Introduction
Colorectal cancer (CRC) is the third most common
malignancy and the fourth most frequent cause of cancer-related
mortality worldwide (1). Despite
numerous advances in treatment, patients with CRC still have an
unfavorable prognosis, and 25–40% of patients develop recurrence
following surgery (2). Therefore,
it is crucial to enhance the understanding of the mechanisms
involved in CRC metastasis and to identify potential prognostic
biomarkers and therapeutic targets.
Epithelial-mesenchymal transition (EMT) is a key
step towards cancer metastasis. Loss of E-cadherin expression is a
hallmark of the EMT process and is likely to be required for
enhanced tumor cell motility (3–5).
Epithelial cells lose their epithelial characteristics and acquire
mesenchymal characteristics by the downregulation of E-cadherin
(6). EMT has a crucial role in
tumor cell invasion and migration in numerous types of cancer,
including CRC (7). Inducers of EMT
include certain protein polypeptides, transcription factors, growth
factors and microRNAs (miRNAs) (8).
miRNAs are small, noncoding RNA gene products of ~22
nucleotides that are found in a variety of organisms. miRNAs have
key roles in regulating the translation and degradation of mRNAs
through base pairing to partially complementary sites,
predominantly in the 3′-untranslated regions (UTRs) of mRNAs
(9–11). It is well known that miRNAs have
important regulatory functions in basic biological processes,
including development, cellular differentiation, proliferation and
apoptosis, which affect major biological systems, including
stemness, immunity and cancer (12,13).
Data suggest that dysregulation of miRNAs is an important step in
the pathogenesis, from initiation to metastasis, of numerous types
of cancer, including CRC (14–16).
Accumulating evidence has demonstrated that the induction of EMT
and aberrant expression of miRNAs are associated with
tumorigenesis, tumor progression, metastasis and relapse in certain
cancers, including CRC (17,18).
Recently, miR-20a has been shown to be deregulated
in several types of cancer (19–21).
In particular, miR-20a has a key role in the regulation of tumor
proliferation, metastasis and EMT (19–24).
Chai et al (25) reported
that the overexpression of miR-20a contributed to the resistance of
colorectal adenocarcinoma to chemotherapeutics. However, the
clinical and biological roles of miR-20a in CRC remain to be fully
elucidated.
The aim of the present study was to assess the
clinical significance of miR-20a in CRC and to investigate the
effects of miR-20a on the migration, invasion and EMT of CRC cells,
and to further discuss the mechanisms of action of miR-20a by
identifying its potential target gene.
Materials and methods
Patients and tissue samples
Paired tissue specimens (tumor and adjacent normal
mucosa) were obtained from 86 patients with primary CRC who
underwent surgery without preoperative treatment at the First
Department of General Surgery, the Affiliated Hospital of North
Sichuan Medical College (Nanchong, China), from 2005 to 2008,
following receipt of their informed consent. All tissue samples
were immediately frozen in liquid nitrogen and stored at −80°C for
subsequent analysis. Specimens were stained with hematoxylin and
eosin and examined histopathologically. Sections that consisted of
>80% carcinoma cells were used to prepare the total RNA.
Clinicopathological information, including age, gender, tumor size,
histological type, depth of invasion, location, lymph node
metastasis, lymphatic invasion and distant metastasis, was
available for all patients. The study was approved by the Medical
Ethics Committee of North Sichuan Medical College.
Cell culture
The human SW480 CRC cell line was obtained from the
American Type Culture Collection (Manassas, VA, USA) and cultured
in Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal
bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin at
37°C in a humidified atmosphere of 5% CO2.
Quantitative polymerase chain reaction
(qPCR)
Total RNA was extracted using TRIzol®
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) for both
miR-20a and SMAD family member 4 (SMAD4) mRNA analyses. For
detection of miR-20a expression, qPCR was performed using the
QuantiMir RT kit (System Biosciences, Mountain View, CA, USA)
according to the manufacturer’s instructions with miR-20a-specific
primers (Applied Biosystems, Foster City, CA, USA). The
amplification profile was denaturation at 95°C for 10 min followed
by 45 cycles of denaturation at 95°C for 15 sec, annealing at 60°C
for 30 sec and extension at 72°C for 1 min. For the detection of
SMAD4 mRNA expression, qPCR was performed using SYBR®
Green PCR Master Mix (Applied Biosystems). The primers for SMAD4
were as follows: 5′-TGGCCCAGGATCAGTAGGT-3′ and
5′-CATCAACACCAATTCCAGCA-3′. The primers for β-actin were:
5′-CCAAGGCCAACCGCGAGAAGATGAC-3′ and
5′-AGGGTACATGGTGGTGCCGCCAGAC-3′. The amplification profile was
denaturation at 95°C for 30 min, followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 10 sec and
extension at 72°C for 1 min. U6 and β-actin were used as internal
controls. Relative gene expression was calculated using the
equation 2−ΔCT. The fold-change of gene expression was
calculated using the equation 2−ΔΔCT. All reactions were
run in triplicate.
Transfection of miRNA
Ectopic expression of miR-20a in cells was achieved
by transfection with Pre-miR™ miR-20a precursor (pre-miR-20a;
Applied Biosystems) using Lipofectamine 2000 (Invitrogen Life
Technologies). A total of 2×105 cells were seeded into
each well of a six-well plate and transfected for 24 or 48 h.
Transfected cells were used in further assays or RNA/protein
extraction.
Migration and invasion assays
To assess cell migration and invasion,
5×104 SW480 cells transfected with either pre-miR-20a or
the negative control miRNA precursor (pre-miR-nc; Applied
Biosystems) were seeded into Transwell chambers (8.0-μm pore size;
Corning, Inc., New York, NY, USA) uncoated or coated with Matrigel™
(BD Biosciences, Bedford, MA, USA). Medium containing 10% fetal
bovine serum in the lower chamber served as the chemoattractant.
Following incubation for 48 h at 37°C in a humidified incubator
with 5% CO2, cells that did not migrate through the
pores were mechanically removed using a cotton swab. The migrated
cells attached to the bottom of the membrane insert were fixed with
methanol at room temperature for 5 min and stained with
hematoxylin. The number of migrated or invaded cells on the lower
surface of the membrane was then counted under a microscope at a
magnification of ×400.
Western blot analysis
To isolate the proteins, cells collected from the
six-well plates were washed twice with phosphate-buffered saline
and lysed in radioimmunoprecipitation assay lysis buffer (ProMab
Biotechnologies, Inc., Richmond, CA, USA). Lysates were kept on ice
for 30 min and then centrifuged at 13,000 × g for 30 min. The
supernatant was collected and then 20 μg of each of the proteins
was separated by 10% SDS-PAGE and transferred to nitrocellulose
membranes. Following blocking in 5% skimmed milk, the membranes
were incubated with the respective antibodies: Mouse
anti-E-cadherin (1:1,000; BD Biosciences, San Jose, CA, USA), mouse
anti-vimentin (1:500; Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA), mouse anti-SMAD4 (1:500; Santa Cruz Biotechnology, Inc.)
and mouse anti-GAPDH (1:1,000; Santa Cruz Biotechnology, Inc.)
overnight at 4°C. Following incubation with the appropriate
secondary antibody, the bands were visualized using ECL-Plus™
reagents (GE Healthcare Bio-Science Corp., Piscataway, NJ, USA).
The density of the SMAD4 and GAPDH bands was measured using Image J
software (National Institutes of Health, Bethesda, MD, USA), and
values were normalized to the densitometric values of GAPDH in each
sample. Fold-changes in the amount of protein were then calculated
for the experimental sets compared with the control.
Luciferase assay
For the luciferase reporter experiments, the
wild-type and mutated 3′UTRs of SMAD4 mRNA were subcloned into the
XhoI and NotI sites of the psicheck-2 vector (Promega
Corp., Madison, WI, USA) and the new wild-type and mutant vectors
were referred to as psicheck-2-SMAD4-WT and psicheck-2-SMAD4-MUT,
respectively. The following primers were used to amplify specific
fragments: WT sense, 5′-CACAACTCGAGCACTGTTCTGCAAAGGTGGC-3′ and WT
antisense, 5′-AAGGAAAAAAGCGGCCGCGACCTT CTGAGCAAGGCAGT-3′; MUT
sense, 5′-CTTTTCGTG AAATGAGTCCAATCTCAGTGATGAGG-3′ and MUT
antisense, 5′-CTCATTTCACGAAAAGAAAAAAAAAAT CTTAAAAATCA-3′. For the
reporter assay, HEK 293T cells (ATCC, Manassas, VA, USA) were
seeded onto 24-well plates at 2×104 cells/well and
transfected with 200 ng psicheck-2-SMAD4-WT or -SMAD4-MUT and 40 nM
pre-miR-20a or -nc using Lipofectamine 2000 (Invitrogen Life
Technologies). Firefly luciferase was used to normalize the Renilla
luciferase. Following transfection for 48 h, cells were harvested
and assayed with the Dual-Luciferase Reporter Assay System (Promega
Corp.) according to the manufacturer’s instructions.
Bioinformatics analysis
The miRNA targets predicted by computer-aided
algorithms were obtained from TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org).
Statistical analysis
Values are expressed as the mean ± standard
deviation from at least three separate experiments performed in
triplicate. The gene expression levels in CRC tissue samples were
compared with those in normal adjacent mucosa using the Wilcoxon
signed-rank test. Measurement data were analyzed using the
two-tailed Student’s t-test, while categorical data were studied
using the χ2 test. The correlation between miR-20a
levels and SMAD4 expression was analyzed using Pearson’s
correlation. The postoperative survival rate was analyzed using the
Kaplan-Meier method, and differences in survival rates were
assessed using the log-rank test. A Cox proportional hazards model
was used for multivariate analysis. The findings were considered
significant at a P-value of <0.05. All statistical analyses were
performed using the SPSS 16.0 software (SPSS, Inc., Chicago, IL,
USA).
Results
Expression of miR-20a in CRC tissue
samples and normal adjacent mucosa
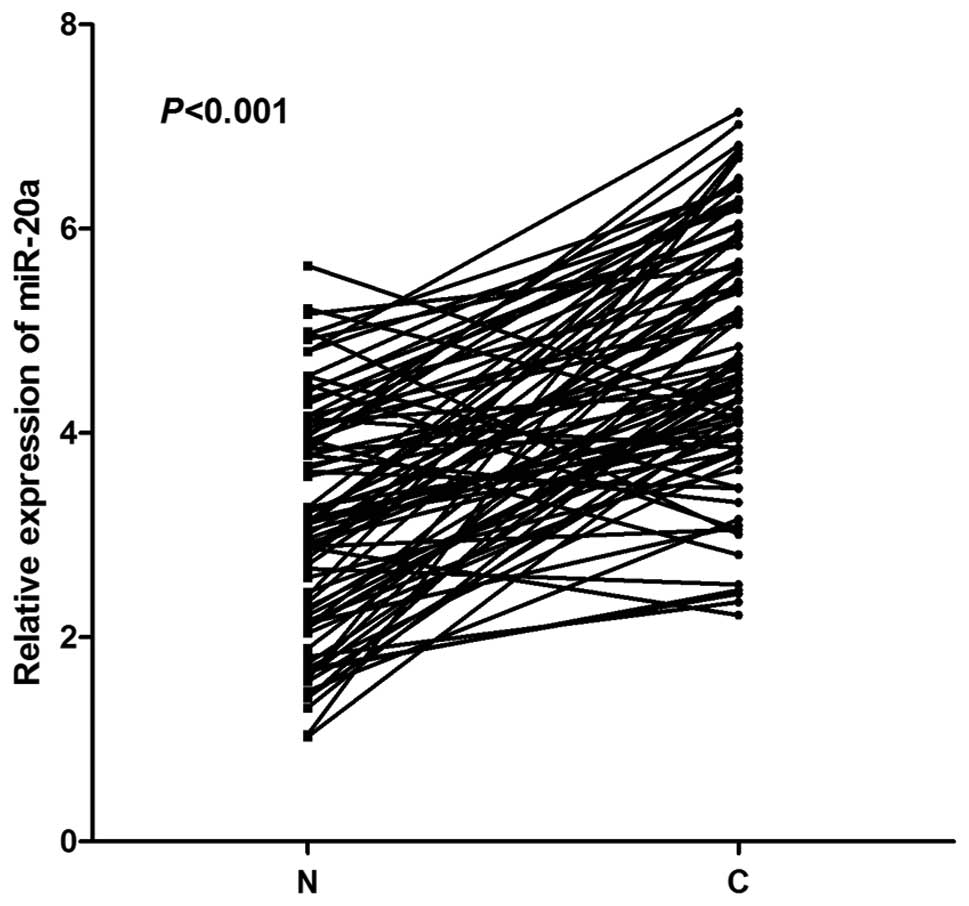
The expression levels of miR-20a in tumor and normal
tissue samples from 86 patients are shown in Fig. 1. Most tumor tissue samples from
patients with CRC (75/86; 87.21%) showed elevated levels of
miR-20a, unlike the corresponding normal tissue samples
(P<0.001).
Correlation between miR-20a expression
and clinicopathological characteristics in CRC
The expression levels of miR-20a were categorized as
low or high relative to the median value (4.65). High expression
rates of miR-20a in CRC tissue samples with respect to several
standard clinicopathological characteristics are listed in Table I. High expression of miR-20a was
significantly correlated with lymph node and distant metastases
(P<0.05, Table I). However, no
significant correlation was observed between miR-20a expression and
other clinicopathological features, including age, gender, tumor
size, histological type, depth of invasion, tumor location and
lymph node invasion (P>0.05, Table
I).
 | Table IAssociation of miR-20a expression with
clinicopathological factors of patients with colorectal cancer. |
Table I
Association of miR-20a expression with
clinicopathological factors of patients with colorectal cancer.
| miR-20a
expression | |
|---|
|
| |
|---|
| Variable | Low, n=43 | High, n=43 | P-value |
|---|
| Age (years) | 62.2±13.4 | 63.3±14.1 | 0.702 |
| Gender | | | 0.506 |
| Male (n) | 25 | 28 | |
| Female (n) | 18 | 15 | |
| Tumor size | | | 0.176 |
| ≤5 cm (n) | 37 | 32 | |
| >5 cm (n) | 6 | 11 | |
| Histological
type | | | 0.181 |
| Well, moderate
(n) | 30 | 24 | |
| Poor, mucinous
(n) | 13 | 19 | |
| Depth of
invasion | | | 0.078 |
| T1, T2 (n) | 21 | 13 | |
| T3, T4 (n) | 22 | 30 | |
| Location | | | 0.651 |
| Colon (n) | 16 | 14 | |
| Rectum (n) | 27 | 29 | |
| Lymph node
metastasis | | | 0.017a |
| Absent (n) | 25 | 14 | |
| Present (n) | 18 | 29 | |
| Lymph node
invasion | | | 0.195 |
| Absent (n) | 26 | 20 | |
| Present (n) | 17 | 23 | |
| Distant
metastasis | | | 0.035a |
| Absent (n) | 40 | 33 | |
| Present (n) | 3 | 10 | |
Correlation between miR-20a expression
and prognosis of patients with CRC
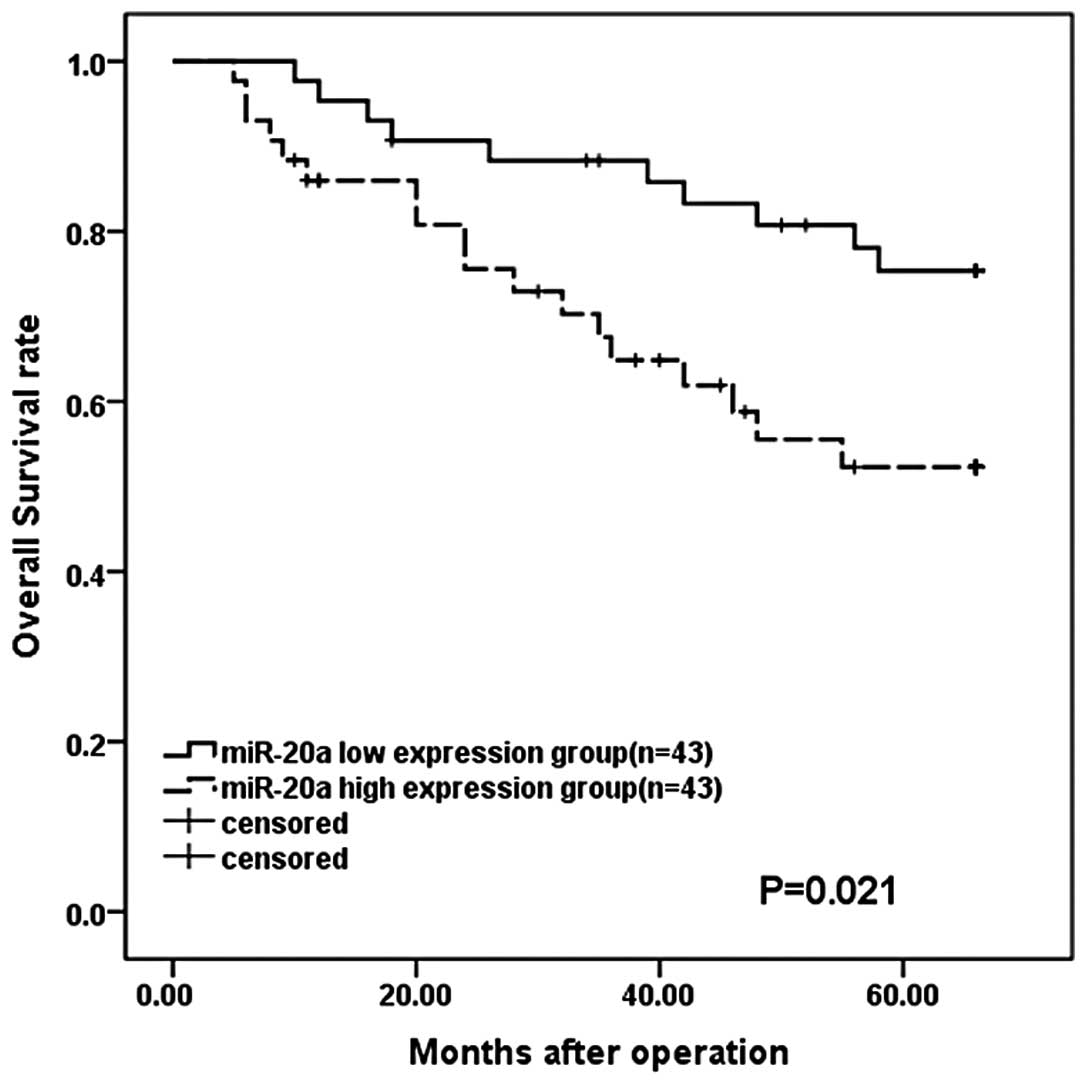
Overall survival curves were plotted according to
miR-20a mRNA expression levels using the Kaplan-Meier method. As
shown in Fig. 2, the overall
survival rate was significantly lower in patients with high miR-20a
expression than that in patients with low expression levels
(P=0.021). Univariate analysis using the Cox proportional hazards
model identified seven prognostic factors: Histological type, tumor
size, depth of invasion, lymph node metastasis, lymph node
invasion, distant metastasis and miR-20a expression. The other
clinicopathological features (age, gender and location) were not
statistically significant prognostic factors (Table II). A multivariate analysis of the
prognostic factors using the Cox proportional hazards model
confirmed that high miR-20a expression was a significant
independent predictor of low survival rates of patients with CRC
(P=0.045) in addition to the presence of lymph node metastasis
(P=0.025) and distant metastasis (P<0.001) (Table III).
 | Table IIUnivariate analysis of
clinicopathological factors for overall survival. |
Table II
Univariate analysis of
clinicopathological factors for overall survival.
| Variable | n | Hazard ratio | 95% CI | P-value |
|---|
| Age (years) | | | 0.715–3.173 | 0.282 |
| ≤60 | 36 | 1 | | |
| >60 | 50 | 1.506 | | |
| Gender | | | 0.380–1.700 | 0.568 |
| Male | 53 | 1 | | |
| Female | 33 | 0.804 | | |
| Tumor size | | | 1.078–5.646 | 0.032a |
| ≤5 cm | 69 | 1 | | |
| >5 cm | 17 | 2.468 | | |
| Histological | | | 1.573–7.160 | 0.002b |
| Well,
moderate | 54 | 1 | | |
| Poor,
mucinous | 32 | 3.356 | | |
| Depth of
invasion | | | 1.064–5.913 | 0.035a |
| T1, T2 | 34 | 1 | | |
| T3, T4 | 52 | 2.187 | | |
| Location | | | 0.796–3.570 | 0.173 |
| Colon | 30 | 1 | | |
| Rectum | 56 | 1.686 | | |
| Lymph node
metastasis | | | 2.447–20.459 | <0.001b |
| Absent | 39 | 1 | | |
| Present | 47 | 7.076 | | |
| Lymph node
invasion | | | 1.072–4.931 | 0.032a |
| Absent | 46 | 1 | | |
| Present | 40 | 2.299 | | |
| Distant
metastasis | | | 4.609–26.247 | <0.001b |
| Absent | 73 | 1 | | |
| Present | 13 | 10.998 | | |
| miR-20a | | | 1.112–5.253 | 0.026a |
| Low | 43 | 1 | | |
| High | 43 | 2.417 | | |
 | Table IIIMultivariate analysis of
clinicopathological factors for overall survival. |
Table III
Multivariate analysis of
clinicopathological factors for overall survival.
| Variable | Hazard ratio | 95% CI | P-value |
|---|
| Tumor size (>5
cm/≤5 cm) | 2.103 | 0.847–5.220 | 0.109 |
| Histological type
(poor, muc/well, mod) | 1.429 | 0.566–3.604 | 0.450 |
| Depth of invasion
(T3, T4/T1, T2) | 1.330 | 0.544–3.250 | 0.532 |
| Lymph node
metastasis (present/absent) | 3.665 | 1.174–11.441 | 0.025a |
| Lymph node invasion
(present/absent) | 1.371 | 0.602–3.121 | 0.452 |
| Distant metastasis
(present/absent) | 6.432 | 2.306–17.937 | <0.001b |
| miR-20a
(high/low) | 2.430 | 1.018–5.799 | 0.045a |
miR-20a regulates CRC cell invasion and
migration in vitro
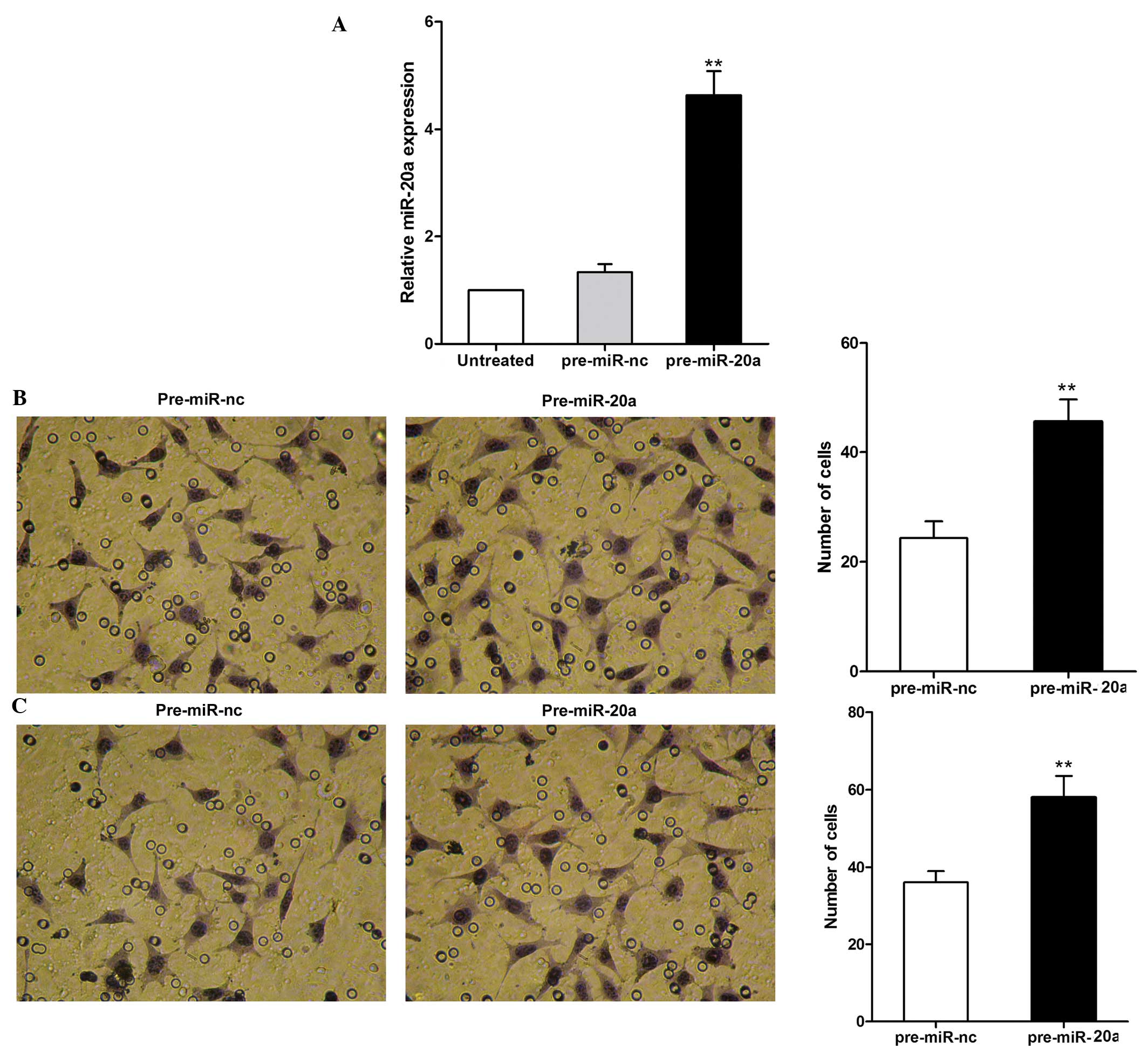
To investigate the role of miR-20a in CRC
metastasis, the role of miR-20a in the migration and invasion of
SW480 cells was examined (Fig. 3).
Levels of miR-20a expression were observed to be significantly
increased following transfection with pre-miR-20a compared with
those following transfection with pre-miR-nc (Fig. 3A). Furthermore, cell migration was
significantly increased following transfection with pre-miR-20a as
compared with that following transfection with the negative control
(P<0.01, Fig. 3B). The effect
of miR-20a on SW480 cell invasion across an extracellular matrix
was then assessed, revealing that the overexpression of miR-20a
markedly enhanced cell invasion as compared with the control
(P<0.01, Fig. 3C). These
observations suggested that miR-20a may have an important role in
promoting migration and the invasive potential of CRC cells.
miR-20a induces EMT-like changes
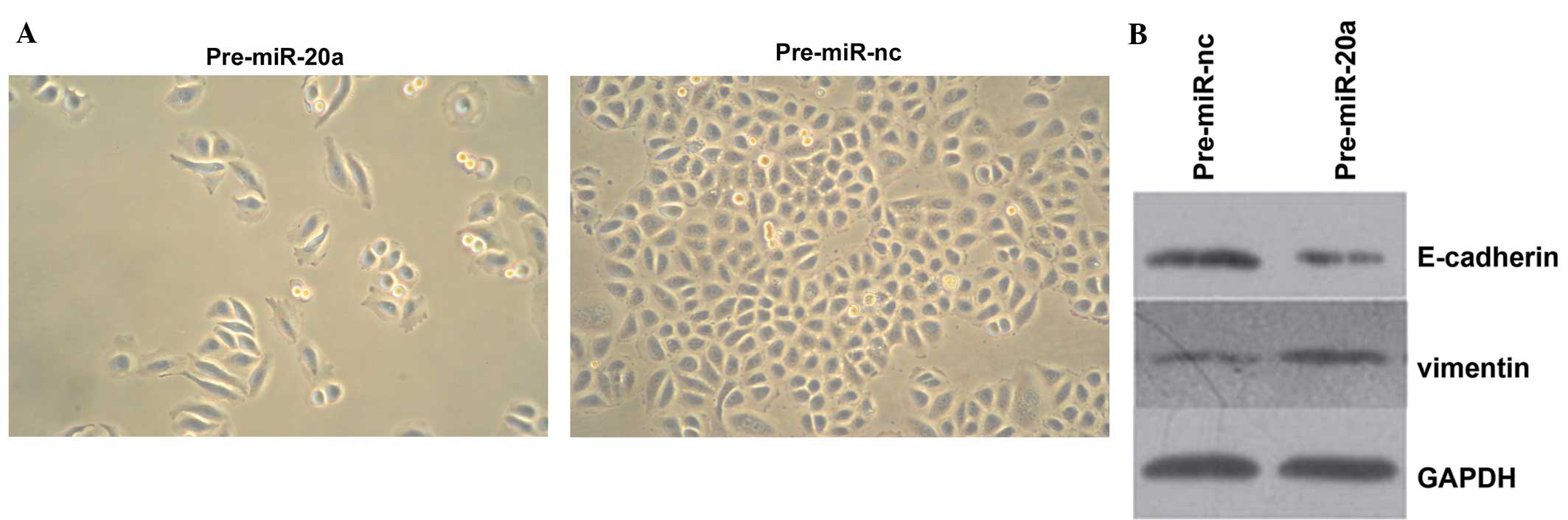
pre-miR-20a-transfected SW480 cells were observed to
undergo EMT-like transformation, as evidenced by the loss of
cell-cell adhesion and alterations in morphology from a round,
compact shape to an elongated, fibroblast-like morphology with
scattered distribution, which may facilitate cell migration
(Fig. 4A). In addition, western
blot analysis indicated that overexpression of miR-20a
downregulated the epithelial marker E-cadherin, while it
upregulated the mesenchymal marker vimentin (Fig. 4B). Therefore, overexpression of
miR-20a may induce EMT in SW480 cells.
SMAD4 is a target of miR-20a
Since miR-20a has a pivotal function in the
metastasis of CRC cells, its putative target genes with metastatic
functions were investigated using the online search tools
TargetScan (http://www.targetscan.org) and
miRanda (http://www.microrna.org). Among these
genes, SMAD4, one of the mediators of transforming growth factor β
(TGF-β) signaling, was predicted to be a theoretical target of
miR-20a based on putative target sequences at position 1,357–1,363
of the SMAD4 3′-UTR (Fig. 5A).
Therefore, it was further investigated whether SMAD4 was the
authentic target gene of miR-20a in CRC.
 | Figure 5Identification of SMAD4 as a potential
target of miR-20a in CRC. (A) WT and MUT 3′UTRs of SMAD4 with the
seed region and base substitutions (bold). (B) HEK 293 cells were
transiently co-transfected with luciferase reporter vectors and
either pre-miR-20a or pre-miR-nc. Luciferase activities were
normalized to the activity of Renilla luciferase. (C) Expression of
SMAD4 protein was assessed using western blot analysis. (D)
Correlation analysis between miR-20a and SMAD4 mRNA levels in CRC
tissue samples (Pearson’s correlation analysis).
**P<0.01, pre-miR-20a versus pre-miR-nc. WT,
Wild-type, MUT, mutated; miR, microRNA; UTR, untranslated region;
SMAD4, SMAD family member 4; CRC, colorectal cancer; pre-miR-nc,
negative control miRNA precursor; pre-miR-20a, miR-20a
precursor. |
To directly address whether miR-20a bound to the
3′-UTR of the target mRNA, a luciferase reporter vector containing
the SMAD4 3′-UTR with the putative miR-20a binding sites was
generated. Correspondingly, a mutant reporter vector containing the
SMAD4 3′-UTR with a mutation at the putative miR-20a binding site
was produced. As shown in Fig. 5B,
a marked reduction in luciferase activity was observed in cells
transfected with pre-miR-20a as compared with
pre-miR-nc-transfected cells (P<0.01). By contrast, no change in
luciferase activity was observed in cells transfected with the
mutant 3′-UTR constructs.
It was then determined whether the overexpression of
miR-20a can lead to downregulation of SMAD4 protein expression.
Western blot analysis showed that SMAD4 protein levels were
markedly reduced in SW480 cells overexpressing miR-20a as compared
with those in the control (P<0.01, Fig. 5C). To further explore the
correlation between miR-20a and SMAD4 expression in CRC tissue
samples, miR-20a and SMAD4 mRNA were examined in 10 sets of CRC
tissue samples. Using Pearson’s correlation analysis, a significant
inverse correlation between miR-20a and SMAD4 mRNA was observed
(P=0.033, Fig. 5D). Taken
together, the results suggest that miR-20a may attenuate the
expression of SMAD4 by directly targeting the 3′UTR of SMAD4.
Discussion
Accumulating evidence has indicated that
dysregulation of miR-20a is associated with the development of
cancer. However, the function of miR-20a in the development and
progression of cancer is controversial. High levels of miR-20a have
been found to correlate with malignant phenotypes and unfavorable
prognosis in hepatocellular and gallbladder carcinoma (19,20).
Recently, it was shown that miR-20a encoded by the miR-17–92
cluster enhances the proliferation and metastatic potential of
ovarian cancer and osteosarcoma cells (21,22).
By contrast, miR-20a overexpression inhibited proliferation and
metastasis in breast and pancreatic cancers (23,24).
These controversial results may reflect the diverse roles of
miR-20a in different types of cancer.
The present study demonstrated that miR-20a was
frequently upregulated in CRC tissue samples as compared with
normal adjacent mucosa, which was consistent with previous studies
(26,27). Furthermore, the patient group with
high miR-20a expression levels showed a greater incidence of lymph
node and distant metastases compared with the low miR-20a
expression group, strongly suggesting that miR-20a may be involved
in the metastasis of CRC. In addition, patients whose tumors had
higher miR-20a expression levels had a significantly lower overall
survival rate, indicating that high miR-20a levels are a marker of
unfavorable prognosis for patients with CRC. Multivariate analysis
indicated that high miR-20a expression was an independent
prognostic factor for survival. However, Yu et al (26) reported that miR-20a was not an
independent prognostic biomarker among the miR-17–92 cluster in
colon cancer. This inconsistency may be due to differences in
sample origin, tumor clinicopathological characteristics or
different detection methods of the studies. Therefore,
multi-institutional, prospective randomized trials are required
before a consensus can be reached.
Given that miR-20a was upregulated in CRC tissue
samples, it was speculated that miR-20a may have an oncogenic
effect in CRC. As expected, ectopic expression of miR-20a promoted
the migration and invasion of SW480 cells. In addition, the present
study revealed that cells with high expression of miR-20a had a
spindle-like morphology and that the upregulation of miR-20a
decreased the expression of E-cadherin, while it increased the
expression of vimentin, suggesting that miR-20a is involved in the
regulation of EMT. The above data suggest that miR-20a is involved
in modulating cell metastasis in CRC.
To address the molecular mechanisms involved in the
miR-20a-mediated changes in biological properties, SMAD4 was
selected for further study as it was predicted to be a target of
miR-20a by bioinformatics analysis. SMAD4 is a member of the
evolutionarily conserved family of SMAD proteins, which are
transmitters of signals from the TGF-β superfamily of cytokines
(28). It has been suggested that
SMAD4 can function as a tumor suppressor gene in gastrointestinal
carcinoma (29,30). The results in the present study
indicated that SMAD4 is a direct target gene of miR-20a in CRC,
since overexpression of miR-20a downregulated SMAD4 protein
expression. Furthermore, miR-20a expression was inversely
correlated with SMAD4 expression in CRC tumors, and overexpression
of miR-20a significantly reduced the activity of a luciferase
reporter containing the 3′UTR sequence of SMAD4. Combining these
experimental results with the bioinformatics analysis led to the
conclusion that SMAD4 is a target gene of miR-20a in CRC.
Loss of SMAD4 is considered to be a genetic step
associated with the advanced stages of the disease, and frequently
occurs in metastatic CRC (31).
SMAD4 inactivation is rarely detected in adenomas; however, it
strongly increases in frequency at late, metastatic stages
(32,33). Deckers et al (34) demonstrated that the knockdown of
SMAD4 in breast cancer cells strongly attenuated the transformation
of epithelial cuboidal cells into parallel aligned
fibroblastic-like cells, as well as the downregulation of
E-cadherin and the upregulation of N-cadherin, suggesting that
SMAD4 acts as an EMT suppressor (34). Further studies have found a
dependency of TGF-β-induced EMT on SMAD4 expression (35,36).
A previous study reported that the invasion suppressor E-cadherin
is a target gene in the SMAD4 signaling network and re-expression
of SMAD4 in SW480 human colon carcinoma induced E-cadherin
expression (37). Pohl et
al (38) reported that loss of
SMAD4 promotes migration and invasion and mediates EMT in the SW480
CRC cell line. In the present study, restoration of miR-20a induced
EMT and promoted CRC cell migration and invasion, which may be
attributed to miR-20a-mediated downregulation of SMAD4
expression.
In conclusion, the present study has provided novel
insights into the role of miR-20a in CRC. The results showed that
miR-20a is an independent prognostic factor for patients with CRC
and provided a potential mechanism for SMAD4 dysregulation and its
contribution to CRC cell migration, invasion and EMT. miR-20a may
function as a potential oncogene in CRC and has a potential
application in cancer therapy.
Acknowledgements
The present study was supported by the Scientific
Research Fund of Sichuan Provincial Education Department of China
(CBY12-A-ZD16).
References
|
1
|
Tenesa A and Dunlop MG: New insights into
the aetiology of colorectal cancer from genome-wide association
studies. Nat Rev Genet. 10:353–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang IP, Tsai HL, Hou MF, et al:
MicroRNA-93 inhibits tumor growth and early relapse of human
colorectal cancer by affecting genes involved in the cell cycle.
Carcinogenesis. 33:1522–1530. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Voulgari A and Pintzas A:
Epithelial-mesenchymal transition in cancer metastasis: mechanisms,
markers and strategies to overcome drug resistance in the clinic.
Biochim Biophys Acta. 1796:75–90. 2009.PubMed/NCBI
|
|
6
|
Du C, Zhang C, Hassan S, et al: Protein
kinase D1 suppresses epithelial-to-mesenchymal transition through
phosphorylation of snail. Cancer Res. 70:7810–7819. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spaderna S, Schmalhofer O, Hlubek F, et
al: A transient, EMT-linked loss of basement membranes indicates
metastasis and poor survival in colorectal cancer.
Gastroenterology. 131:830–840. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yao D, Dai C and Peng S: Mechanism of the
mesenchymal-epithelial transition and its relationship with
metastatic tumor formation. Mol Cancer Res. 9:1608–1620. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee Y, Jeon K, Lee JT, et al: MicroRNA
maturation: stepwise processing and subcellular localization. EMBO
J. 21:4663–4670. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee Y, Ahn C, Han J, et al: The nuclear
RNase III Drosha initiates microRNA processing. Nature.
425:415–419. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lund E, Güttinger S, Calado A, et al:
Nuclear export of microRNA precursors. Science. 303:95–98. 2004.
View Article : Google Scholar
|
|
12
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
13
|
Li C, Nguyen HT, Zhuang Y, et al:
Comparative profiling of miRNA expression of lung adenocarcinoma
cells in two-dimensional and three-dimensional cultures. Gene.
511:143–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moriyama T, Ohuchida K, Mizumoto K, et al:
MicroRNA-21 modulates biological functions of pancreatic cancer
cells including their proliferation, invasion, and chemoresistance.
Mol Cancer Ther. 8:1067–1074. 2009. View Article : Google Scholar
|
|
15
|
Bhaumik D, Scott GK, Schokrpur S, et al:
Expression of microRNA-146 suppresses NF-kappaB activity with
reduction of metastatic potential in breast cancer cells. Oncogene.
27:5643–5647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang G, Xia S, Tian H, et al: Clinical
significance of miR-22 expression in patients with colorectal
cancer. Med Oncol. 29:3108–3812. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Paterson EL, Kazenwadel J, Bert AG, et al:
Down-regulation of the miRNA-200 family at the invasive front of
colorectal cancers with degraded basement membrane indicates EMT is
involved in cancer progression. Neoplasia. 15:180–191.
2013.PubMed/NCBI
|
|
18
|
Zhang GJ, Xiao HX, Tian HP, et al:
Upregulation of microRNA-155 promotes the migration and invasion of
colorectal cancer cells through the regulation of claudin-1
expression. Int J Mol Med. 31:1375–1380. 2013.PubMed/NCBI
|
|
19
|
Fan MQ, Huang CB, Gu Y, et al: Decrease
expression of microRNA-20a promotes cancer cell proliferation and
predicts poor survival of hepatocellular carcinoma. J Exp Clin
Cancer Res. 32:212013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang Y, Liu C, Yang J, et al: MiR-20a
triggers metastasis of gallbladder carcinoma. J Hepatol.
59:518–527. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fan X, Liu Y, Jiang J, et al: miR-20a
promotes proliferation and invasion by targeting APP in human
ovarian cancer cells. Acta Biochim Biophys Sin (Shanghai).
42:318–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang G, Nishimoto K, Zhou Z, et al:
miR-20a encoded by the miR-17–92 cluster increases the metastatic
potential of osteosarcoma cells by regulating Fas expression.
Cancer Res. 72:908–916. 2012.
|
|
23
|
Li JY, Zhang Y, Zhang WH, et al:
Differential distribution of miR-20a and miR-20b may underly
metastatic heterogeneity of breast cancers. Asian Pac J Cancer
Prev. 13:1901–1906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan H, Wu J, Liu W, et al: MicroRNA-20a
overexpression inhibited proliferation and metastasis of pancreatic
carcinoma cells. Hum Gene Ther. 21:1723–1734. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chai H, Liu M, Tian R, et al: miR-20a
targets BNIP2 and contributes chemotherapeutic resistance in
colorectal adenocarcinoma SW480 and SW620 cell lines. Acta Biochim
Biophys Sin (Shanghai). 43:217–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu G, Tang JQ, Tian ML, et al: Prognostic
values of the miR-17-92 cluster and its paralogs in colon cancer. J
Surg Oncol. 106:232–237. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Motoyama K, Inoue H, Takatsuno Y, et al:
Over- and under-expressed microRNAs in human colorectal cancer. Int
J Oncol. 34:1069–1075. 2009.PubMed/NCBI
|
|
28
|
Massagué J and Chen YG: Controlling
TGF-beta signaling. Genes Dev. 14:627–644. 2000.PubMed/NCBI
|
|
29
|
Wang LH, Kim SH, Lee JH, et al:
Inactivation of SMAD4 tumor suppressor gene during gastric
carcinoma progression. Clin Cancer Res. 13:102–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Powell SM, Harper JC, Hamilton SR, et al:
Inactivation of Smad4 in gastric carcinomas. Cancer Res.
57:4221–4224. 1997.PubMed/NCBI
|
|
31
|
Miyaki M, Iijima T, Konishi M, et al:
Higher frequency of Smad4 gene mutation in human colorectal cancer
with distant metastasis. Oncogene. 18:3098–3103. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maitra A, Molberg K, Albores-Saavedra J
and Lindberg G: Loss of Dpc4 expression in colonic adenocarcinomas
correlates with the presence of metastatic disease. Am J Pathol.
157:1105–1111. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Riggins GJ, Kinzler KW, Vogelstein B and
Thiagalingam S: Frequency of Smad gene mutations in human cancers.
Cancer Res. 57:2578–2580. 1997.PubMed/NCBI
|
|
34
|
Deckers M, van Dinther M, Buijs J, et al:
The tumor suppressor Smad4 is required for transforming growth
factor beta-induced epithelial to mesenchymal transition and bone
metastasis of breast cancer cells. Cancer Res. 66:2202–2209. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Valcourt U, Kowanetz M, Niimi H, et al:
TGF-beta and the Smad signaling pathway support transcriptomic
reprogramming during epithelial-mesenchymal cell transition. Mol
Biol Cell. 16:1987–2002. 2005. View Article : Google Scholar
|
|
36
|
Levy L and Hill CS: Smad4 dependency
defines two classes of transforming growth factor {beta}
(TGF-{beta}) target genes and distinguishes TGF-{beta}-induced
epithelial-mesenchymal transition from its antiproliferative and
migratory responses. Mol Cell Biol. 25:8108–8125. 2005.PubMed/NCBI
|
|
37
|
Müller N, Reinacher-Schick A, Baldus S, et
al: Smad4 induces the tumor suppressor E-cadherin and P-cadherin in
colon carcinoma cells. Oncogene. 21:6049–6058. 2002.PubMed/NCBI
|
|
38
|
Pohl M, Radacz Y, Pawlik N, et al: SMAD4
mediates mesenchymal-epithelial reversion in SW480 colon carcinoma
cells. Anticancer Res. 30:2603–2613. 2010.PubMed/NCBI
|