Introduction
The development of transgenic animals is a promising
technology in numerous fields; however, low production efficiency
is a significant limiting factor (1). Rapid and efficient screening of
positive clones of genetically modified cells may accelerate the
development of transgenic animals. To produce transgenic animals,
exogenous DNA must first enter the cell and then the nucleus, prior
to integrating into the chromosomal DNA. To achieve safe and
efficient delivery of plasmid DNA using a non-viral gene delivery
system, the most important cell barrier to overcome is the nuclear
membrane (2,3). The functional diameter of the nuclear
pore is ~10 nm (4). The efficiency
of nuclear import is considered to be an important kinetic block
that influences transgenic expression efficiency (5). Due to the low efficiency rate of the
integration of plasmid DNA into chromosomal DNA, focus has been
placed on improving transfection efficiency by enhancing the
efficiency of transport through the nuclear pore (6).
In order to enhance transfection efficiency, one
strategy is to attach a nuclear localization sequence (NLS) peptide
to the plasmid DNA to facilitate nuclear localization and therefore
improve gene expression efficiency in target tissues (7). Subramanian et al (8) demonstrated that, by conjugating an
NLS peptide to DNA, there was an increase in reporter gene
expression (8). However, other
previous studies that have chemically conjugated NLS to DNA to
guide transport into the nucleus have found that this does not
significantly improve transfection efficiency, and in certain cases
the efficiency was even observed to decrease due to chemical
modification of the DNA, leading to reduced transcription
efficiency in the nucleus (9,10).
In an attempt to solve this problem, bi- (11) and tri-functional (12) synthetic peptides have been designed
to improve gene transfer efficiency.
Succinimidyl-[4-(psoralen-8-yloxy)]-butyrate (SPB), a DNA
intercalating reagent, has been used to non-covalently modify NLS
moieties to enhance transgenic expression efficiency.
The optimal conditions for SPB-NLS peptides to
enhance gene transfer efficiency have yet to be elucidated. Only
upon entering the nucleus is a plasmid able to be expressed. The
aim of the present study was to confirm the function of the NLS
peptide and the peptide derivative SPB-NLS in enhancing the
transfection efficiency of large DNA molecules (plasmids pIN and
pGN). The elucidation of this function may reduce the problems
associated with low screen efficiency in the development of
transgenic animals.
Materials and methods
Materials and reagents
SPB was obtained from Pierce Biotechnology Inc.
(Rockford, IL, USA). Dulbecco’s Modified Eagle’s Medium (DMEM),
DMEM/F-12, fetal bovine serum (FBS) and liposome (Lipofectamine
2000) were obtained from Invitrogen Life Technologies (Carlsbad,
CA, USA). Radioimmunoprecipitation assay (RIPA) buffer [0.1% (w/v)
SDS, 1% (v/v) Triton X-100, 1% (w/v) sodium deoxycholate in
Tris-buffered saline (TBS; 25 mM Tris/HCl, pH 7.5 and 150 mM
NaCl)], and phosphatase and protease inhibitors [1 mM
phenylmethylsulfonyl fluoride (PMSF), 1 mM
Na3VaO4 and 25 mM NaF] were obtained from the
Beyotime Institute of Biotechnology (Haimen, China). Horseradish
peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG)
and rabbit anti-mouse IgG were obtained from Bioworld Technology,
Inc. (St. Louis Park, MN, USA). Antibodies specific to green
fluorescent protein (GFP) and monoclonal bovine β-actin were
obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Mouse monoclonal antibody against insulin-like growth factor 1
(IGF-1) was obtained from Abcam (Cambridge, MA, USA).
Phalloidin-fluorescein isothiocyanate, DAPI and RNaseA were
obtained from Sigma (St. Louis, MO, USA). All other chemicals and
reagents used were of analytical grade. The present study was
approved by the ethics committee of Nanjing Agriculture University
(Nanjing, Jiangsu, China) and the Jiangsu Provincial Academy of
Agricultural Sciences. The license number was SCXK (Su)
2002-0029.
Plasmid DNA
Plasmid DNA (pAC-GFP-N1) was provided by the College
of Life Sciences, Nanjing Agricultural University (Nanjing, China).
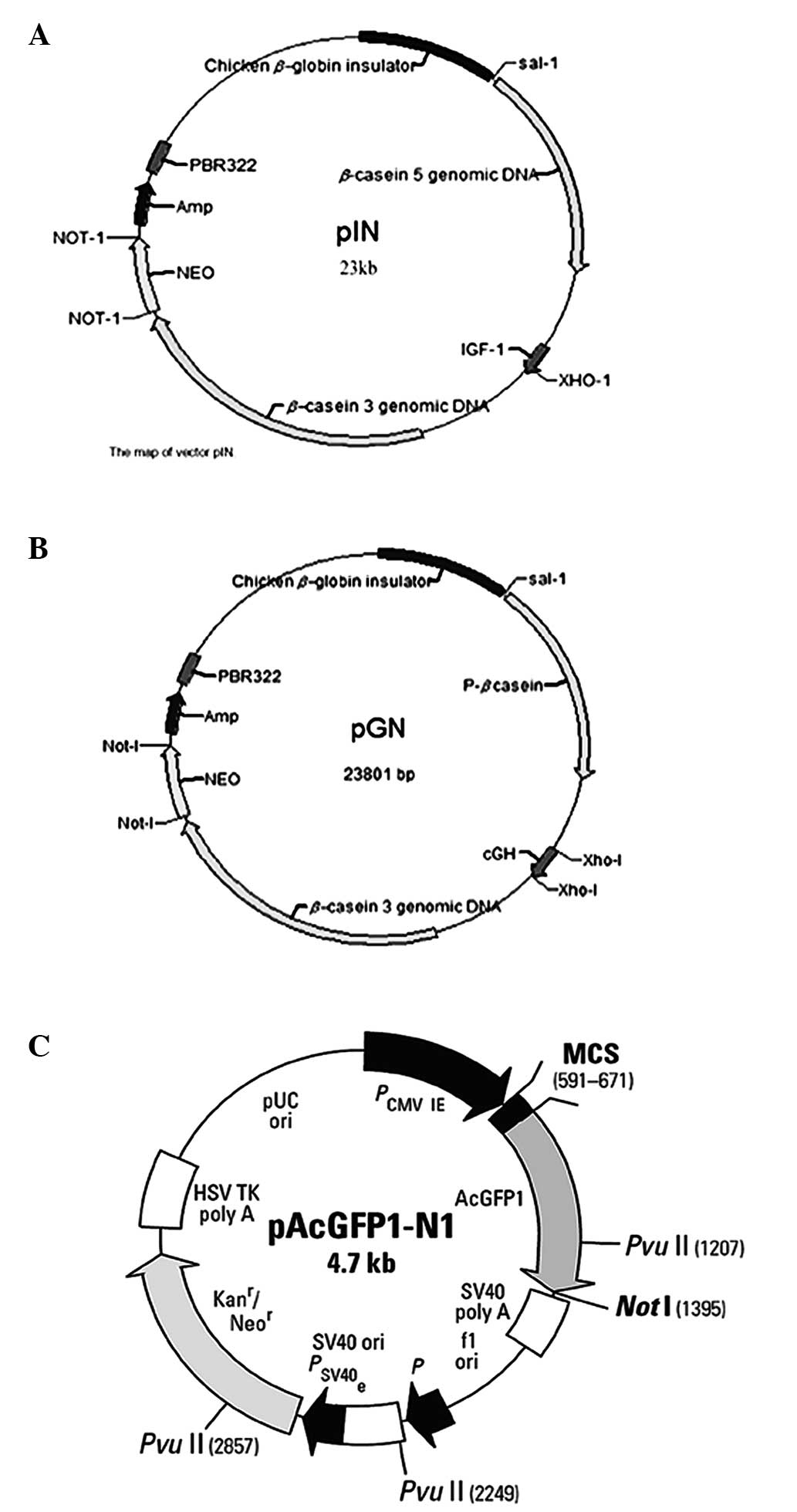
Plasmid DNA pGN and pIN (13) were
constructed in our lab (Key Laboratory of Animal Physiology and
Biochemistry, Nanjing, China). A map of the vectors and a basic
description are shown in Fig. 1.
Plasmid DNA was amplified by Escherichia coli DH5a and
purified using E.Z.N.A® Endo-Free Plasmid Mini Kit I
(Omega Bio-tek®, Norcross, GA, USA). The purity of the
plasmids was determined using electrophoresis on a 1.0% agarose
gel, and the concentration of DNA was determined by measuring the
ultraviolet absorbance at 260 and 280 nm.
Cell line
The human breast cancer cell line Bcap-37 (estrogen
receptor negative, p53 mutated) was obtained from Shanghai Cell
Collection, Chinese Academy of Sciences (Shanghai, China). The
Bcap-37 cell line is difficult to transfect, and was therefore
appropriate for the study of gene transfer efficiency. Bcap-37
cells were grown in DMEM supplemented with 10% FBS at 37°C in a
humidified atmosphere of 5% CO2.
Mammary epithelial cell isolation and
culture
Mammary gland tissues were obtained from Saanen
dairy goats (Capra hircus). Cells were grown in DMEM/F-12
with 10% FBS in the presence of penicillin (100 U/ml) and
streptomycin (100 mg/ml) under standard culture conditions (5%
CO2, 37°C). Mammary epithelial cell culture was
performed as described previously (14).
Preparation of complex formation and
evaluation of optimal conditions
SPB conjugation with NLS
To take advantage of cellular import machinery NLS
peptide and peptide derivative SPB-NLS were synthesized by Sangon
Biotech Shanghai Co., Ltd. (Shanghai, China) using the following
sequences: CGGPKKKRKVP (classical NLS) and SPB-PKKKRKV.
Amine-reactive SPB was used during NLS peptide synthesis to obtain
SPB-terminated NLS conjugates. Purification was performed using
reverse-phase chromatography. High, pure grades of conjugates were
selected with ≥98% high-performance liquid chromatography purity in
accordance with the manufacturer’s own quality control.
Preparation of NLS/DNA and SPB-NLS/DNA
complexes
A total of 2 mg NLS (1.67 μmol; molecular weight,
1,195.53) was first dissolved in 167 μl dimethyl sulfoxide (DMSO)
solution, and 1,500 μl 20 mM
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES)
solution was then added and agitated for 30 min; the final
concentration of NLS solution was 1 pM. In addition, 2 mg SPB-NLS
(1.73 μmol; molecular weight, 1,153.42) was dissolved in 173 μl
DMSO solution, and 1500 μl 20 mM HEPES solution was then added and
agitated for 30 min; the final concentration of SPB-NLS solution
was 1 pM. NLS/DNA and SPB-NLS/DNA complexes were prepared at
various molar ratios by adding different amounts of NLS or SPB-NLS
solution into fixed DNA (plasmid pGN) solution (1 μg). The molar
ratio of the NLS/DNA and SPB-NLS/DNA complexes ranged between
5×102 and 5×104. Mixtures were gently
agitated at room temperature for 30 min to initiate formation.
Gel retardation assay
Different amounts of NLS and SPB-NLS solutions
(molar ratio, between 5×102 and 5×104) were
combined with DNA solution (1 μg plasmid pGN). The function of
condensing plasmid DNA by NLS or SPB-NLS was measured using
electrophoresis. Gels were prepared with 1% agarose in
Tris-acetate-EDTA buffer with ethidium bromide. Electrophoresis was
performed for 30 min at 150 V. The volume of samples loaded was 10
μl.
Analysis of the transfection efficiency
of NLS and SBP-NLS Quantitative polymerase chain reaction (qPCR)
analysis
Bcap-37 cells were incubated at 37°C for 48 h prior
to transfection. Transfection was performed using plasmid pGN
mediated by NLS and SPB-NLS to evaluate the transfection
efficiency. Primers used are listed in Table I. NLS/DNA and SPB-NLS/DNA complexes
were prepared as mentioned above. Liposome solution was added to
complexes and incubated for a further 20 min at room temperature
prior to transfection. Medium was changed to complete medium 6 h
after transfection. Growth hormone (GH) gene expression level was
analyzed using qPCR at 48 h. All transfection experiments were
performed in triplicate.
 | Table IqPCR primers used to detect plasmid
pGN. |
Table I
qPCR primers used to detect plasmid
pGN.
| Gene | Sense primer | Antisense primer | Product length
(bp) |
|---|
| GH |
gagaagctgaaggacctgga |
tacgtctccgtcttgtgcag | 194 |
| Bcap-37 β-actin |
gatcattgctcctcctgagc |
tgtggacttgggagaggact | 385 |
Fluorescence microscopy and flow
cytometric analysis
The plasmid pAC-GFP-N1 (expressing GFP as a reporter
gene) was transfected into Bcap-37 cells with the aid of NLS or
SPB-NLS to evaluate their bioactivity. Approximately 10,000
cells/population were plated on 12-well dishes (Costar®;
Corning, Inc., Corning, NY, USA) and incubated at 37°C for 24 h
prior to transfection. Transfection was performed in accordance
with the aforementioned method. Following transfection, cells were
incubated for 48 h at 37°C and then washed twice with
phosphate-buffered saline (PBS). Images were captured by standard
fluorescence microscopy (Olympus, Tokyo, Japan).
Preliminary results from the fluorescence microscopy
revealed high levels of GFP expression in the SPB-NLS-mediated
group. Therefore, the relative increase in expression was
calculated using flow cytometry, since this method allows the rapid
analysis a large number of cells. Transfection of Bcap-37 cells was
performed in accordance with the aforementioned method using the
plasmid pAC-GFP-N1. Cells were harvested 48 h after transfection,
washed three times with PBS and then suspended in 500 μl cold PBS
and examined using a BD FACSCalibur™ flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA) equipped with an argon laser
(488 nm).
Western blot analysis
Bcap-37 cells were transfected with plasmid
pAC-GFP-N1 to further investigate the function of SPB-NLS.
Transfection was performed in accordance with the aforementioned
method. Cells were harvested 24 and 48 h after transfection and
lysed using RIPA buffer containing phosphatase and protease
inhibitors (1 mM PMSF) on ice for 30 min. Cell lysates were then
centrifuged at 14,000 × g for 10 min at 4°C and the concentration
of the proteins in the supernatant was determined using a
bicinchoninic acid protein assay kit (Pierce Biotechnology Inc.).
The proteins were separated using 4–15% SDS-PAGE and transferred
onto nitrocellulose membranes. The membranes were probed using an
anti-GFP primary antibody (1:2,500), followed by a goat anti-mouse
IgG-HRP secondary antibody. Protein expression was detected using
an enhanced chemiluminescence detection system (Amersham Pharmacia
Biotech, Inc., Amersham, UK). β-actin was used as a loading
control. All bands from the western blotting were analyzed using
Quantity One software (Bio-Rad, Hercules, CA, USA) to verify their
relative expression.
Confocal microscopy to investigate
plasmid DNA localization
To investigate whether NLS and SPB-NLS enhanced
plasmid transfection to the nucleus, plasmid pGN was labeled using
a Cy™3 labeling kit (Mirus Bio LLC, Madison, WI, USA). Transfection
was performed in accordance with the aforementioned method. Bcap-37
cells were divided into five groups [blank, liposome, NLS, SPB-NLS
and wheat-germ agglutinin (WGA)/SPB-NLS]. WGA was used to
specifically block the nuclear pores. Cells were washed three times
with PBS 2 h after transfection and then fixed using 4%
paraformaldehyde for 30 min. Cells were then washed three times
with PBS and dyed with DAPI for 5 min, prior to being washed a
further three times and analyzed using a confocal microscope
(CarlZeiss LSM 710, Carl Zeiss, Oberkochen, Germany). Excitation
and emission wavelengths were 539 nm for Cy3 and 460 nm for DAPI,
respectively.
Cell cycle analysis of cytotoxicity
To evaluate cell cytotoxicity of peptide NLS and
peptide derivative SPB-NLS, the cell cycle of different
transfection groups was analyzed. Transfection was performed in
accordance with the aforementioned method. Cells were harvested and
washed three times with PBS 48 h after transfection. Cells were
then treated with 5 μg/ml RNaseA for 10 min at room temperature,
and stained with 5 μg/ml propidium iodide, a DNA-binding dye, for 2
h at room temperature. Flow cytometry was then used to analyze the
cell cycle.
Validation of the function of SPB-NLS in
goat mammary epithelial cells (GMECs)
Induction of GMECs and cell
transfection
GMECs were seeded in six-well dishes (Costar), and
grown to 50–60% confluence. Prior to experiment, cells were
cultured in induction media for one week to promote the synthesis
of milk protein and fat. The induction media contained 1%
Insulin-Transferrin-Selenium Supplement (Invitrogen Life
Technologies), 5 mg/ml progesterone (ProSpec, East Brunswick, NJ,
USA), 10-7 mol/l hydrocortisone (R&D Systems, Minneapolis, MN,
USA), 10 ng/ml ovine epithelial growth factor (ProSpec) and 5 mg/ml
bovine estradiol (Sigma-Aldrich).
Transfection was performed using plasmid pIN,
mediated by SPB-NLS. Cells were divided into three groups: a
control group (non-transfection), a transfection group (transfected
with plasmid pIN) and an SPB-NLS group (transfected with pIN
mediated by SPB-NLS). The culture medium was changed to DMEM/F-12
containing 10% FBS 6 h after transfection. Cells were collected
after 48 h for subsequent analysis.
Western blot analysis of IGF-1 protein
expression
GMECs were lysed with RIPA buffer containing
phosphatase and protease inhibitors (1 mM PMSF) on ice for 30 min.
Cell lysates were centrifuged at 14,000 × g for 10 min at 4°C. The
proteins in the supernatant were separated using 4–15% SDS-PAGE,
and transferred onto nitrocellulose membranes. Membranes were
blocked with TBS and Tween 20 buffer containing 5% goat serum at
room temperature for 2 h. Membranes were subsequently incubated
with mouse anti-human IGF-1 monoclonal antibody at 4°C for 18 h and
then rabbit anti-mouse IgG-HRP secondary antibody. Protein
expression was detected using an enhanced chemiluminescence
detection system (Amersham Pharmacia Biotech, Inc.), using β-actin
as a loading control. All bands from the western blotting were
analyzed using Quantity One software (Bio-Rad) to verify their
relative expression.
Statistical analysis
The transfection efficiency and cell cytotoxicity
were analyzed using one-way analysis of variance followed by a
Least Significant Difference post hoc test. P<0.05 was
considered to indicate a statistically significant difference. All
results are expressed as the mean ± standard error of the mean.
Results
Gel retardation assay
A prerequisite for transfection-enhancing agents is
their ability to bind nucleic acids. In order to determine the
optimal conditions for complex formation between plasmid DNA (pGN,
1 μg) and NLS peptide or peptide derivative SPB-NLS, different
molar ratios (between 5×102 and 5×104) of NLS
and SPB-NLS were used. The migration of the complexes was detected
using a gel retardation assay (Fig.
2). Compared with naked plasmid DNA (pGN, lane 1), the NLS/DNA
and SPB-NLS/DNA showed varying degrees of retardation (lanes 2–7).
The results from the gel electrophoresis demonstrated that the
complexes (NLS/DNA and SPB-NLS/DNA) were strongly retarded at molar
ratios of 5×104 and 2×104, respectively,
whilst the complexes at molar ratios of 5×102 and
1×103 exhibited no or little retardation. At a molar
ratio 5×103 the complexes showed optimal retarded
mobility. Additionally, NLS and SPB-NLS at a molar ratio of
5×103 did not affect the mobility of plasmid DNA.
Therefore, a molar ratio of 5×103 was used in subsequent
experiments.
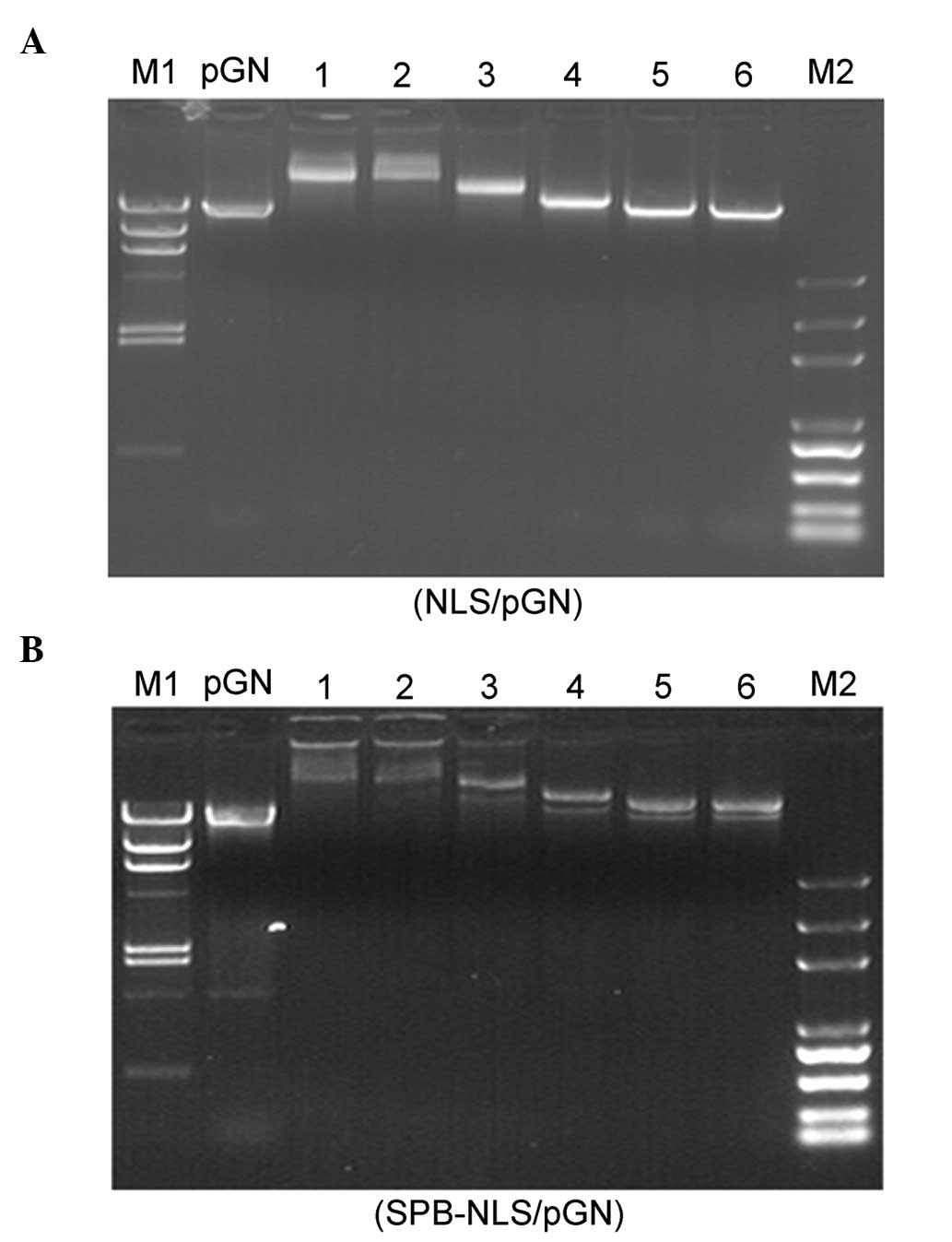 | Figure 2Gel retardation assay. Electrophoresis
of different concentrations of (A) NLS and (B) SPB-NLS binding to
plasmid pGN. (A) Lane 1, naked DNA; lanes 2–7, varying molar ratios
of NLS/pGN (5×104, 2×104, 1×104,
5×103, 1×103 and 5×102); M1,
Marker λ DNA/HindIII; M2, Marker DNA/Trans2K Plus. (B) Lane
1, naked DNA; lanes 2–7, varying molar ratios of SPB-NLS/pGN
(5×104, 2×104, 1×104,
5×103, 1×103 and 5×102); M1,
Marker λ DNA/HindIII; M2, Marker DNA/Trans2K Plus. NLS,
nuclear localization sequence; SBP-NLS,
succinimidyl-[4-(psoralen-8-yloxy)]-butyrate-NLS. |
Analysis of the transfection efficiency
of NLS and SPB-NLS qPCR analysis
Transfection efficiency of NLS and SPB-NLS was
analyzed with plasmid pGN. The results from the qPCR showed that
the expression levels of GH mRNA were significantly higher in the
liposome/pGN group (11.006±1.909) than those in the blank
(1.039±0.349) and control (2.182±0.329) groups. GH mRNA expression
was also significantly higher in the NLS-mediated group
(18.644±1.534) and the SPB-NLS-mediated group (47.648±4.620). The
SPB-NLS-mediated group exhibited peak levels of GH mRNA. The
expression levels of GH mRNA in the NLS and SPB-NLS groups were 69%
and 330% greater than those in the liposome/pGN group, respectively
(Table II).
 | Table IIRelative GH mRNA expression levels in
different groups. |
Table II
Relative GH mRNA expression levels in
different groups.
| Treatment |
|---|
|
|
|---|
| Parameter | Blank | Control
(liposome) | Liposome + pGN | Liposome + NLS +
pGN | Liposome + SPB-NLS
+ pGN |
|---|
| Relative GH mRNA
expression | 1.039±0.349 | 2.182±0.329 | 11.006±1.909 |
18.644±1.534a |
47.648±4.620b |
Fluorescence microscopy and flow
cytometry
Achieving high transgenic expression efficiency was
the ultimate aim of the present study. An in vitro gene
transfer assay was performed with plasmid pAC-GFP-N1, and
fluorescence microscopy was used to evaluate the transfection
efficiency of NLS and SPB-NLS. Untreated cells served as the blank
group and cells transfected with plasmid pGN served as the negative
control group. The results from the fluorescence microscopy showed
that there was no GFP expression in the blank and negative groups.
Compared with the blank group, only small areas of fluorescence
were observed in the NLS group, similar to those in the group
treated with liposome/pAC-GFP-N1. However, large areas of
fluorescence were observed in the SPB-NLS group, indicating that
GFP was highly expressed in the SPB-NLS group (Fig. 3A).
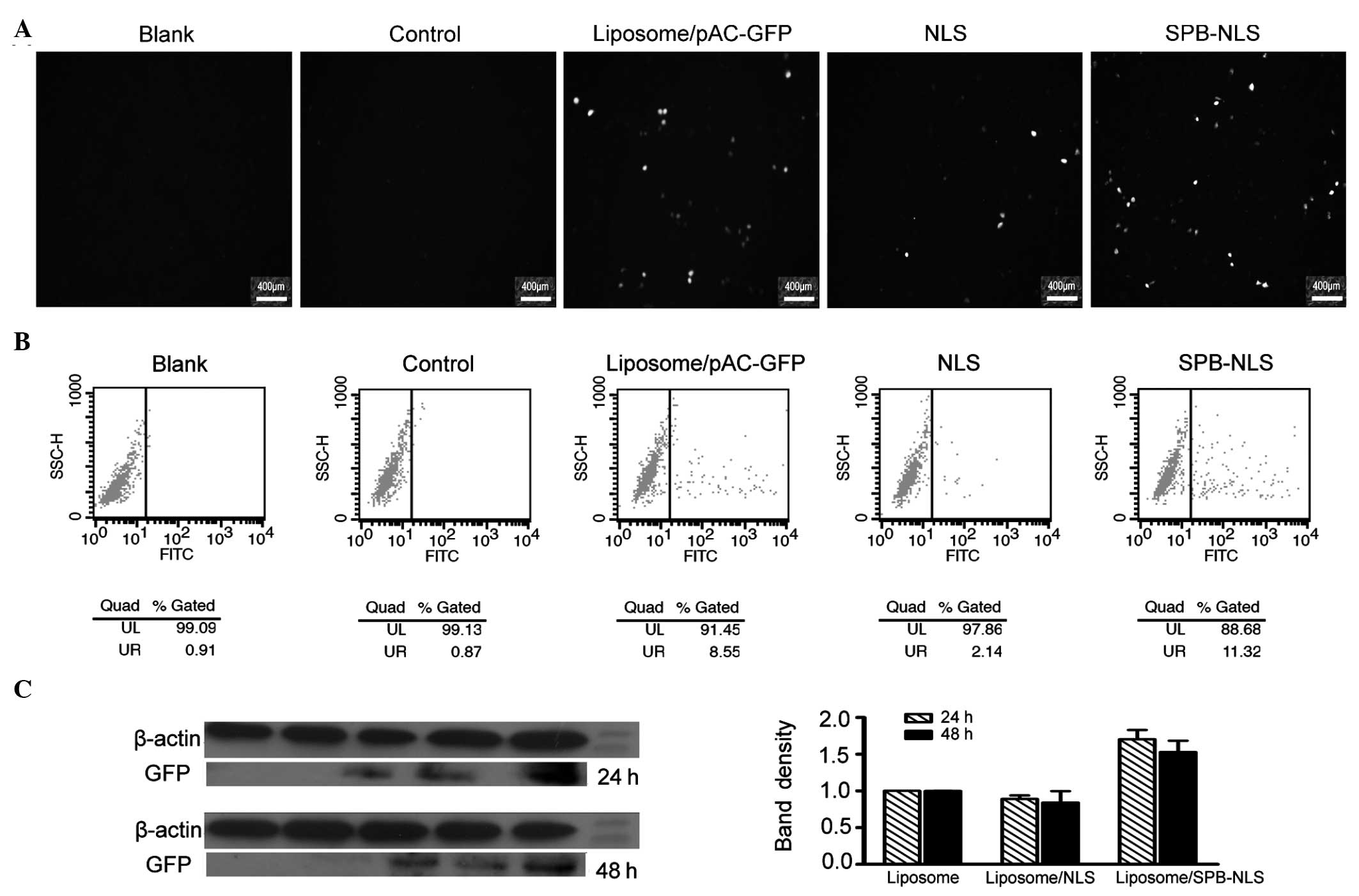 | Figure 3Analysis of transfection efficiency.
(A) Fluorescence microscopy results of transfection efficiency in
blank, control, liposome, NLS and SPB-NLS groups. (B) Flow
cytometry results of transfection efficiency in the blank,
liposome, NLS and SPB-NLS groups. (C) Western blot analysis of GFP
expression levels. Lane 1, blank group; lane 2, negative control
(cells transfected with plasmid pGN); lane 3, liposome group (cells
transfected with plasmid pAC-GFP-N1 by liposome); lane 4,
liposome/NLS group (cells transfected with plasmid pAC-GFP-N1 by
liposome and NLS); lane 5, liposome/SPB-NLS group (cells
transfected with plasmid pAC-GFP-N1 by liposome and SPB-NLS). The
data are presented as the mean ± the standard error of the mean of
triplicate wells from three independent experiments.
*P<0.05 vs. liposome group. GFP, green fluorescent
protein; NLS, nuclear localization sequence; SBP-NLS,
succinimidyl-[4-(psoralen-8-yloxy)]-butyrate-NLS; FITC, fluorescein
isothiocyanate; SSC-H, side scatter pulse height; UL, upper left;
UR, upper right. |
Further analysis using flow cytometry demonstrated
that the percentage of cells expressing GFP in the blank, negative
(liposome) and control (liposome/pGN) groups was 0.91, 1.32 and
0.87%, respectively. The percentage of cells expressing GFP in the
positive group (liposome/pAC-GFP-N1) was 8.55%. The percentage of
positive cells was greater in the SPB-NLS group (11.32%) than that
in the positive group. However, the percentage of positive cells in
the NLS group (2.14%) was lower than that in the positive group
(Fig. 3B). Data analysis
demonstrated that the number of cells positively expressing GFP in
the SPB-NLS group was increased by 32.4%, but decreased by 75% in
the NLS group, compared with the positive group. These results are
consistent with the results from the fluorescence microscopy
analysis.
Western blot analysis
The results from the fluorescence microscopy and
flow cytometry demonstrated that SPB-NLS has an important role in
promoting gene transfer. To confirm these results, the expression
of GFP in the SPB-NLS and NLS groups was investigated. The results
of the western blot analysis demonstrated a significant increase in
GFP expression in the SPB-NLS group (relative band density,
1.70±0.13, 24 h; 1.53±0.16, 48 h) compared with that in the
liposome group (relative band density, 1.00±0.001, 24 h;
1.00±0.001, 48 h) (P<0.05; Fig.
3C). By contrast, the expression of GFP in the NLS group
(relative band density, 0.89±0.048, 24 h; 0.84±0.16, 48 h) was
slightly lower than that in the liposome group (Fig. 3C).
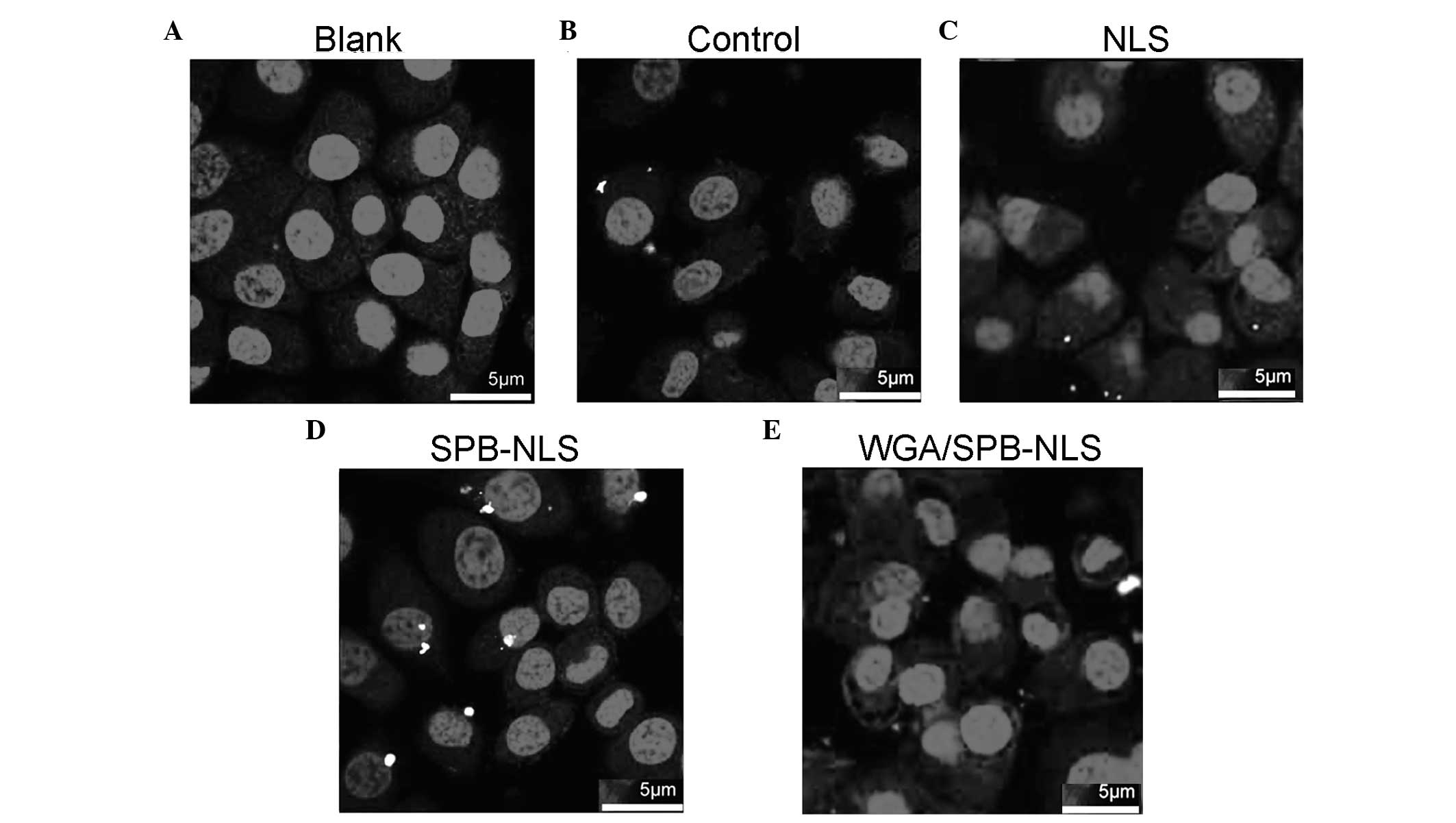
Confocal microscopy to identify the
delivery of plasmid DNA
Since nuclear import efficiency greatly influences
transgenic expression efficiency, it was investigated whether NLS
and SPB-NLS facilitated nuclear entry of plasmid DNA. Confocal
microscopy analysis demonstrated that no plasmid DNA (red) was
present in the nucleus in the control group (Fig. 4A). Merged images with Cy3 and DAPI
staining revealed that a small number of red plasmids were present
in the perinuclear area in the liposome/pGN group (Fig. 4B). In the NLS group, there was an
increase in the number of plasmids in the cell, indicating that NLS
enhanced plasmid transport into the cytoplasm. However, in the NLS
group few or no plasmids entered the nucleus (Fig. 4C). By contrast, in the SPB-NLS
group there was an increase in the number of plasmids in the
nucleus, demonstrating that SPB-NLS enhanced entry of plasmid DNA
into the nucleus (Fig. 4D). To
confirm whether SPB-NLS was interacting with the nuclear pore
complex (NPC), WGA was used to specifically block the nuclear
pores. When cells were treated with WGA, few plasmids were observed
in the cytoplasm and none were observed in the nucleus. Therefore,
peptide derivative SPB-NLS increases transfection efficiency by
interacting with the NPC (Fig.
4E).
Cells cytotoxicity analysis
An optimized gene delivery system should be able to
improve transfection efficiency without any toxic effects. The cell
cycle is a good indicator of cell viability. Flow cytometric
analysis revealed marked differences in the cell cycle between the
blank and liposome/pGN groups in the G1 and S phases (Table III). Data analysis demonstrated
that liposome affected the G1, S and G2 phases, indicating that
liposome treatment may have a slight cytotoxic effect on cells. The
apoptosis percentages of the SPB NLS and NLS groups revealed
similarities with the liposome group, indicating that SPB NLS and
NLS may also have a slight toxic effect.
 | Table IIICell cycle analysis of
cytotoxicity. |
Table III
Cell cycle analysis of
cytotoxicity.
| Treatment | G1 (%) | S (%) | G2 (%) | Apoptosis (%) |
|---|
| Blank | 61.41±0.42 | 31.58±0.89 | 7.01±0.47 | 1.11±0.29 |
| Liposome/pGN | 56.79±0.87 | 38.93±2.30 | 4.29±1.43 | 2.21±0.59 |
|
Liposome/SPB-NLS/pGN | 57.19±0.13 | 38.70±1.04 | 4.12±0.91 | 1.74±0.29 |
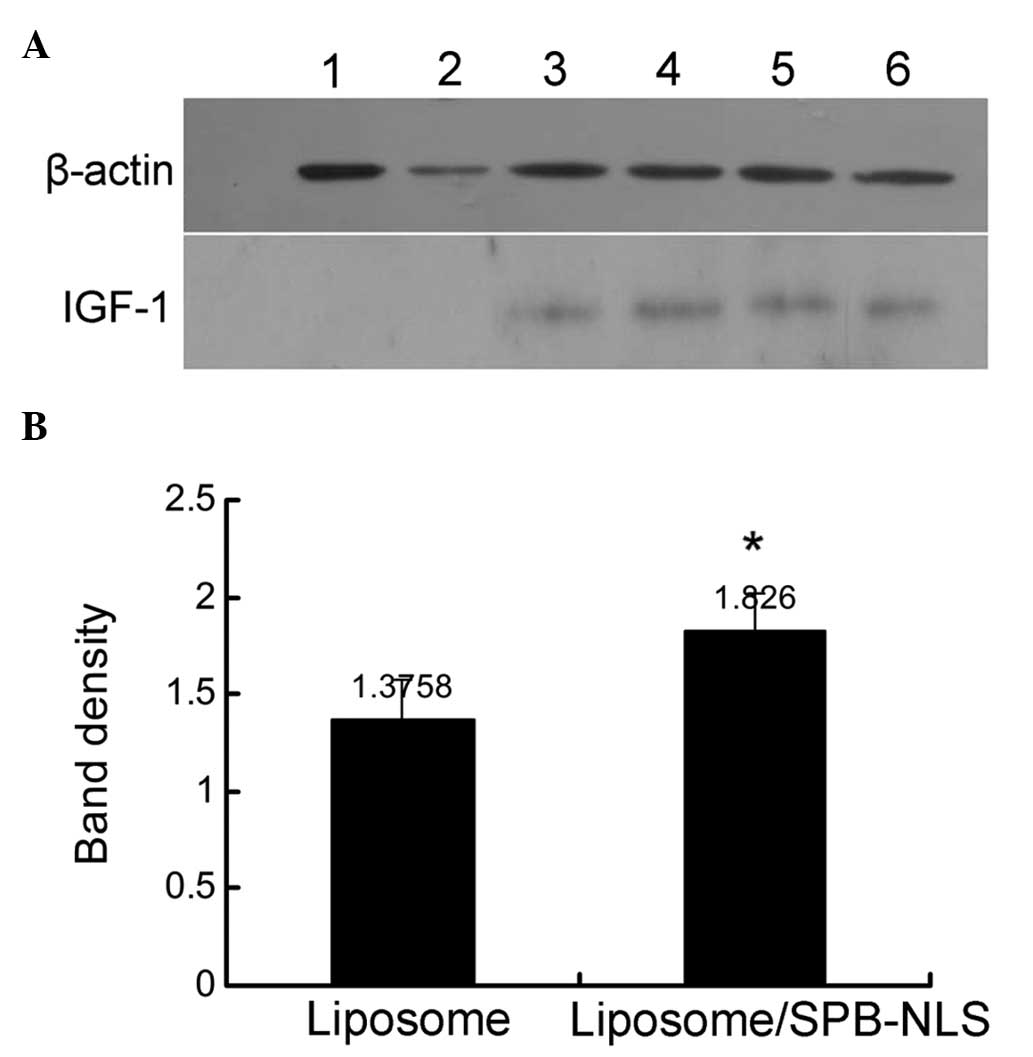
Analysis of SPB-NLS function in
GMECs
Previous experiments demonstrated that SPB-NLS
improved the transfection efficiency in the Bcap-37 cell line. To
investigate whether SPB-NLS may be used to alleviate problems in
the development of transgenic animals, plasmid pIN was transfected
into GMECs to evaluate the transfection efficiency. Transfection of
pIN into GMECs was confirmed using immunohistochemistry and PCR
(data not shown). IGF-1 protein was found to be expressed in all
plasmid pIN-transfected groups. In addition, the SPB-NLS group
(relative band density, 1.826±0.191) exhibited greater IGF-1
protein expression than the liposome group (relative band density,
1.376±0.204) (Fig. 5). These
results further indicated that SPB-NLS enhances transfection of
large DNA molecules (plasmid pIN).
Discussion
The present study aimed to investigate methods to
improve gene transfer efficiency and to characterize the mechanism
of plasmid import into transfected cells in order to improve DNA
delivery in vivo. The study also aimed to develop a
convenient delivery system for functional gene transfer and
efficient screening for the development of transgenic animals. All
macromolecules enter or exit the nucleus through the NPC (15). Similarly, plasmid DNA is
transported into the nuclei of non-dividing cells via the NPC. DNA
fragments >1 kb are unable to enter the nuclei, whilst those
<1 kb readily pass through the NPC (16,17).
In contrast to what has previously been demonstrated in intact
cells, plasmids ≤15 kb are transported into the nuclei upon cell
division (18). It is well
established that the NLS has an important role in DNA transport
into the nucleus. In this study it was investigated whether large
DNA molecules (such as plasmids pIN, 23 kb, and pGN, 23 kb) were
able to pass in and out of the nucleus with the aid of NLS/SPB-NLS
to allow continued expression.
The interaction between the transfection complex,
NLS/SPB-NLS, and DNA was first evaluated. Two peptides were
designed to condense DNA and enhance delivery into the nucleus.
Previous studies have found that NLS peptides do not markedly
enhance plasmid DNA nuclear entry efficiency and that chemical
modifications of DNA even reduce the transcription efficiency or
stability in the nucleus (10).
Consistent with this, the results from the present study
demonstrated that NLS peptide downregulated gene expression
efficiency: Fluorescence microscopy and western blot analysis
showed that the NLS group expressed less GFP than the liposome/pGN
group (10). The low expression
efficiency in the NLS group may have been due to a lack of binding.
Enhancing the binding of DNA with NLS was therefore the first
challenge. A previous study by Lechardeur and Lukacs (2) found that single- and double-stranded
plasmids were degraded with an apparent half-life of between 50 and
90 min. Several different methods have been used to attach NLS to
plasmid DNA, including ionic interaction (19), covalent attachment (7) and peptide nucleic acid clamp
(20), and have been shown to
significantly improve nuclear transport efficiency. However,
increased transport into the nucleus does not necessarily correlate
with increased levels of protein expression (21). Ciolina et al (22) demonstrated that microinjected DNA
with covalently attached NLS raised transfection efficiency by
≤160% compared with unmodified DNA (22); however, expression decreased by 60%
with plasmid bearing 43 NLS peptides. The results from the present
study showed that the expression levels of GH mRNA in the NLS group
were increased by 69% compared with those in the liposome/pGN
group, but the percentage of positive cells (2.14%) and the GFP
protein expression levels (relative band density, 0.89±0.048, 24 h;
0.84±0.16, 48 h) were lower than those in the liposome/pGN group.
In combination these results suggest that covalent binding of NLS
to DNA leads to transcriptional inactivation, possibly due to
over-tight binding (10).
Therefore, non-covalent modification of DNA may be an alternative
(23). It has been suggested that
SPB has the potential to non-covalently modify target molecules
without affecting gene expression (24). In the present study it was
demonstrated using flow cytometry and western blot analysis that
GFP expression was elevated in the SPB-NLS group, indicating that
SPB-NLS has high biological activity.
qPCR was performed to quantitatively evaluate the
transfection efficiency of NLS and SPB-NLS. The results revealed
that SPB-NLS notably improved gene transfer efficiency. The mRNA
expression levels of GH in the NLS and SPB-NLS groups were higher
than those in the liposome/pGN group, which is consistent with the
results by Jain and Gewirtz (25).
The results from the present study demonstrate that the NLS group
had higher mRNA expression levels of GH than the liposome/pGN
group, indicating that NLS has an important role in gene transfer.
The exact mechanism of SPB-NLS-mediated gene delivery has yet to be
elucidated; however, the results of the present study show that the
addition of NLS and SPB-NLS may affect gene transcription
efficiency. Further investigation is required, however, to
understand the mechanism by which SPB-NLS affects transfection.
In order to assess the effect of NLS and SPB-NLS on
gene expression, GFP expression levels were analyzed using
fluorescence microscopy, flow cytometry and western blotting. Flow
cytometry quantitatively estimates gene expression and accurately
determines fluorescent intensity. Similar results were found using
the different methods. It was found that GFP expression was
significantly increased in the SPB-NLS group but decreased in the
NLS group compared with that in the liposome/pGN group. Similarly,
high GH mRNA transcription efficiency along with low GFP expression
efficiency was observed in the NLS group. This phenomenon may have
been a result of the content of the DNA transferred into the
nucleus and due to the plasmid DNA being prevented from being
degraded by nucleases.
Lipid-mediated transfection requires endocytosis.
Unprotected DNA in the cytoplasm is degraded by resident cytosolic
nucleases (26). Much of the
transferred DNA is retained in endosomes, escapes to the cytoplasm
and enters into the nucleus at low rates, which limits the
efficiency of liposome-mediated gene transfer (27). The results of the present study
demonstrated that in the liposome/pGN group plasmids were located
around the nuclear membrane; however, in the SPB-NLS group, plasmid
DNA was observed in the nucleus. Since the nuclear envelope is an
important barrier in transfection, the mechanism underlying the
interaction of SPB-NLS with the nuclear envelope was investigated.
The position of labeled plasmid DNA was analyzed using confocal
microscopy (28). The results
revealed that the labeled plasmids were delivered to the nucleus in
the SPB-NLS group. In the liposome/pGN group, however, the plasmids
remained around the nuclear envelope. To determine whether SPB-NLS
was interacting with the NPC, WGA was used. WGA cross-links with
phenylalanine-rich repeat motifs in the NPC, specifically
inhibiting the exchange of material between the nucleus and the
cytoplasm. Following WGA treatment, no labeled plasmid was observed
in the nucleus, further demonstrating that SPB-NLS affects the
formation of the NPC by interacting with nuclear envelope.
Contradictory to our study, a previous report suggested that a
random-walk diffusion of DNA molecules was likely to be inefficient
and slow (29). However, naked DNA
does not remain free in the nucleus as histones rapidly assemble
transferred DNA into chromatin-like structures, thus providing a
mechanism for pulling and condensing the filamentous molecule into
the nucleus.
Unless plasmid DNA enters the nucleus, no gene
integration or replication of any plasmid DNA occurs. NLS mediates
the trafficking from the cytoplasm into the nucleus. In the present
study it was found that SPB-NLS significantly enhanced gene
transfer and expression efficiency. Although high transfection
efficiency was demonstrated, the mechanism by which SPB-NLS
enhances transfection has yet to be fully elucidated. In
conclusion, in this study it was shown that SPB-NLS, as a
transfection-enhancing agent, may improve the transport of large
molecular DNA into the nucleus and provides a ‘fixed target’ in
nuclear trafficking. In the future this may provide a safe and
alternative strategy for rapid and efficient screening of
transgenic positive clones.
Acknowledgements
This study was supported by grants from the National
Animal Transgenic Breeding Grand Project (nos. 2011ZX08008-004 and
2013ZX08008-004) and the Priority Academic Program Development of
Jiangsu Higher Education Institutions (PAPD).
References
|
1
|
Whitelaw CB, Farini E and Webster J: The
changing role of cell culture in the generation of transgenic
livestock. Cytotechnology. 31:3–8. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lechardeur D and Lukacs GL: Intracellular
barriers to non-viral gene transfer. Curr Gene Ther. 2:183–194.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leong KW, Mao HQ, Truong-Le VL, Roy K,
Walsh SM and August JT: DNA-polycation nanospheres as non-viral
gene delivery vehicles. J Control Release. 53:183–193. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ludtke JJ, Sebestyén MG and Wolff JA: The
effect of cell division on the cellular dynamics of microinjected
DNA and dextran. Mol Ther. 1:579–588. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Munkonge FM, Dean DA, Hillery E,
Griesenbach U and Alton EW: Emerging significance of plasmid DNA
nuclear import in gene therapy. Adv Drug Deliv Rev. 55:749–760.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kamiya H, Tsuchiya H, Yamazaki J and
Harashima H: Intracellular trafficking and transgene expression of
viral and non-viral gene vectors. Adv Drug Deliv Rev. 52:153–164.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zanta MA, Belguise-Valladier P and Behr
JP: Gene delivery: a single nuclear localization signal peptide is
sufficient to carry DNA to the cell nucleus. Proc Natl Acad Sci
USA. 96:91–96. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Subramanian A, Ranganathan P and Diamond
SL: Nuclear targeting peptide scaffolds for lipofection of
nondividing mammalian cells. Nat Biotechnol. 17:873–877. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brandén LJ, Mohamed AJ and Smith CI: A
peptide nucleic acid-nuclear localization signal fusion that
mediates nuclear transport of DNA. Nat Biotechnol. 17:784–787.
1999.PubMed/NCBI
|
|
10
|
Tanimoto M, Kamiya H, Minakawa N, Matsuda
A and Harashima H: No enhancement of nuclear entry by direct
conjugation of a nuclear localization signal peptide to linearized
DNA. Bioconjug Chem. 14:1197–1202. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Man N, Yu L, Zheng F, Li Y and Wen LP:
Efficient gene transfer to rat fetal osteoblastic cells by
synthetic peptide vector system. Protein Pept Lett. 16:368–372.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smith J, Guidry J and Wittung-Stafshede P:
Novel ‘three-in-one’ peptide device for genetic drug delivery.
Protein Pept Lett. 10:1–7. 2003.
|
|
13
|
Lin J, Yu Q, Zhang Q and Yang Q:
Construction of mammary gland specific expression plasmid pIN and
its expression in vitro and in vivo. Afr J Biotechnol.
11:7038–7045. 2012.
|
|
14
|
Hu H, Wang J, Bu D, et al: In vitro
culture and characterization of a mammary epithelial cell line from
Chinese Holstein dairy cow. PLoS One. 4:e76362009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gasiorowski JZ and Dean DA: Mechanisms of
nuclear transport and interventions. Adv Drug Deliv Rev.
55:703–716. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ludtke JJ, Zhang G, Sebestyén MG and Wolff
JA: A nuclear localization signal can enhance both the nuclear
transport and expression of 1 kb DNA. J Cell Sci. 112:2033–2041.
1999.PubMed/NCBI
|
|
17
|
Sebestyén F, Szendrei G, Mák M, et al:
Coloured peptides: synthesis, properties and use in preparation of
peptide sub-library kits. J Pept Sci. 4:294–299. 1998.PubMed/NCBI
|
|
18
|
Wildeman AG: Regulation of SV40 early gene
expression. Biochem Cell Biol. 66:567–577. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Akita H, Tanimoto M, Masuda T, et al:
Evaluation of the nuclear delivery and intra-nuclear transcription
of plasmid DNA condensed with micro (mu) and NLS-micro by
cytoplasmic and nuclear microinjection: a comparative study with
poly-L-lysine. J Gene Med. 8:198–206. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Díaz-Mochón JJ, Bialy L, Watson J,
Sánchez-Martín RM and Bradley M: Synthesis and cellular uptake of
cell delivering PNA-peptide conjugates. Chem Commun (Camb).
3316–3318. 2005.PubMed/NCBI
|
|
21
|
Leahy P, Carmichael GG and Rossomando EF:
Novel biotinylated plasmid expression vectors retain biological
function and can bind streptavidin. Bioconjug Chem. 7:545–551.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ciolina C, Byk G, Blanche F, Thuillier V,
Scherman D and Wils P: Coupling of nuclear localization signals to
plasmid DNA and specific interaction of the conjugates with
importin alpha. Bioconjug Chem. 10:49–55. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boulanger C, Di Giorgio C and Vierling P:
Synthesis of acridine-nuclear localization signal (NLS) conjugates
and evaluation of their impact on lipoplex and polyplex-based
transfection. Eur J Med Chem. 40:1295–1306. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grosse S, Thévenot G, Monsigny M and Fajac
I: Which mechanism for nuclear import of plasmid DNA complexed with
polyethylenimine derivatives? J Gene Med. 8:845–851. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jain PT and Gewirtz DA: Enhancement of
liposomal gene delivery in human breast cancer cells by dimethyl
sulfoxide. Int J Mol Med. 1:609–611. 1998.PubMed/NCBI
|
|
26
|
Braun K, von Brasch L, Pipkorn R, et al:
BioShuttle-mediated plasmid transfer. Int J Med Sci. 4:267–277.
2007. View Article : Google Scholar
|
|
27
|
Mehier-Humbert S, Bettinger T, Yan F and
Guy RH: Ultrasound-mediated gene delivery: kinetics of plasmid
internalization and gene expression. J Control Release.
104:203–211. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou M, Liu H, Xu X, et al: Identification
of nuclear localization signal that governs nuclear import of BRD7
and its essential roles in inhibiting cell cycle progression. J
Cell Biochem. 98:920–930. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cereghini S and Yaniv M: Assembly of
transfected DNA into chromatin: structural changes in the
origin-promoter-enhancer region upon replication. EMBO J.
3:1243–1253. 1984.PubMed/NCBI
|