Introduction
Gastric cancer (GC) is the second leading cause of
cancer-associated mortality in males and the third leading cause in
females worldwide (1). The
majority of patients are diagnosed at an advanced stage and, thus,
the overall treatment response is poor and the five-year survival
rate is low. Understanding the molecular pathophysiology of GC is
essential for determining methods to effectively inhibit tumor
progression.
Recent studies have demonstrated that epigenetic
mechanisms are closely associated with the development, progression
and metastasis of GC. These epigenetic changes include DNA
methylation, histone modifications and miRNA expression (2). Aberrant DNA methylation and histone
modification occurs in the promoter CpG island of tumor suppressor
genes (TSGs) where DNA is transcribed into RNA (3). EFEMP1 has antiangiogenic activity via
suppression of endothelial cell sprouting (4), suggesting that it has a critical role
in cancer development. However, the association between EFEMP1
deregulation and cancer remains a matter of debate. In support of a
possible tumor-suppressive role, downregulation of EFEMP1 gene
and/or EFEMP1 promoter methylation occurs in lung, liver, prostate,
breast and nasopharyngeal carcinoma (5–9). In
addition, EFEMP1 may have a potential cancer-promoting function in
cervical cancer (10), breast
carcinoma (11) and pancreatic
cancer (12). These studies have
demonstrated that EFEMP1 has contrasting roles in cancer, depending
on the tissue in which it is expressed. The 5′-end of the EFEMP1
gene contains numerous CpG islands, suggesting that its expression
may be controlled by DNA methylation and histone modification.
Hypermethylation of the EFEMP1 promoter has been reported in
various human malignancies. This finding prompted us to examine
whether epigenetic silencing of EFEMP1 was involved in
carcinogenesis.
To the best of our knowledge, the role of EFEMP1 in
GC has not been examined to date. The present study aimed to verify
whether decreased EFEMP1 expression in GC is associated with DNA
methylation and histone modification. Four GC cell lines were used
to examine EFEMP1 mRNA expression, DNA methylation and histone
modification of EFEMP1. In addition, 45 GC specimens and 45
corresponding non-malignant gastric tissues were recruited to
observe the mRNA expression and DNA methylation of EFEMP1 as well
as its clinical significance.
Materials and methods
Cell culture and treatment with
5-aza-2′-deoxycytidine (DAC) and trichostatin A (TSA)
Four human GC cell lines, MKN45, SGC7901, BGC823 and
AGS were obtained from the Institute of Biochemistry and Cell
Biology, Chinese Academy of Sciences (Shanghai, China). One
immortalized normal gastric cell line, GES1, was obtained from the
Oncology Institute of China Medical University (Shenyang, China).
These cells were cultured in RPMI-1640 medium (Gibco-BRL, Grand
Island, NY, USA) supplemented with 10% fetal bovine serum
(Gibco-BRL) and incubated at 37°C in a humidified 5% CO2
atmosphere. MKN45, SGC7901, BGC823 and AGS cells were incubated for
24 h prior to treatment as follows: (i) DAC group, 5 μM DAC (Sigma,
St. Louis, MO, USA) was added and cells were incubated for three
days and the medium containing DAC was refreshed every day; (ii)
TSA group, 0.3 μM TSA (Sigma) for 24 h; (iii) DAC + TSA group, 5 μM
DAC for 48 h followed by 0.3 μM TSA for an additional 24 h; (iv)
control group, control cells of the same batch were incubated
without DAC or TSA, with replacement of fresh medium on the same
schedule as that used for the drug-treated cells. The time, dose
and sequence of DAC and/or TSA are based on those of previous
studies (13,14).
Tissue samples
Human GC samples were collected from 45 patients who
underwent a gastrectomy at the Department of Surgery, China Medical
University (Shenyang, China) between January 2009 and June 2011.
All GC cases were pathologically confirmed. Non-malignant gastric
tissues that were at least 5 cm away from the tumor, were obtained
from the patients.
Wound healing assay
The cells were plated in 6-well plates and
maintained in RPMI-1640 medium containing 10% fetal calf serum. A
wound was created in the center of the cell monolayer by a sterile
plastic pipette tip. The cells were allowed to migrate for 24 h.
The images were captured at 0, 12 and 24 h following wounding to
assess the ability of the cells to migrate into the wound area
using an inverted microscope (IX-71; Olympus, Tokyo, Japan).
Matrigel invasion assay
Approximately 5×104 cells cultured in 200
μl serum-free RPMI-1640 medium were seeded onto Matrigel-coated
8-μm pore size Transwell filters (Corning Life Sciences, Corning,
NY, USA) in the upper chambers. A total of 500 μl RPMI-1640
containing 10% fetal calf serum was added to the lower chambers as
a chemoattractant. The cells were incubated at 37°C in a humidified
5% CO2 atmosphere for 24 h. The cells that had
successfully invaded through the inserts were fixed in 4%
paraformaldehyde for 30 min and stained with methylrosanilinium
chloride. The invaded cells were counted from five pre-selected
microscopic fields of view at a magnification of ×200. The results
of the Matrigel invasion assay were obtained from three independent
experiments.
Cell counting kit-8 (CCK-8) assay
The cell proliferative ability was evaluated by a
CCK-8 assay (C0037; Beyotime Institute of Biotechnology, Shanghai,
China). The cells were seeded in 96-well plates
(3×103/well). The cells were washed with PBS, and the
medium was replaced with fresh medium containing DAC (5 μmol/l)
and/or TSA (300 nmol/l). Following culture, the CCK-8 solution (10
μl/100 μl medium) was added to each well and the cells were
incubated for 1 h at 37°C. The absorbance was measured at 450 nm
using a Synergy2 Multi-Mode Microplate reader (BioTek, Winooski,
VT, USA). The GC cells cultured in RPMI-1640 without DAC or TSA
were used as the controls. The assay was conducted in five
replicate wells for each sample and three parallel experiments were
performed.
RNA extraction and quantitative
polymerase chain reaction (qPCR)
The total RNA was extracted from GC cells, human GC
tissues and the corresponding non-cancerous tissues from the same
patients. Extraction was performed using TRIzol reagent (Invitrogen
Life Technologies, Carlsbad, CA, USA) according to the
manufacturer’s instructions and reversely transcribed into cDNA
using an Expand Reverse Transcriptase kit (Takara, Dalian, China).
The expression of EFEMP1 mRNA was detected using qPCR with the
following program: 95°C for 30 sec, 35 cycles of 95°C for 5 sec and
60°C for 30 sec. The reaction mixture contained 12.5 μl SYBR Green
(Takara), 1 μl each primer, 2 μl cDNA and 8.5 μl
diethylpyrocarbonate (DEPC)-treated water. The primers used were as
follows: Sense: 5′-CGCCAGCACATTGTGAATGAC-3′ and antisense:
5′-TTTGAGTTGCACTCCACCACG-3′ for EFEMP1; and sense:
5′-CATGAGAAGTATGACAACAGCCT-3′ and antisense:
5′-AGTCCTTCCACGATACCAAAGT-3′ for GADPH. The negative control used
DEPC-treated water to replace the cDNA templates for every PCR. The
EFEMP1 level was expressed as Ct following normalization to the
levels of GAPDH mRNA.
Chromatin immunoprecipitation (ChIP)
assay
ChIP assays were performed as described previously
(15) to measure the levels of
histone methylation and acetylation at EFEMP1 promoter regions in
GES1, MKN45, SGC7901, BGC823 and AGS cell lines. The cells were
fixed to crosslink the nuclear protein to DNA by adding
formaldehyde, resuspended in lysis buffer and sonicated to generate
~500–1,000 bp DNA fragments using a sonicator (Shanghai Bilon
Instrument Co., Ltd., Shanghai, China). The main soluble chromatin
fraction was immunoprecipitated using antibodies against Lys-9
trimethylated histone H3 antibody (05–1242; Millipore, Billerica,
MA, USA) or Lys-9 acetylated histone H3 (07–352; Millipore). The
remaining soluble fraction was incubated with normal rabbit IgG
(negative control). In addition, 1/100 of the soluble fraction
collected prior to adding the antibody was used as an internal
control for the quantity of input DNA. The crosslinking between DNA
and proteins was reversed by heating the samples at 65°C for 4 h,
followed by proteinase K digestion. The DNA was then extracted with
phenol/chloroform. A total of 2 μl of immunoprecipitated DNA, DNA
input control and negative control were used for PCR. The following
primer set for PCR were designed to amplify the overlapping
fragments of 131 bp along the EFEMP1 promoter: Sense:
5′-ATCCCTTGATGGACACTT-3′ and antisense:
5′-TCTCATTTCTGGGTATTTACT-3′. PCR products were subjected to 2.5%
agarose gel electrophoresis at 120 V for 40 min and quantified
using the Fluor Chen 2.0 system (Bio-Rad, Hercules, CA, USA). The
levels of histone modification in each immunoprecipitation were
calculated by quantifying the intensity of the PCR product in the
immunoprecipitated DNA vs. the DNA input control. The ChIP
experiments were repeated three times.
Methylation-specific PCR (MSP)
Genomic DNA (2 μg) was treated with NaOH (2 M) at
42°C for 20 min. Following denaturation, the DNA was incubated with
hydroquinone and sodium bisulfate at 54°C for 16 h in the dark. The
DNA was purified using a DNA clean up kit (Promega Corporation,
Madison, WI, USA), followed by incubation with 3 M NaOH at 37°C for
15 min, and precipitation with ammonium acetate and 100% ethanol at
−20°C overnight. The following day, DNA was washed with 70% ethanol
and dissolved in 15 μl Tris-EDTA buffer. The primers used for MSP
were located in the promoter region of the EFEMP1 gene. The CpG map
of the EFEMP1 promoter and the location of primers used in the
present study were based on a previous investigation (16). The primers for the methylated
EFEMP1 CpG island were: Sense: 5′-TTTTTTCGTAGGGCGTTTTTTATC-3′ and
antisense: 5′-TTATAATCTACGATCGAACCTCGATT-3′. The primers for the
unmethylated EFEMP1 CpG islands were sense:
5′-GCGGATTGTTTCGGGAGATC-3′ and antisense:
5′-CAAAAAACGAAAATAAAACGACGAC-3′. Peripheral blood cell DNA from
healthy adults treated with SssI methyltransferase (New England
Biolabs, Ipswich, MA, USA) and untreated DNA were used as the
positive and negative controls, respectively. Water blanks were
used as a negative control. PCR products were separated by
electrophoresis on 2% agarose gels and quantified using the Fluor
Chen 2.0 system.
Statistical analysis
All statistical analyses were performed using SPSS
version 17.0 (SPSS, Inc., Chicago, IL, USA). The χ2 and
Fisher’s exact tests were used for categorical variables, and
Student’s t-test or one-way analysis of variance for continuous
variables. The relative mRNA expression levels (EFEMP1/GAPDH) were
calculated from the quantified data. Data are expressed as the mean
± standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
Effect of DAC and TSA on migration,
invasion and proliferation of MKN45 cells in vitro
To investigate the inhibitory effect of DAC and TSA
on migration of MKN45 cells, a wound-healing assay was performed.
At 24 h following establishing the wound, the control group
achieved almost complete wound closure (80.00±0.9129%), compared
with 64.75±0.8539% in the TSA group and only 52.25±0.6292 and
53.25±1.109% in the DAC and DAC + TSA groups, respectively. These
results demonstrated that DAC reduced cell migration significantly.
Furthermore, TSA had a moderate effect on the migration of MKN45
cells. Combined treatment with the two agents reduced cell
migration similar to that with DAC (P<0.01; Fig. 1A and B).
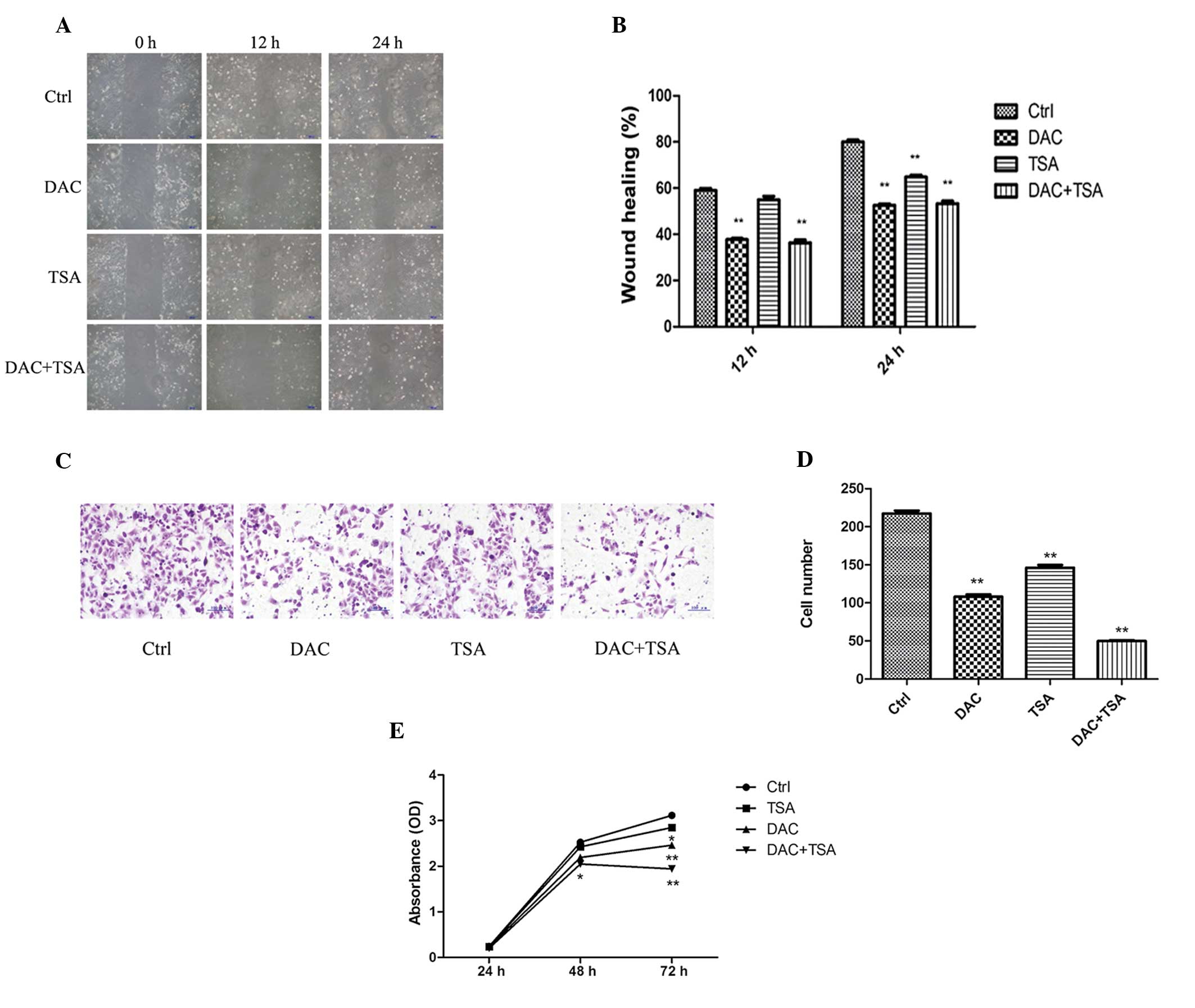 | Figure 1Effect of DAC and/or TSA on MKN45 cell
migration, invasion and proliferation. (A) Wound healing assay of
the MKN45 cells. The images were captured at 0, 12 and 24 h
following the wound incision (magnification, ×100). (B) The
percentage of wound closure was measured in at least three randomly
selected regions (mean ± SD). (C) The invasion ability of MKN45
cells was observed by Matrigel invasion assay following treatment
with DAC and/or TSA. Representative images of the treated and
untreated cells are presented (magnification, ×200). (D) The
columns indicate the number of cells invading at the 24 h time
point. The values represent the mean ± SD. (E) MKN45 cells were
treated with DAC and/or TSA and proliferation was estimated at 24 h
intervals up to 72 h using a cell counting kit-8 assay. Data are
presented as the mean ± SD. *P<0.05,
**P<0.01, compared with the Ctrl. SD, standard
deviation; DAC, 5-aza-2′-deoxycytidine; TSA, trichostatin A; Ctrl,
control. |
In order to examine whether DAC and TSA regulate GC
invasion, the invasive capability of MKN45 cells was investigated
using a Matrigel invasion assay. The numbers of cells penetrating
Matrigel and adhering to the membrane in the control, DAC, TSA and
DAC + TSA groups was 217.4±3.723, 108±3.033, 146.2±3.455 and
49.8±1.158, respectively. A decrease in cell numbers in the DAC,
TSA and DAC + TSA groups was observed, compared with the control
group (P<0.01; Fig. 1C and
D).
To determine the effect of DAC and TSA on MKN45 cell
growth in vitro, following exposure to DAC (5 μmol/l) and/or
TSA (300 nmol/l) for 24, 48 and 72 h, the cell proliferation was
analyzed by a CCK-8 assay. The results indicated that MKN45 cells
treated with DAC and combined DAC and TSA exhibited growth
retardation at 48 and 72 h, compared with the controls. TSA only
had a significant inhibitory effect on MKN45 cell proliferation
following 72 h. However, there was no significant difference in the
cell proliferation activity between the treated cells and control
groups with 24 h exposure. (Fig.
1E).
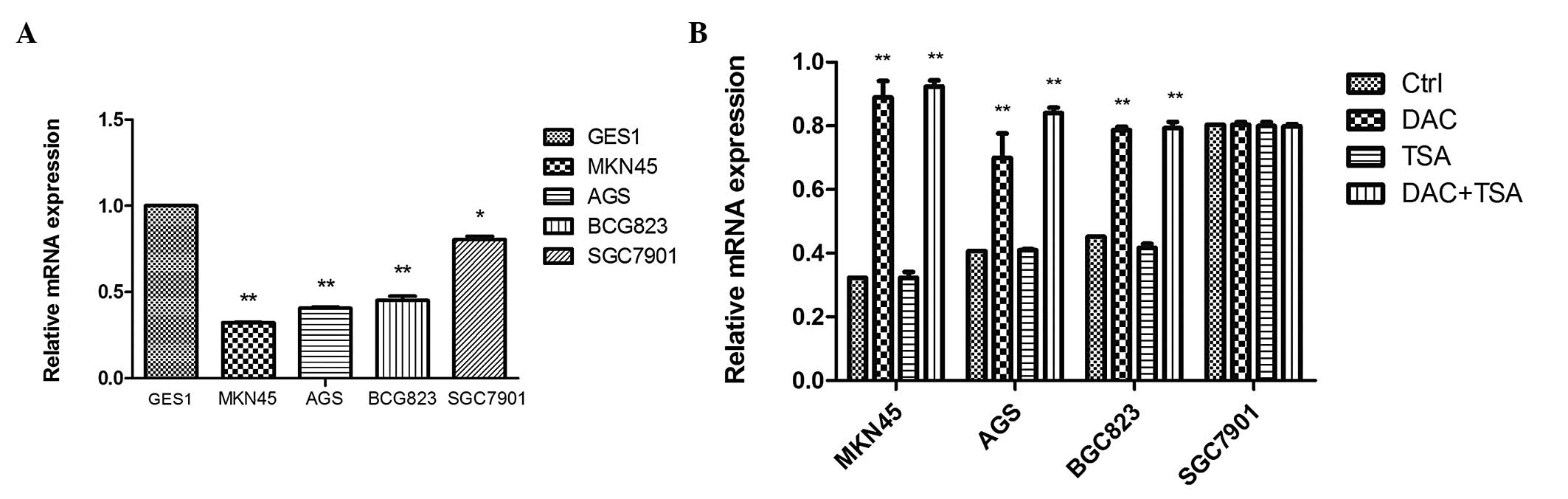
EFEMP1 expression is downregulated in GC
cells and tissues
qPCR was performed to assess the mRNA expression of
EFEMP1, which was substantially downregulated in MKN45
(0.323±0.002), AGS (0.407±0.008), BGC823 (0.454±0.024) and SGC7901
(0.806±0.018) cells compared with the normal mucosa line, GES1
(1-fold as the control; P<0.05; Fig. 2A). In order to examine whether
epigenetic agents were able to reverse EFEMP1 silencing in GC
cells, the cells were treated with the DNA methyltransferase (DNMT)
inhibitor DAC, the histone deacetylase (HDAC) inhibitor TSA and
combined treatment with the two agents. qPCR demonstrated that DAC
and TSA had different effects on EFEMP1 expression in the GC cell
lines. In the cells with low expression of EFEMP1 (MKN45, AGS and
BGC823), DAC alone restored EFEMP1 expression. TSA had no effect on
EFEMP1 expression. Combined treatment restored EFEMP1 expression
similar to that with DAC. In the EFEMP1-positive cell lines
(SGC7901), treatment with DAC and TSA, alone or in combination, had
no significant effect on the expression of EFEMP1 (Fig. 2B).
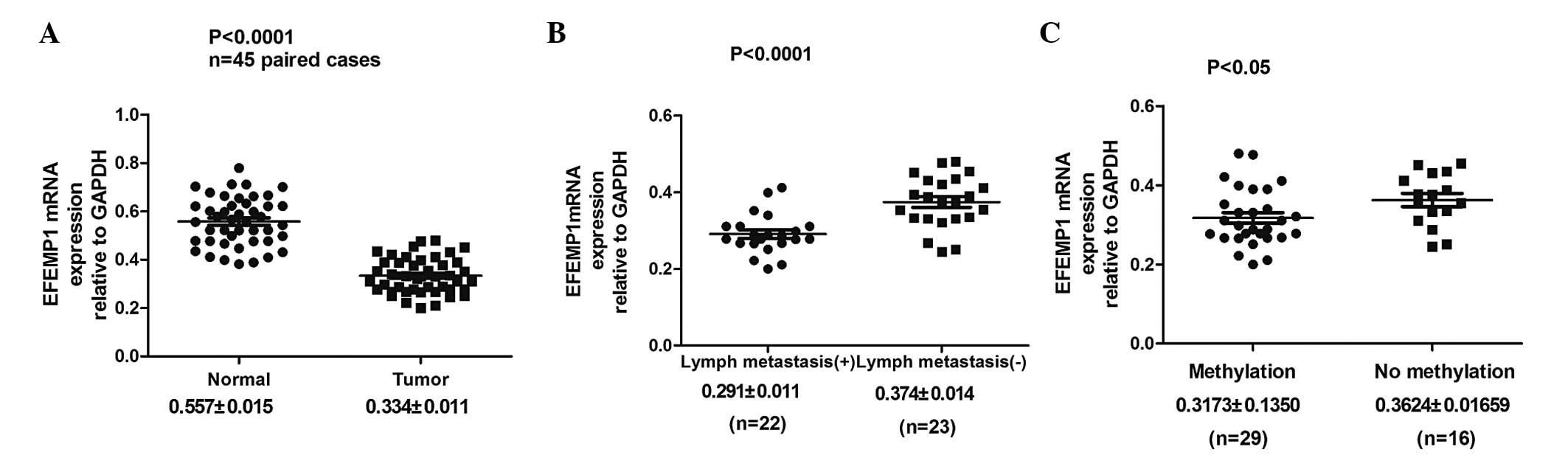
EFEMP1 expression was analyzed in 45 paired GC
specimens and corresponding normal tissues by qPCR. It was
identified that EFEMP1 mRNA expression was significantly lower in
the GC tissues than in their corresponding normal tissues
(0.334±0.011 vs. 0.557±0.015; P<0.0001; Fig. 3A; Table I). Furthermore, the correlation
between EFEMP1 mRNA expression and the clinicopathological factors
of GC were examined. The EFEMP1 mRNA expression level was
associated with tumor differentiation, depth of tumor invasion and
lymph node metastasis (Fig. 3B;
Table I).
 | Table ICorrelation between the
clinicopathological features and EFEMP1 mRNA expression in 45 GC
patients. |
Table I
Correlation between the
clinicopathological features and EFEMP1 mRNA expression in 45 GC
patients.
| Variable | Patients (n) | EFEMP1 mRNA
expression relative to GAPDH | P-value |
|---|
| Normal | 45 | 0.557±0.015 | <0.01a |
| Tumor | 45 | 0.334±0.011 | |
| Age (years) |
| <65 | 23 | 0.331±0.015 | 0.828 |
| ≥65 | 22 | 0.336±0.016 | |
| Gender |
| Male | 27 | 0.325±0.013 | 0.499 |
| Female | 18 | 0.340±0.019 | |
| Tumor
differentiation |
| Well/moderate | 26 | 0.358±0.014 | <0.05a |
| Poor | 19 | 0.300±0.015 | |
| Invasion depth |
| T1 + T2 | 23 | 0.368±0.014 | <0.01a |
| T3 + T4 | 22 | 0.300±0.013 | |
| Tumor location |
| Upper +
middle | 16 | 0.334±0.018 | 0.803 |
| Lower | 29 | 0.331±0.013 | |
| Size (cm) |
| <3 | 28 | 0.332±0.013 | 0.868 |
| ≥3 | 17 | 0.336±0.020 | |
| Lymph node
metastasis |
| No | 23 | 0.374±0.014 | <0.01a |
| Yes | 22 | 0.291±0.011 | |
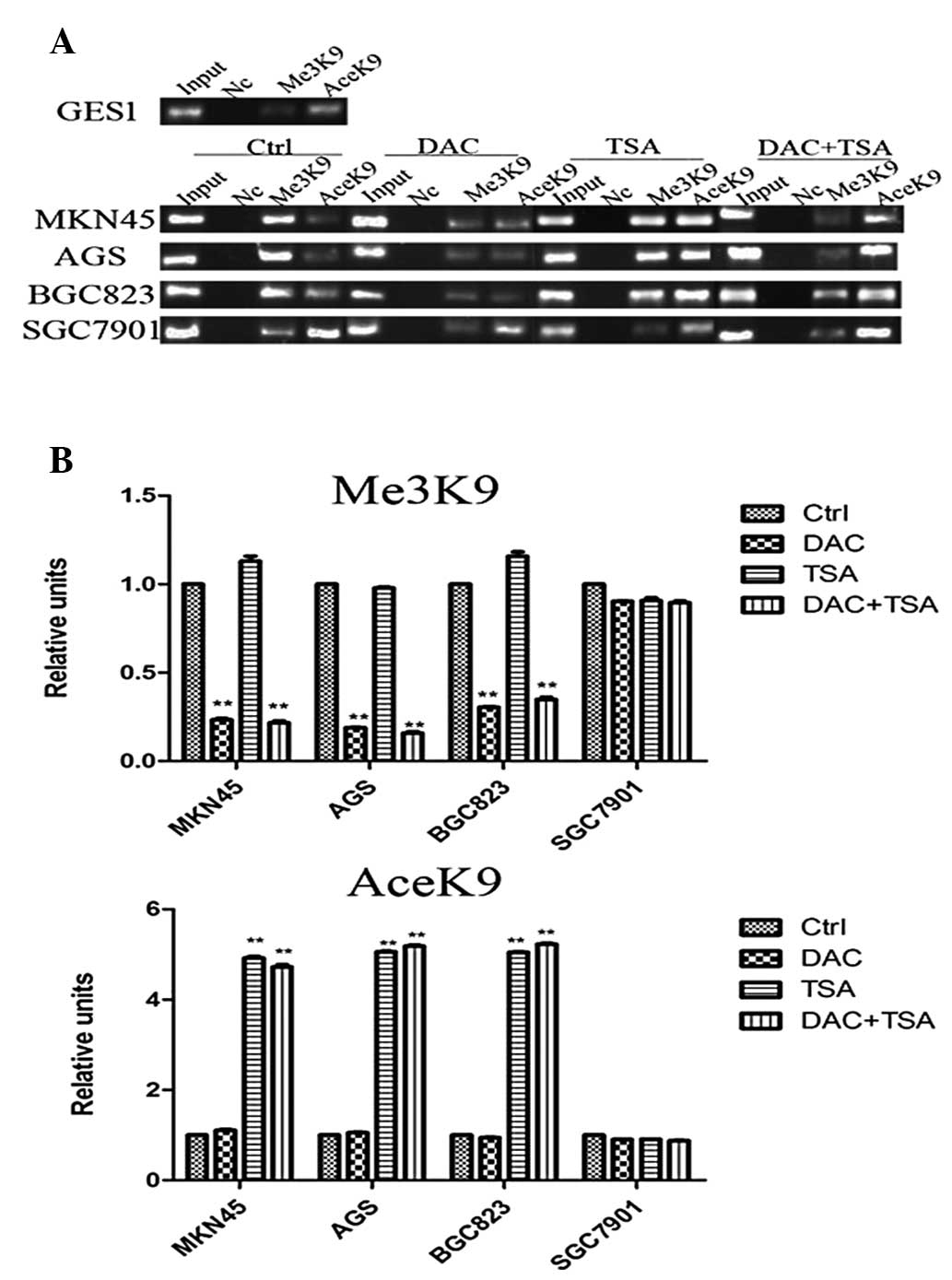
Low expression of EFEMP1 is associated
with DNA methylation
To identify whether DNA methylation was responsible
for the downregulation of EFEMP1 expression, the DNA methylation
status of EFEMP1 in GC cell lines and GES1 cells was assessed using
MSP. The EFEMP1-positive cell lines (GES1 and SGC7901) demonstrated
unmethylated bands (neither allele was methylated), which was in
agreement with the observed high levels of EFEMP1 expression. By
contrast, the MKN45 and AGS cells only demonstrated methylated
bands (hypermethylated, both alleles methylated), BGC823 cells
exhibited methylated and unmethylated bands (partially methylated,
only one allele methylated), which was in agreement with the
observed low levels of EFEMP1 expression (Fig. 4A). In hypermethylated MKN45, AGS
and BGC823 cells, treatment with DAC resulted in DNA demethylation.
TSA had no effect on DNA methylation, and treatment with the two
agents had no additional effect on DNA demethylation beyond that
produced by treatment with DAC alone. In unmethylated SGC7901
cells, treatment with DAC, TSA or the two agents had no significant
effect on DNA methylation (Fig.
4B).
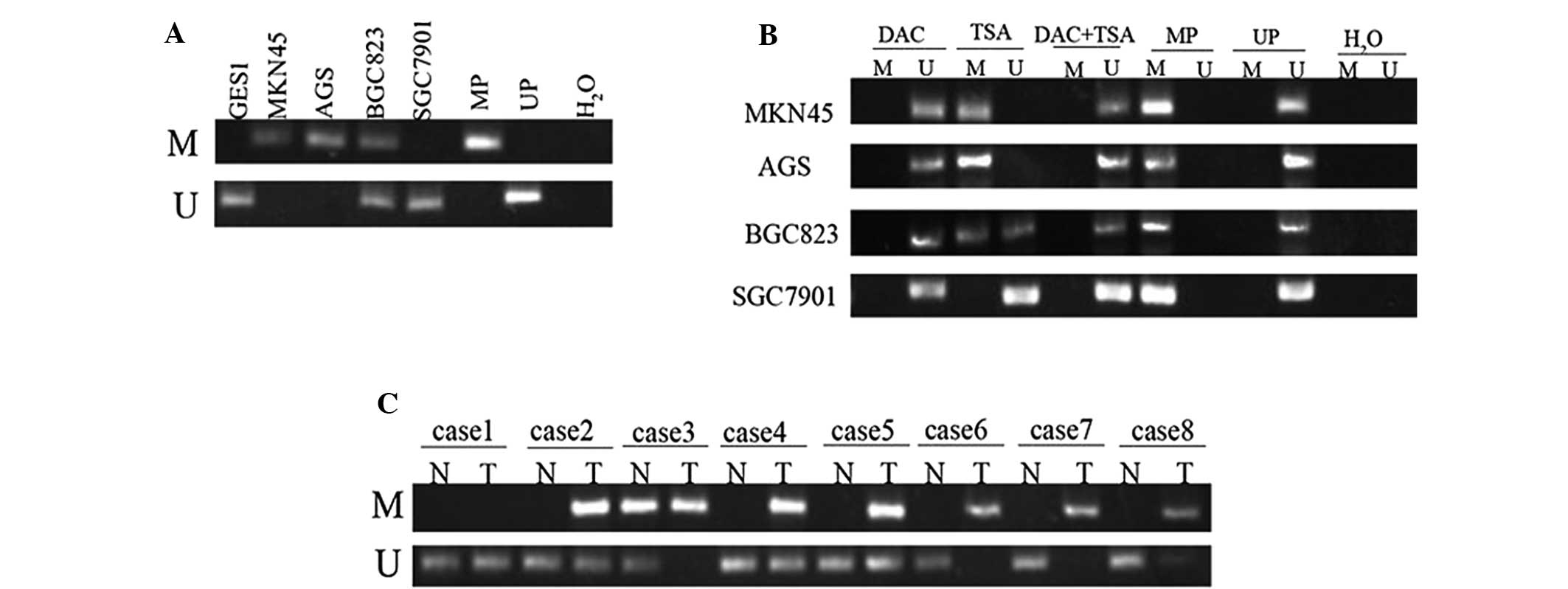 | Figure 4MSP analysis of DNA methylation at the
EFEMP1 promoter region in human GC cells and tissues. (A) EFEMP1
was hypermethylated in MKN45 and AGS, and partially methylated in
BGC823, but not methylated in GES1 and SGC7901 cells. (B) MSP
analysis of DNA methylation at the EFEMP1 promoter region prior to
and following treatment with DAC, TSA or DAC+TSA. (C) DNA
methylation of EFEMP1 in GC specimens and corresponding
non-malignant gastric tissues. Lane M indicates the presence of
methylated alleles; lane U indicates the presence of unmethylated
alleles. At least three independent experiments were performed with
similar results. U, unmethylated; M, methylated; UP,
non-methylation positive control; MP, methylation positive control;
N, non-malignant gastric tissue; T, tumor specimens; MSP,
methylation-specific PCR; EFEMP1, epidermal growth
factor-containing fibulin-like extracellular matrix protein 1; DAC,
5-aza-2′-deoxycytidine; TSA, trichostatin A; GC, gastric
cancer. |
EFEMP1 methylation in gastric specimens, including
45 tumor and 45 corresponding non-malignant gastric tissues, was
then examined by MSP. Hypermethylation of the EFEMP1 gene was
detected in 29 (64.44%) of the 45 primary gastric carcinomas, while
methylation in the non-malignant gastric tissue was only identified
in 11 cases (24.44%). No methylation was observed in 16 (35.56%)
primary GC tissues and 34 (75.56%) non-malignant gastric tissues
(Fig. 4C). The difference in
methylation status of EFEMP1 between the primary GC and
non-malignant gastric tissue specimens was significant (P<0.001;
Table II). It was also identified
that EFEMP1 mRNA expression was significantly lower in the GC
tissues with DNA methylation of EFEMP1 than that in GC tissues
without DNA methylation of EFEMP1 (0.3173±0.1350 vs.
0.3624±0.01659; P<0.05; Fig.
3C). In addition, the correlation between gastric tumor EFEMP1
methylation status and the clinicopathological features of the
patients was analyzed. The DNA methylation status of EFEMP1 was
associated with tumor invasion depth and differentiation, but there
was no correlation with the other clinicopathological features,
including age, gender and tumor location (Table III).
 | Table IIMethylation status of EFEMP1 between
T and N. |
Table II
Methylation status of EFEMP1 between
T and N.
| Group | Case | Methylation
(%) | No methylation
(%) | P-value |
|---|
| T | 45 | 29 (64.44) | 16 (35.56) | <0.001a |
| N | 45 | 11 (24.44) | 34 (75.56) | |
 | Table IIIClinicopathological parameters of GC
samples and EFEMP1 methylation. |
Table III
Clinicopathological parameters of GC
samples and EFEMP1 methylation.
| | EFEMP1
methylation | |
|---|
| |
| |
|---|
| Variable | Patients (n) | M (%) | U (%) | P-value |
|---|
| Age (years) |
| <65 | 22 | 13 (59.1) | 9 (40.9) | 0.104 |
| ≥65 | 23 | 16 (69.6) | 7 (30.4) | |
| Gender |
| Male | 27 | 17 (62.9) | 10 (37.1) | 0.553 |
| Female | 18 | 12 (66.7) | 6 (33.3) | |
| Tumor
differentiation |
| Well/moderate | 26 | 11 (42.3) | 15 (57.7) | <0.01a |
| Poor | 19 | 18 (94.7) | 1 (5.3) | |
| Invasion depth |
| T1 + T2 | 23 | 10 (43.4) | 13 (56.6) | <0.01a |
| T3 + T4 | 22 | 19 (86.3) | 3 (13.7) | |
| Tumor location |
| Upper +
middle | 16 | 9 (56.3) | 7 (43.7) | 0.058 |
| Lower | 29 | 20 (69.0) | 9 (31.0) | |
| Size (cm) |
| <3 | 28 | 19 (67.9) | 9 (32.1) | 0.186 |
| ≥3 | 17 | 10 (58.8) | 7 (41.2) | |
| Lymph node
metastasis |
| No | 23 | 15 (65.2) | 8 (34.8) | 0.883 |
| Yes | 22 | 14 (63.6) | 8 (36.4) | |
Abnormal histone modification is
associated with EFEMP1 gene silencing in GC cell lines
The results demonstrated that EFEMP1 was
downregulated in GC. However, the mechanism by which the expression
of EFEMP1 was inhibited remains unknown. To elucidate whether
abnormal histone modification was associated with the loss of
EFEMP1, the basic levels of H3-K9 trimethylation and H3-K9
acetylation in the EFEMP1 promoters were compared in the GES1 and
GC cells with different EFEMP1 expression, using ChIP. As revealed
in Fig. 4A, in the EFEMP1-positive
cell lines (GES1 and SGC7901), H3-K9 trimethylation of the promoter
regions was minimal. H3-K9 trimethylation levels in the EFEMP1 gene
promoter were higher in cells with low expression of EFEMP1 (MKN45,
AGS and BGC823). By contrast, H3-K9 acetylation at the EFEMP1
promoter region was significantly higher in GES1 and SGC7901 cells
than in the MKN45, AGS and BGC823 cells.
To clarify whether epigenetic agents may affect
epigenetic modifications, the GC cells were treated with DAC and
TSA. It was identified that the levels of H3-K9 trimethylation in
the EFEMP1 promoter in MKN45, AGS and BGC823 cells was decreased
significantly following treatment with DAC, and TSA marginally
reduced histone H3-K9 trimethylation. The effects of combined
treatment with DAC and TSA on H3-K9 trimethylation were similar to
those of DAC alone. H3-K9 acetylation at EFEMP1 promoter regions
was analyzed using ChIP assays to determine whether DAC and TSA may
affect H3-K9 acetylation in GC cells. In MKN45, AGS and BGC823
cells, treatment with TSA alone significantly increased H3-K9
acetylation, but DAC alone had no effect on H3-K9 acetylation. The
effects of combined treatment with DAC and TSA on H3-K9 acetylation
were similar to that of TSA alone. Treatment with both DAC and TSA
had no significant effect on H3-K9 trimethylation and H3-K9
acetylation in SGC7901 cells, in which EFEMP1 was expressed
(Fig. 5A and B).
Discussion
Epigenetics attempts to explain how heritable
changes in gene expression occur without altering nucleotide
sequence, and how epigenetic alterations have an important role in
silencing TSGs (17). In GC, a
growing number of TSGs have been identified as undergoing aberrant
methylation. When DNA is methylated in the promoter region of the
genes, where transcription is initiated, they are typically
inactivated and silenced (18–20).
Modification of the histone tail is another epigenetic regulatory
mechanism. The acetylation of lysine residues on histone H3-K9
leads to the formation of an open chromatin structure and allows
regulatory factors to access the chromatin, which is an active
marker, but methylation of histone H3-K9 is a marker of gene
inactivity (21). Increasing
evidence now indicates that DNA methylation and histone
modifications appear to be linked to each other. DNA methylation
acts synergistically with repressive histone modifications,
including dimethylation or trimethylation of H3-K9, to consolidate
gene transcriptional silencing (22). However, it is not clearly
understood how the formation of histone modifications may affect
DNA methylation and which genes are involved with GC formation.
EFEMP1, also known as fibulin-3, is located on human
chromosome 2p16, and is one of seven members of the fibulin gene
family of extracellular glycoproteins. It contains 11 exons and
encodes a 54-kDa protein. EFEMP1 regulates cell proliferation and
cell-to-cell and cell-to-matrix communication, providing
organization and stability to extracellular matrix structures
(23). The precise mechanism
underlying the role of EFEMP1 in the progression of tumors remains
largely unknown. Hu et al (24) found that the overexpression of
EFEMP1 inhibited glioma cell development and suppressed
angiogenesis, vascular endothelial growth factor receptor A
expression and cell proliferation. The EGF receptor level was
reduced and AKT signaling activity attenuated following treatment
with exogenous EFEMP1. Kim et al (25) reported that overexpression of
EFEMP1 may inhibit non-small cell lung cancer cell invasion by
downregulating cellular matrix metalloproteinase (MMP)-7 and MMP-2.
However, the mechanism by which the expression of EFEMP1 was
inhibited remains unclear. Frequent DNA methylation of the EFEMP1
promoter has been detected in lung cancer (5), hepatocellular carcinoma (6), prostate cancer (7), sporadic breast cancer (8) and colon tumors (26), but has not been reported in GC.
In the present study it was identified that EFEMP1
expression was significantly reduced in GC cell lines and tissues
compared with normal gastric cells and tissues. It was demonstrated
that EFEMP1 may function as a tumor suppressor in GC. In addition,
two mechanisms underlying the decreased expression of EFEMP1 were
identified, including DNA hypermethylation of the EFEMP1 promoter
and hypermethylation of H3-K9 attached to the promoter.
In the present study, the EFEMP1 gene was
hypermethylated in MKN45 and AGS cells, partially methylated in
BGC823 cells, but not methylated in SGC7901 cells. The differential
EFEMP1 expression and the methylation status of the gene between
the four GC cell lines may be associated with the cell type. By
contrast, aberrant methylation of EFEMP1 gene was also observed in
primary gastric carcinomas. Hypermethylation of the EFEMP1 gene was
detected in 29 (64.44%) of the 45 primary gastric carcinomas, while
methylation in non-malignant gastric tissues was only identified in
11 cases (24.44%). Furthermore, it was demonstrated that GC with
invasion depth at T3 and T4 had a notably higher methylation
frequency than that with invasion depth at T1 and T2. Methylation
frequency of EFEMP1 was also negatively correlated with tumor
differentiation. These results suggest that the degree of
malignancy of GC may be enhanced when the methylation frequency of
EFEMP1 is high. These results are consistent with that of a study
by Tong et al (26), who
reported that aberrant methylation caused EFEMP1 downregulation in
colorectal cancer, and EFEMP1 downregulation was correlated with
lymph node metastasis, tumor stage and poor survival. These results
are also supported by Sadr-Nabavi et al (8), who demonstrated that the level of
EFEMP1 expression decreased in sporadic breast cancer due to its
aberrant promoter methylation and was correlated with poor survival
as an antagonist of angiogenesis. Yang et al (27) found EFEMP1 hypermethylation in
65/97 (67%) endometrial carcinoma tissues compared with 4/40 (10%)
normal tissues. Their results demonstrated that the downregulation
of EFEMP1 was associated with promoter hypermethylation.
Histone modification is another critical epigenetic
process that facilitates the control of chromatin structure and
gene regulation, which is associated with DNA methylation status in
regulating gene expression. Unmethylated CpG islands are enriched
in activated chromatin, but methylated DNA is associated with
repressed chromatin (28). Using
ChIP techniques in four GC cell lines, the present study
demonstrated that H3-K9 trimethylation in the EFEMP1 promoter
region was also closely associated with DNA methylation and acted
as a marker of gene silencing. It was revealed that H3-K9
trimethylation of the EFEMP1 gene promoter correlated markedly with
DNA methylation status. H3-K9 trimethylation levels in the EFEMP1
gene promoter were lower in the unmethylated GC cell lines
(SGC7901) with high expression of EFEMP1; however, H3-K9
trimethylation levels in the EFEMP1 gene promoter were higher in
the hypermethylated GC cell lines (MKN45, AGS and BGC823) with low
expression of EFEMP1. In contrast to H3-K9 trimethylation, it was
demonstrated that H3-K9 acetylation was inversely correlated with
DNA methylation status.
To further examine the correlation between
epigenetic alteration and EFEMP1 mRNA expression, four GC cell
lines were treated with DAC and TSA. DAC, a pyrimidine analog with
the 2′-deoxycytidine fifth carbon atom replaced by nitrogen, is
able to form a complex with DNMT1 following binding to DNA during
replication and subsequently inhibits transmethylation activity of
this enzyme. TSA is an HDAC inhibitor and causes DNA histone
hyperacetylation and induces p21(WAF1/CIP1) gene
expression (29). Consistent with
previous studies (30,31), DAC inhibited migration invasion and
proliferation of MKN45 cells, but TSA had a weaker effect on the
biological behavior of MKN45 cells. The present study also
identified that DAC restored EFEMP1 expression in MKN45, AGS and
BGC823 cells, which have low expression of EFEMP1 mRNA. In addition
to its effect on DNA methylation, DAC reduced the level of H3-K9
trimethylation in the EFEMP1 promoter, but DAC had no significant
effect on H3-K9 acetylation. TSA alone significantly increased
H3-K9 acetylation but it did not restore EFEMP1 mRNA expression. A
combination of DAC and TSA markedly increased Lys-9 acetylation and
decreased Lys-9 methylation, and was most effective in restoring
EFEMP1 gene expression in MKN45, AGS and BGC823 cells. However, in
SGC7901 cells, which express EFEMP1 mRNA, treatment with DAC and
TSA, alone or in combination, had no significant effect on the
expression of EFEMP1. These results indicated that promoter DNA
methylation and H3-K9 trimethylation, but not H3-K9 acetylation,
are involved in the repression of EFEMP1 gene expression in human
GC cells. It was also identified that DAC not only demethylated DNA
promoters, but also altered existing histone H3-K9 trimethylation.
DAC-induced changes in histone modifications were limited in the
DNA hypermethylated cells (MKN45, AGS and BGC823). The mechanism
underlying the modifications of histone methylation by DAC remain
unclear. One simple possibility is that DNMTs, together with the
methyl-CpG-binding protein MECP2, are able to recruit histone
H3-K9-specific methyltransferases SUV39H1. Therefore, epigenetic
information embodied in methylated residues flows from DNA to
histone and back. DNMT and SUV39H1 form complexes to regulate
EFEMP1 gene expression. The decreased expression of DNMT1 induced
by DAC may then lead to histone demethylation via disruption of
these silencing complexes (32).
These results are in agreement with previous studies that the
ability of DAC to reactivate the expression of DSC3 and MASPIN
genes tracked closely with the reductions of H3-K9 methylation
levels in their promoter regions (33). Therefore, DNA methylation and
histone modification may function together to regulate gene
expression (34,35). In addition, as epigenetic
alterations are reversible, they are considered useful therapeutic
targets. Recently, DAC was demonstrated to synergize with
progesterone therapy to inhibit endometrial cancer cell growth and
invasion (36). These findings
suggest that chemotherapeutic drugs combined with epigenetic agents
may be potentially utilized for future cancer therapy.
In conclusion, the present study demonstrated that
EFEMP1 was downregulated in GC, which was mainly caused by aberrant
DNA methylation and histone H3-K9 trimethylation. DAC acts via
epigenetic alterations to reactivate EFEMP1 expression. The mRNA
expression of EFEMP1 gene and EFEMP1 methylation were associated
with invasion and metastasis, which may be potential prognostic
factors for GC. These findings provide a foundation for the role of
EFEMP1 gene in GC and its potential as a biomarker for early
diagnosis, and may lead to the identification of novel targets for
pharmacological intervention. Therefore, further in vitro
and in vivo studies are required to detect the function of
EFEMP1 in the progression of GC.
Acknowledgements
The present study was supported in part by a grant
from the National Natural Science Foundation of China (grant no.
30572162), the Foundation of Liaoning Province Science and
Technology Plan Project (grant no. 2013225021) and the Higher
Specialized Research Fund for Doctoral Program of Ministry of
Education of China (grant no. 20102104110001).
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Allis CD, Berger SL, Cote J, et al: New
nomenclature for chromatin-modifying enzymes. Cell. 131:633–636.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cantone I and Fisher AG: Epigenetic
programming and reprogramming during development. Nat Struct Mol
Biol. 20:282–289. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Albig AR, Neil JR and Schiemann WP:
Fibulins 3 and 5 antagonize tumor angiogenesis in vivo. Cancer Res.
66:2621–2629. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yue W, Dacic S, Sun Q, et al: Frequent
inactivation of RAMP2, EFEMP1 and Dutt1 in lung cancer by promoter
hypermethylation. Clin Cancer Res. 13:4336–4344. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nomoto S, Kanda M, Okamura Y, et al:
Epidermal growth factor-containing fibulin-like extracellular
matrix protein 1, EFEMP1, a novel tumor suppressor gene detected in
hepatocellular carcinoma using double combination array analysis.
Ann Surg Oncol. 17:923–932. 2010. View Article : Google Scholar
|
|
7
|
Kim YJ, Yoon HY, Kim SK, et al: EFEMP1 as
a novel DNA methylation marker for prostate cancer: array-based DNA
methylation and expression profiling. Clin Cancer Res.
17:4523–4530. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sadr-Nabavi A, Ramser J, Volkmann J, et
al: Decreased expression of angiogenesis antagonist EFEMP1 in
sporadic breast cancer is caused by aberrant promoter methylation
and points to an impact of EFEMP1 as molecular biomarker. Int J
Cancer. 124:1727–1735. 2009. View Article : Google Scholar
|
|
9
|
Hwang CF, Chien CY, Huang SC, et al:
Fibulin-3 is associated with tumour progression and a poor
prognosis in nasopharyngeal carcinomas and inhibits cell migration
and invasion via suppressed AKT activity. J Pathol. 222:367–379.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
En-lin S, Sheng-guo C and Hua-qiao W: The
expression of EFEMP1 in cervical carcinoma and its relationship
with prognosis. Gynecol Oncol. 117:417–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Davidson B, Stavnes HT, Holth A, Chen X,
Yang Y, et al: Gene expression signatures differentiate
ovarian/peritoneal serous carcinoma from breast carcinoma in
effusions. J Cell Mol Med. 15:535–544. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seeliger H, Camaj P, Ischenko I, et al:
EFEMP1 expression promotes in vivo tumor growth in human pancreatic
adenocarcinoma. Mol Cancer Res. 7:189–198. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fahrner JA, Eguchi S, Herman JG and Baylin
SB: Dependence of histone modifications and gene expression on DNA
hypermethylation in cancer. Cancer Res. 62:7213–7218.
2002.PubMed/NCBI
|
|
14
|
Cameron EE, Bachman KE, Myöhänen S, Herman
JG and Baylin SB: Synergy of demethylation and histone deacetylase
inhibition in the re-expression of genes silenced in cancer. Nat
Genet. 21:103–107. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kuo MH and Allis CD: In vivo cross-linking
and immunoprecipitation for studying dynamic Protein:DNA
associations in a chromatin environment. Methods. 19:425–433. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang R, Zhang YW and Chen LB: Aberrant
promoter methylation of FBLN-3 gene and clinicopathological
significance in non-small cell lung carcinoma. Lung Cancer.
69:239–244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar
|
|
18
|
Meng CF, Zhu XJ, Peng G and Dai DQ: Role
of histone modifications and DNA methylation in the regulation of
O6-methylguanine-DNA methyltransferase gene expression
in human stomach cancer cells. Cancer Invest. 28:331–339. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Meng CF, Zhu XJ, Peng G and Dai DQ:
Promoter histone H3 lysine 9 di-methylation is associated with DNA
methylation and aberrant expression of p16 in gastric cancer cells.
Oncol Rep. 22:1221–1227. 2009.PubMed/NCBI
|
|
20
|
Chang X, Zhang S, Ma J, et al: Association
of NDRG1 gene promoter methylation with reduced NDRG1 expression in
gastric cancer cells and tissue specimens. Cell Biochem Biophys.
66:93–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grewal SI and Moazed D: Heterochromatin
and epigenetic control of gene expression. Science. 301:798–802.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peters AH, Mermoud JE, O’Carroll D, et al:
Histone H3 lysine 9 methylation is an epigenetic imprint of
facultative heterochromatin. Nat Genet. 30:77–80. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y and Marmorstein LY: Focus on
molecules: fibulin-3 (EFEMP1). Exp Eye Res. 90:374–375. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu Y, Pioli PD, Siegel E, et al: EFEMP1
suppresses malignant glioma growth and exerts its action within the
tumor extracellular compartment. Mol Cancer. 10:1232011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim EJ, Lee SY, Woo MK, et al: Fibulin-3
promoter methylation alters the invasive behavior of non-small cell
lung cancer cell lines via MMP-7 and MMP-2 regulation. Int J Oncol.
40:402–408. 2012.PubMed/NCBI
|
|
26
|
Tong JD, Jiao NL, Wang YX, Zhang YW and
Han F: Downregulation of fibulin-3 gene by promoter methylation in
colorectal cancer predicts adverse prognosis. Neoplasma.
58:441–448. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang T, Qiu H, Bao W, et al: Epigenetic
inactivation of EFEMP1 is associated with tumor suppressive
function in endometrial carcinoma. PLoS One. 8:e674582013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: how the genome integrates intrinsic
and environmental signals. Nat Genet. 33(Suppl): 245–254. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nawrocki ST, Carew JS, Douglas L, et al:
Histone deacetylase inhibitors enhance lexatumumab-induced
apoptosis via a p21Cip1-dependent decrease in survivin levels.
Cancer Res. 67:6987–6994. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang X, Li Z, Ma J, et al: DNA
methylation of NDRG2 in gastric cancer and its clinical
significance. Dig Dis Sci. 58:715–723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhi Y, Chen J, Zhang S, et al:
Down-regulation of CXCL12 by DNA hypermethylation and its
involvement in gastric cancer metastatic progression. Dig Dis Sci.
57:650–659. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fuks F, Hurd PJ, Deplus R and Kouzarides
T: The DNA methyltransferases associate with HP1 and the SUV39H1
histone methyltransferase. Nucleic Acids Res. 31:2305–2312. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wozniak RJ, Klimecki WT, Lau SS, Feinstein
Y and Futscher BW: 5-Aza-2′-deoxycytidine-mediated reductions in
G9A histone methyltransferase and histone H3 K9 di-methylation
levels are linked to tumor suppressor gene reactivation. Oncogene.
26:77–90. 2007.
|
|
34
|
Lin W and Dent SY: Functions of
histone-modifying enzymes in development. Curr Opin Genet Dev.
16:137–142. 2006. View Article : Google Scholar
|
|
35
|
Martin C and Zhang Y: The diverse
functions of histone lysine methylation. Nat Rev Mol Cell Biol.
6:838–849. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu Q, Yu L, Chen R, et al:
5-aza-2′-deoxycytidine improves the sensitivity of endometrial
cancer cells to progesterone therapy. Int J Gynecol Cancer.
22:951–959. 2012.
|