-deficient form; in
1960, Mattmand (14) explained its
biological characteristics in detail. It was demonstrated that the
changes of the L-form were evident due to the absence of the cell
wall either partly or completely, thus affecting the biological
characteristics, drug sensitivity and DNA. The L-form is considered
a type of mutation. This mutation may be induced by a number of
factors, including chemotherapeutics, lysozymes and bacteriophages.
Once Mycobacterium tuberculosis L form variation occurs, it
continues to possess pathogenicity, causing chronic transformation
of the disease process, which leads to a worse prognosis. This has
resulted in problems in the diagnosis and treatment of tuberculosis
(15).
In the present study, the results of the PCR
analysis demonstrated that there were 29 mutational strains of the
rpoB gene within the 31 RFP-resistant strains; the mutation rate
was 93.55% (29/31). In the resistant strains, the mutations were
mainly located in codons 531 (51.61%; 16/31) and 526 (32.26%;
10/31), occurring by base substitutions. The former site included
10 strains of Ser→Leu (TCG→TTG) and six strains of Ser→Trp
(TCG→TGG), while the latter contained mainly Gln→Asp (CAC→GAC)
substitutions. These results revealed that unit point mutations of
the rpoB gene of Mycobacterium tuberculosis L-form are
located in codons 531 and 526 (83.87%; 26/31), as mentioned in
previous studies (10,16). The point mutation of codon 516,
which has not been previously reported, was a same-sense mutation.
A total of 11 RFP-sensitive strains were not mutations of the ropB
gene.
Furthermore, two strains of two-site point mutations
were identified, one at codons 526 (Gln→Leu) and 531 (Ser→Trp) and
the other at codons 511 (Leu→Pro) and 526 (Gln→Leu). The mutation
rate was 6.45% (2/31), appreciably lower than the rate of mutation
reported inland and overseas (10,16).
This demonstrated that the type of conjoined mutation was a point
mutation, and no same-sense mutation was detected. The possible
cause may be related to the novel mutational characteristics of the
Mycobacterium tuberculosis L-form and the effects of
sampling errors, and thus further studies on a greater sample size
are required in order to verify this conclusion.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81172778) and the
Natural Science Foundation of Anhui Province (grant nos. KJ2010A087
and KJ2012A081).
References
|
1
|
Li YL: Microbiological examination method
of common pathogenic bacteria infection. The Book of Chinese
Medical Laboratory Science. First Edition. People’s Medical
Publishing House Co. Ltd; Beijing: pp. 1119–1120. 2000, (In
Chinese).
|
|
2
|
du Toit LC, Pillay V and Danckwerts MP:
Tuberculosis chemotherapy: current drug delivery approaches. Respir
Res. 7:1182006.PubMed/NCBI
|
|
3
|
Telenti A and Iseman M: Drug-resistant
tuberculosis: what do we do now? Drugs. 59:171–179. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Valim AR, Rossetti ML, Ribeiro MO and Zaha
A: Mutations in the rpoB gene of multidrug-resistant
Mycobacterium tuberculosis isolates from Brazil. J Clin
Microbiol. 38:3119–3122. 2000.PubMed/NCBI
|
|
5
|
Ahmad S, Mokaddas E and Fares E:
Characterization of rpoB mutations in rifampin-resistant clinical
Mycobacterium tuberculosis isolates from Kuwait and Dubai.
Diagn Microbiol Infect Dis. 44:245–252. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng XD, Yu WB, Su MQ, Bie LF, et al:
Study on M. tuberculosis isoniazid-resistant isolates by
multi-PCR. Journal of Fourth Military Medical University.
24:849–851. 2003.(In Chinese).
|
|
7
|
Pozzi G, Meloni M, Iona E, et al: ropB
mutations in multidrug-resistant strains of Mycobacterium
tuberculosis isolated in Italy. J Clin Microbiol. 37:1197–1199.
1999.PubMed/NCBI
|
|
8
|
Morris S, Bai GH and Suffys P: Molecular
mechanisms of multiple drug resistance in clinical isolates of
Mycobacterium tuberculosis. J Infect Dis. 171:954–960. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu X, Zhuang Y, Zhang J, et al: Study on
the molecular mechanism of multi-drug resistance in clinical
isolates of Mycobacterium tuberculosis. Zhonghua Jie He He
Hu Xi Za Zhi. 20:332–335. 1997.(In Chinese).
|
|
10
|
Isakova ZhT, Pak OA, Iusupova Elu, et al:
Use of biological microchips in the determination of
drug-resistance of Mycobacterium tuberculosis to rifampicin.
Probl Tuberk Bolezn Legk. 8:50–53. 2005.(In Russian).
|
|
11
|
Heep M, Rieger U, Beck D and Lehn N:
Mutations in the beginning of the rpoB gene can induce resistance
to rifamycins in both Helicobacter pylori and
Mycobacterium tuberculosis. Antimicrob Agents Chemother.
44:1075–1077. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kapur V, Li LL, Iordanescu S, et al:
Characterization by automated DNA sequencing of mutations in the
gene (rpoB) encoding the RNA beta subunit in rifampin-resistant
Mycobacterium tuberculosis strains from New York City and
Texas. J Clin Microbiol. 32:1095–1098. 1994.PubMed/NCBI
|
|
13
|
Wang H and Chen Z: Observations of
properties from the L-form of M. tuberculosis induced by the
antituberculosis drugs. Zhonghua Jie He He Hu Xi Za Zhi. 24:52–55.
2001.(In Chinese).
|
|
14
|
Mattmand LH: Lvariation in mycobacteria.
Am Rev Respir Dis. 202:82–85. 1960.
|
|
15
|
Lu J, Ye S and Li CP: Application of
PCR-SSCP technique on detecting Drug Resistant Genetic Mutation in
M. tuberculosis L-forms among pneumoconiosis patients complicated
with tuberculosis in Huainan mining district. Chin J Ind Hyg Occup
Dis. 25:369–370. 2007.(In Chinese).
|
|
16
|
Cao LX, Wu XQ, Liang JQ, et al: Rapid
detection of rpoB mutations in Mycobacterium tuberculosis by
gene array. Chin Tuberc Respir Dis. 27:332–335. 2004.(In
Chinese).
|