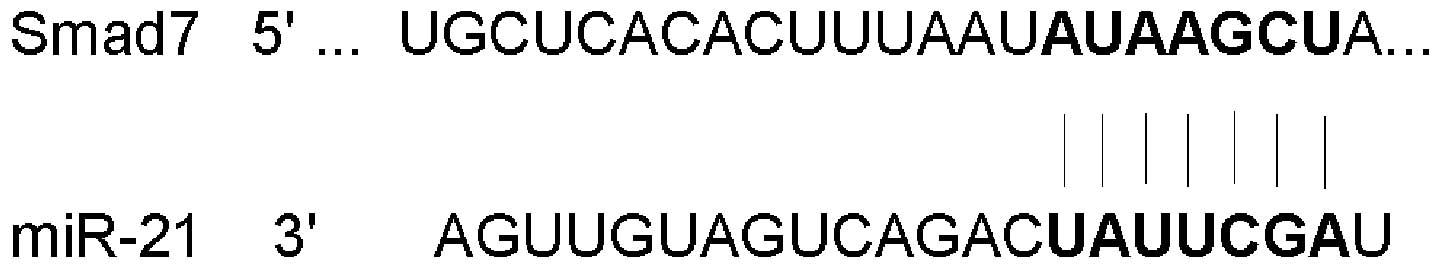Introduction
The identification of microRNA (miRNA) in the 1990s
was prominent in biological research (1). miRNAs are a class of small non-coding
single-stranded RNA, 21–25 nucleotides in length, with diverse
biological functions (2,3). miRNA has been implicated in
regulating cell division, differentiation, development,
oncogenesis, apoptosis and other processes through translational
arrest and/or messenger RNA (mRNA) degradation (4–6). It
has been shown that a single miRNA may be directly responsible for
the repression of hundreds of proteins, and indirectly for the
regulation of thousands of others (4). Abnormal expression of miRNA has been
implicated in various diseases, including cancer, diabetes, and
cardiovascular, neurodegenerative and psychological disorders
(5–8). Despite the involvement of miRNA in
numerous biological and pathological processes, the large number of
candidate targets and cellular pathways, which are regulated by
miRNAs, remain to be understood.
miRNA-21 (miR-21) is a small RNA, which is
ubiquitously expressed in normal tissues and cells (9). Previous studies have predominantly
focused on the association between miR-21 and tumors, since miR-21
has been consistently identified to be overexpressed in numerous
tumor samples (10), including
glioblastomas (11), lung
(12,13), stomach (14), prostate (14) and colon cancer (15,16),
and cholangiocarcinoma (17)
samples.
miR-21 has numerous predicted target genes; however,
only a few have been validated in an experimental study (18). Despite the widespread expression of
miR-21 in normal tissues, little is known regarding its
physiological functions and regulatory mechanisms (19).
Transforming growth factor (TGF)-β is a
multifunctional growth factor that functions in the initiation and
termination of tissue repair. TGF-β is considered to be a key
mediator in disease pathogenesis (20–22).
SMAD7 is a negative regulator of Smad signaling and inhibits TGF-β
activity through the prevention of the phosphorylation of Smad2/3
(23,24). However, the association between
TGF-β and miR-21 in renal disease, in particular diabetic
nephtropathy, remains to be elucidated.
The aim of the present study was to investigate the
functional interaction between miR-21 and the TGF-β signaling
pathway, in order to eventually determine its involvement in
diabetic nephropathy.
Materials and methods
Cell lines and culture conditions
Rat renal tubular epithelial cells and HEK 293T
cells were obtained from the Shanghai Academy Sciences Cell Bank
(Shanghai, China). Cell lines were cultured in Dulbecco’s modified
Eagle’s medium (DMEM) high glucose (25 mmol/l) and low glucose (5
mmol/l), respectively (Gibco-BRL, Carlsbad, CA, USA). The low
glucose group served as a control, while the high glucose group
represented the environment of renal cells in a patient with
diabetes. The media were supplemented with 10% fetal bovine serum
(FBS; Gibco-BRL). All cells were grown in a humidified incubator in
5% CO2 at 37°C.
Bioinformatic analyses
TargetScan 4.1 (http://www.targetscan.org), PicTar (http://pictar.mdc-berlin.de/) and miRNA.org
(http://www.microrna.org/), GenBank (http://www.ncbi.nlm.nih.gov/gene/) online
databases were used to predict the targets of miR-21. These three
programs used a different algorithm to identify highly
complementary sites of miRNA. SMAD7 was consistently identified as
a target of miR-21 with all three programs (Fig. 1).
Dual luciferase reporter assay
Luciferase reporter gene constructs containing
either the wild-type or mutated full-length 3′ untranslated region
(UTR) of SMAD7, were constructed using the luciferase reporter
vector psi-CHECK2 (Promega Corporation, Madison, WI, USA). The
following primers were used for site direct mutagenesis plasmid
construction: Forward:
5′-CACACTTTAATGCGGTTCATTTTTCTAACTACAAAGGTTT-3′; and reverse:
5′-AGTTAGAAAAATGAACCGCATTAAAGTGTGAGCATGCTCA-3′ for MutSmad7.
ATAAGCT was mutated to GCGGTTC. The cells were transiently
cotransfected into 24-well plates using Lipofectamine™ reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA). Mutations to the
seed region of miR-21 mimics, which can result in high levels of
miR-21 expression, and miR-21 negative controls (NC) which have no
function were compounded. These were cotransfected into HEKT cells
with the WT or mutant 3′UTR SMAD7. Reporter assays were performed
at 48 h posttransfection using the Dual-luciferase Reporter Assay
system (E1910; Promega Corporation) according to manufacturer’s
instructions. All experiments were performed in triplicate and the
standard deviation was calculated.
miR-21 transfection in renal tubular
epithelial cells
Rat renal tubular epithelial cells were transfected
with 1.2 μl lentivirus overexpressing miR-21 or empty virus
(Shanghai Jikai Gene Chemical Co., Ltd, Shanghai, China). Flow
cytometry (FACSVantage; BD Biosciences, San Jose, CA, USA) was used
36 h after transfection in order to assess transfection
efficiencies. Total RNA was then extracted from the cells and the
miR-21 levels were assessed using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Cell growth inhibition assay
Following transfection, the cells were seeded into
96-well plates at a density of 10,000 cells/well. Four plates were
used in total, and were incubated for 0, 24, 48, or 72 h. The
cellular proliferation was analyzed at each time point using an MTT
Cell Proliferation Assay kit (Invitrogen Life Technologies) over
four days.
RT-qPCR
Cells were collected following transfection for 72 h
and total RNA was extracted using TRIzol™ reagent (Invitrogen Life
Technologies). cDNA was synthesized from the extracted RNA using a
First-strand cDNA Synthesis kit (Promega Corporation) according to
the manufacturer’s instructions. The miRNA reverse
transcription-PCR primers for miR-21, and endogenous control U6RT
were purchased from Applied Biosystems (Applied Biosystems,
Invitrogen Life Technologies). The primer sequences used in the
experiment were designed as follows: Forward:
5′-ACACTCCAGCTGGGTAGCTTATCAGACTGAT-3′; and reverse:
5′-ACTGGTGTCGTGGAGTCG-3′ for Rno-mir-21. qPCR analysis was
performed using an ABI PRISM® 7500 Sequence Detection
system (Applied Biosystems). The conditions for the PCR were as
follows: 95°C for 5 min, 95°C for 15 sec and 62°C for 30 sec, for a
total of 40 cycles. Each sample was run in triplicate. The
comparative threshold cycle (CT) method was used to evaluate the
relative gene expression.
Western blot analysis
Western blot analysis was performed according to
standard protocols. Briefly, the cells were solubilized in lysis
buffer (containing posphatase inhibitors, protease inhibitors and
PMSF) and the protein was extracted using a Protein Extraction
reagent (Sigma, St. Louis, MO, USA). Protein concentrations were
determined using a Bio-Rad Protein Assay kit (Bio-Rad, Hercules,
CA, USA). The proteins were separated by SDS-PAGE and transferred
to nitrocellulose membranes. Then the membranes were blocked and
incubated overnight at 4°C with a rabbit anti-SMAD7 antibody
(Abcam, Cambridge, MA, USA). The membranes were then washed and
incubated with horseradish peroxidase-conjugated goat anti-rabbit
secondary antibody (Abcam) at 37°C for two hours. Proteins were
visualized by an enhanced chemiluminescence detection system
(Amersham Pharmacia Biotech, Piscataway, NJ, USA). When required,
the membranes were stripped and reprobed with β-actin, as an
internal loading control.
Statistical analysis
Statistical analyses were performed using a one-way
analysis of variance or a Kruskal-Wallis non-parametric test,
according to the Gaussian or non-Gaussian distribution of the data.
P<0.05 was considered to indicate a statistically significant
difference in miRNA expression profiling. All data are presented as
the mean ± standard deviation. Statistical analysis was performed
using SPSS software 17.00 (SPSS Inc., Chicago, IL, USA).
Results
SMAD7 is a predicted target of
miR-21
In order to determine whether SMAD7 was a predicted
target of miR-21, the miRNA GenBank was used. Using the TargetScan
software, it was predicted that SMAD7 was a target of miR-21. One
binding site for miR-21 was identified in the 3′UTR of the SMAD7
gene, complementary to the 3′ region of miR-21. The predicted
binding site was located at 1192–1198 and contained seven
conservative target sites (Fig.
1).
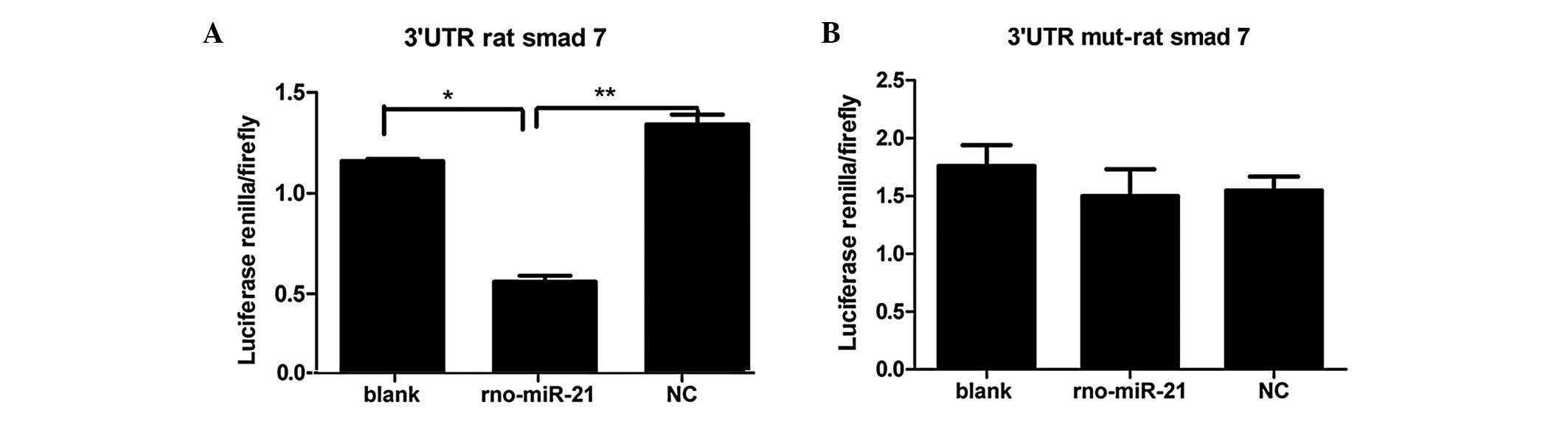
Luciferase assay confirms that SMAD7 is
regulated by miR-21
To confirm whether SMAD7 could be regulated by
miR-21 in vitro, wild-type and mutant plasmids expressing
the 3′UTR region predicted to bind miR-21, were constructed. Then
miR-21-mimics with a mutated seed region and miR-21-negative
control (NC) were synthetized (Shanghai Jima Company, Shanghai,
China). Subsequently, the two luciferase reporter vectors with
miR-21 response were cotransfected into HEK 293T cells using a
Dual-Luciferase Reporter Assay system. As shown in Fig. 2A, following transfection in the HEK
293T cells, a significant difference was observed in luciferase
activity between the miR-21 and blank groups (P<0.05) and
between the miR-21 and NC groups (P<0.01). These data showed
that miR-21 could interact with the 3′UTR of SMAD7, which was not
observed in the NC and blank groups (P>0.05). As shown in
Fig. 2B, in HEK 293T cells
cotransfected with the mutated 3′UTR rat SMAD7 gene, there was no
statistical significance among the miR-21, blank and NC groups
(P>0.05). These data indicate that the SMAD7 3′UTR could not
interact with miR-21 when mutated. Therefore, these results confirm
that the SMAD7 3′UTR may be regulated by miR-21.
Protein level of SMAD7 inversely
correlates with the expression level of miR-21 in rat renal tubular
epithelial cells
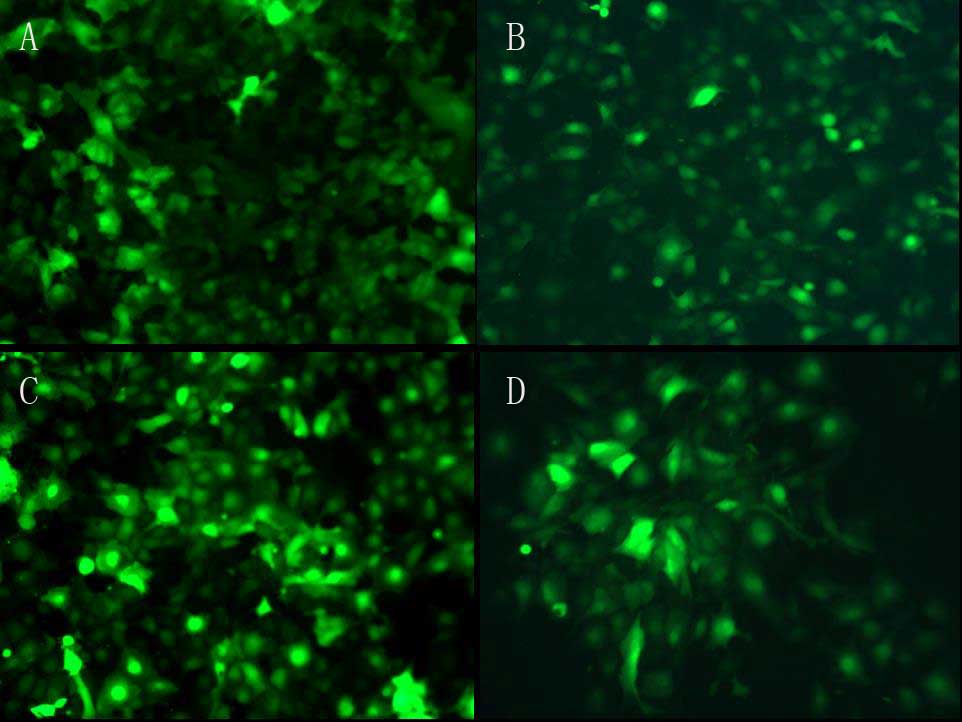
To further confirm whether miR-21 directly targets
the 3′UTR of SMAD7, lentiviral vectors overexpressing miR-21 and
empty lentiviral vectors were constructed. The plasmids were
transfected into rat renal tubular epithelial cells respectively
(Fig. 3) and the transfection
efficiency was analyzed using flow cytometry (Fig. 3E). The RNA and protein was
extracted from renal tubular epithelial cells, 72 h
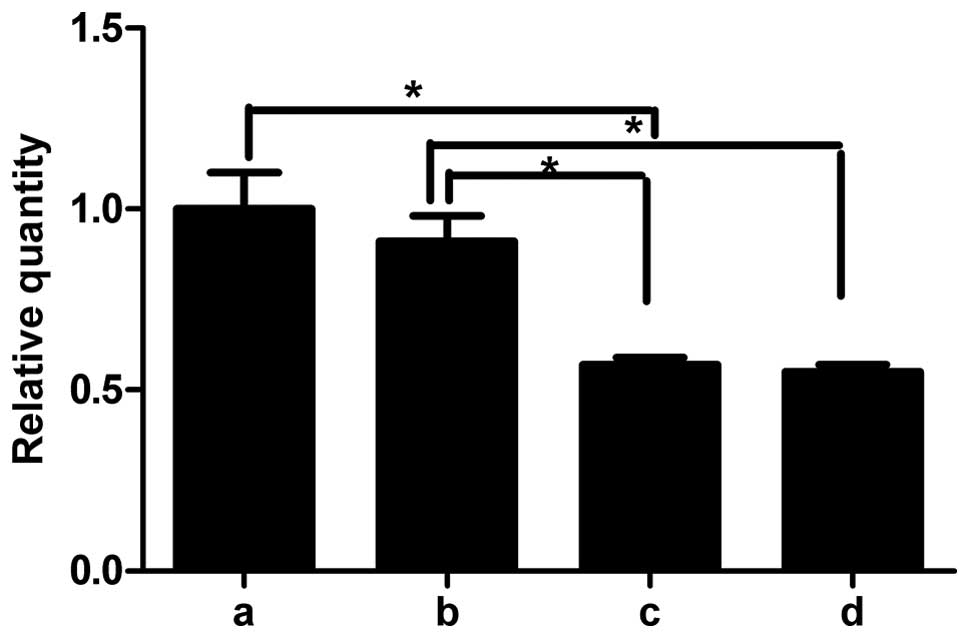
post-transfection. The results of RT-qPCR demonstrated that the
expression of miR-21 was higher in the lentivirus-transfected rat
renal tubular epithelial cells (Fig.
4b) cultured in high glucose compared with untransfected cells
cultured in high glucose (Fig.
4c). There was no difference between the transfected and
untransfected cells in the low glucose groups (data not shown). By
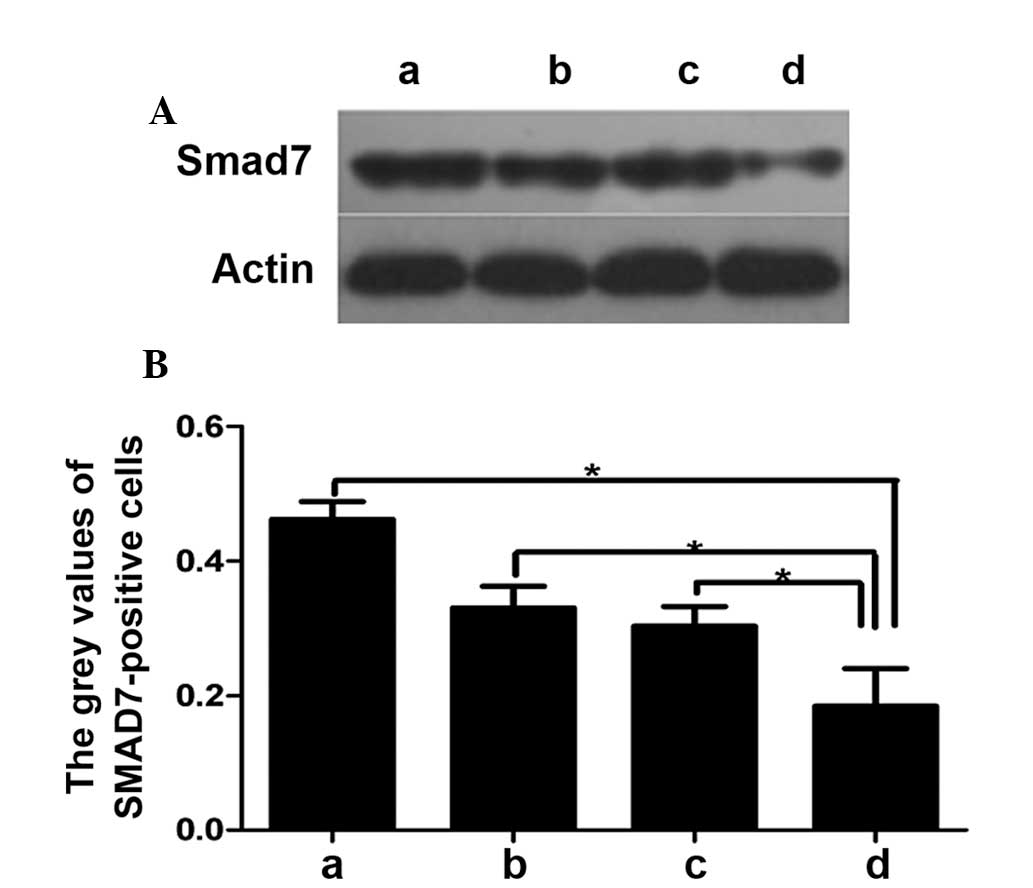
western blot analysis, it was shown that the protein expression of
SMAD7 was increased in the low glucose groups. In the high glucose
groups, the SMAD7 expression was lower compared with that in the
low glucose group. When lentivirus expressing miR-21 and empty
lentivirus were transfected the protein expression of SMAD7 was
lower in the miR-expressing groups in high and low glucose
conditions (Fig. 5) when compared
with the cells transfected with empty lentivirus.
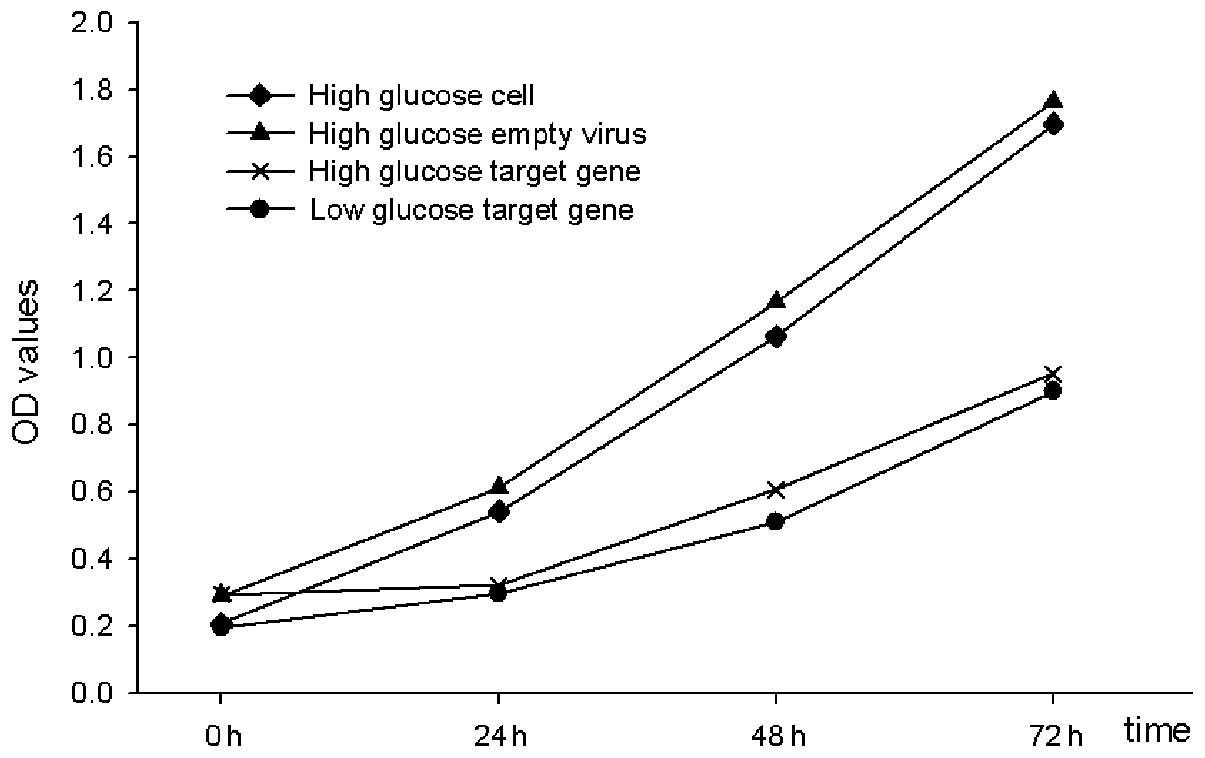
miR-21 inhibits the proliferation of rat
renal tubular epithelial cells
To investigate the effects of miR-21 on renal
tubular epithelial cells, miR-21 overexpressing- and empty
lentiviruses were transfected into the rat renal tubular epithelial
cells and were cultured under high and low glucose conditions.
Following transfection for 24, 48 and 72 h, it was detected that
the cellular proliferation in the miR-21-transfected lentivirus
group was significantly decreased compared with the empty
lentivirus and untransfected groups (P<0.01). These data
indicated that overexpression of miR-21 may inhibit rat renal
tubular epithelial cellular proliferation (Fig. 6).
Discussion
miRNA are a class of non-coding, highly conserved
RNA molecules, with numerous biological functions in various
processes, including development, differentiation, cell
proliferation and apoptosis (25,26).
TGF-β is a member of a large family of structurally
related cytokines, including activins, Nodal and bone morphogenetic
proteins, which transmit cellular signals through the Smad
signaling pathway (27). TGF-β1
binds to TGF-β receptor II (TβRII), to initiate intracellular
signaling. This results in activation of TβRI kinase, which
phosphorylates and activates Smad2 and 3. Activated Smad2 and 3
form heteromeric complexes, together with Smad4, and translocate to
the nucleus to regulate target gene activity. Inhibitory Smads
(SMAD6 and SMAD7) provide negative feedback and repress TGF-β
superfamily signaling through various molecular mechanisms
(28).
miRNA-21 is a conserved miRNA that has been
associated with various types of tumors. Less research has been
performed on the cellular pathways involving miR-21 in renal
diseases. Certain studies have shown that miRNAs are associated
with TGF-β signaling in other diseases (29,30).
This study has explored the possible role of miR-21 in TGF-β
signaling, and confirmed the results of previous studies that
showed SMAD7 to be a direct target of miR-21, with predicted target
sites in its 3′UTR. To further confirm whether SMAD7 could be
regulated by miR-21, a luciferase assay was used to assess the
SMAD7 3′UTR reporter activity when co-transfected with miR-21.
Mutation in the SMAD7 miR-21 target site reduced the inihibitory
effects on SMAD7 expression. When miR-21 lentivirus was transfected
into rat renal tubular epithelial cells, the protein level of SMAD7
was lower than that of the empty lentivirus-transfected cells, in
untransfected low glucose and high glucose cells conditions.
These data confirmed that SMAD7 is a direct target
of miR-21, and that miR-21 can repress the expression of SMAD7
proteins and inhibit the proliferation of rat renal tubular
epithelial cells by targeting TGF-β/SMAD signaling in vitro.
Preliminary data suggested that miR-21 affects proliferation of rat
renal tubual epithelial cells and further studies should
investigate whether miR-21 is abberantly expressed in renal
disease. Further studies using a diabetic nephropathy animal model
are required in order to determine the function of miR-21
regulation of SMAD7 in diabetes.
Acknowledgements
The authors would like to thank the teachers,
including Professor Weixue Tang, of the First Affiliated Hospital
Chongqing Medical University Experiment Center, for their
assistance in the experimental studies.
References
|
1
|
Baek D, Villén J, Shin C, et al: The
impact of microRNAs on protein output. Nature. 455:64–71. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eulalio A, Huntzinger E and Izaurralde E:
Getting to the root of miRNA-mediated gene silencing. Cell.
132:9–14. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
et al: Frequent deletions and down-regulation of microRNA genes
miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl
Acad Sci USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA, Ferracin M, Cimmino A, Di Leva
G, et al: A MicroRNA signature associated with prognosis and
progression in chronic lymphocytic leukemia. N Engl J Med.
353:1793–1801. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Michael MZ, O’Conner SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
8
|
Pfeffer S, Zavolan M, Grasser FA, Chien M,
et al: Identification of virus-encoded microRNAs. Science.
304:734–736. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hwang H and Mendell J: MicroRNAs in cell
proliferation, cell death and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen C, Li L, Lodish H and Bartel D:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yanaihara N, Caplen N, Bowman E, Seike M,
et al: Unique microRNA molecular profiles in lung cancer diagnosis
and prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Markou A, Tsaroucha EG, Kaklamanis L, et
al: Prognostic value of mature microRNA-21 and microRNA-205
overexpression in non-small cell lung cancer by quantitative
real-time RT-PCR. Clin Chem. 54:1696–1704. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Volinia S, Calin GA, Liu CG, Ambs S, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
et al: Altered expression of miR-21, miR-31, miR-143 and miR-145 is
related to clinicopathologic features of colorectal cancer.
Oncology. 72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, et al: MicroRNA expression profiles associated with prognosis
and therapeutic outcome in colonadenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meng F, Henson R, Lang M, Wehbe H, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Papadopoulos GL, Reczko M, Simossis VA,
Sethupathy P and Hatzigeorgiou AG: The database of experimentally
supported targets: a functional update of TarBase. Nucleic Acids
Res. 37:D155–D158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Selcuklu SD, Donoghue MT and Spillane C:
miR-21 as a key regulator of oncogenic processes. Biochem Soc
Trans. 37:918–925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ashcroft GS, Yang X, Glick AB, Weinstein
M, et al: Mice lacking Smad3 show accelerated wound healing and an
impaired local inflammatory response. Nat Cell Biol. 1:260–266.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Attisano L and Wrana JL: Smads as
transcriptional co-modulators. Curr Opin Cell Biol. 12:235–243.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Attisano L and Wrana JL: Signal
transduction by the TGF-beta superfamily. Science. 296:1646–1647.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hayashi H, Abdollah S, Qiu Y, et al: The
SMAD-related protein SMAD7 associates with the TGF beta receptor
and functions as an antagonist of TGF beta signaling. Cell.
89:1165–1173. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kavsak P, Rasmussen RK, Causing CG, et al:
SMAD7 binds to Smurf 2 to form an E3 ubiquitin ligase that targets
the TGF beta receptor for degradation. Mol Cell. 6:1365–1375. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Karp X and Ambros V: Encountering
microRNAs in cell fate signaling. Science. 310:1288–1289. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Heldin CH, Miyazono K and ten Dijke P:
TGF-beta signaling from cell membrane to nucleus through SMAD
proteins. Nature. 390:465–471. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Massague J and Wotton D: Transcriptional
control by the TGF-beta/Smad signaling system. EMBO J.
19:1745–1754. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu H, Li Y, Qu S, et al: MicroRNA
expression abnormalities in limited cutaneous scleroderma and
diffuse cutaneous scleroderma. J Clin Immunol. 32:514–522. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei J, Feng L, Li Z, Xu G and Fan X:
MicroRNA-21 activates hepatic stellate cells via PTEN/Akt
signaling. Biomed Pharmacother. 67:387–392. 2013. View Article : Google Scholar : PubMed/NCBI
|