Introduction
Zipper-interacting protein kinase (ZIPK) is a novel
serine/threonine (Ser/Thr) protein kinase, which was initially
cloned and identified in 1998 (1,2).
ZIPK is a member of the large family of death-associated protein
kinases and has been associated with the regulation of numerous
cellular processes, including cell death (1), cell motility (3) and mitotic processes (4), as well as smooth muscle contraction
(5,6). In addition, ZIPK interacts with the
signal transducer and activator of transcription 3 (STAT3), a
latent cytoplasmic transcription factor that plays a role in cell
growth and apoptosis. ZIPK has been shown to phosphorylate STAT3 on
Ser-727, thus enhancing the transcriptional activity of STAT3
(7). Vetterkind et al
(8) have demonstrated that
prostate apoptosis response-4 (Par-4), which is characterized
mainly as a proapoptotic protein, targets ZIPK to the cytoskeleton
in nonmuscle cells, leading to apoptosis. By contrast, in smooth
muscle cells, Par-4 supports contractility by targeting ZIPK to the
cytoskeleton (9). A previous
study, conducted by the authors of the present study, demonstrated
that incubation of smooth-muscle cells with glucose induced a time-
and dose-dependent increase in the protein expression levels of
ZIPK (unpublished data). These findings indicate that ZIPK plays a
key role in cellular function.
However, only a limited number of studies have
focused on the effects of ZIPK on cell death, motility and mitotic
processes. To date, the interacting partners of ZIPK, particularly
those associated with the cell cycle, have been rarely identified
and the regulatory network of ZIPK is not fully understood. The aim
of the present study was to investigate the interaction of ZIPK
with the human cell division cycle 14A (HsCdc14A) phosphatase in
vitro, and identify whether ZIPK plays a role in the cell cycle
regulation.
Materials and methods
Yeast two-hybrid assay
The study was approved by the ethics committee of
Huashan Hospital, Fudan University (Shanghai, China). Yeast
two-hybrid interaction screening was performed as described in
previous studies (10–12). Full-length HsCdc14A was introduced
into the GAL4 DNA (Clontech Laboratories, Inc., Mountain View, CA,
USA) binding domain as a bait. Its interaction partner was a human
testis cDNA library (Clontech Laboratories, Inc.) integrated with
GAL4 transactivation domain. The bait plasmids and the cDNA library
plasmids were used to transform a yeast strain (AH109; Clontech
Laboratories, Inc.), containing HIS3 and LacZ reporter genes, using
a lithium acetate method (13).
The transformed samples were spread onto plates with a synthetic
defined (SD)/Leu/Trp/His medium. Subsequently they were spread onto
a SD/-Leu/-Trp/-His/-Ade/X-α-Gal medium for further selection.
Positive clones (those with diameter >3 mm) were co-transformed
into the AH109 yeast with bait plasmids. Following extraction of
plasmids from yeast, the plasmids were used to transform
Escherichia coli (Tiandz, Inc., Beijing, China). The
interacting protein was verified by sequencing the plasmid
following extraction from Escherichia coli.
Glutathione S-transferase (GST) pull-down
assay
HsCdc14A cDNA [full length, amino acid (aa) 1–623;
deletion mutant, aa1-348N terminal; or aa349-623C terminal] was
fused with a 6-His tag (pET-28a; Tiandz Inc., Beijing, China). ZIPK
cDNA (full length, aa1-454) was cloned into a pGEX-5X-3 vector
(Amersham Biosciences, Piscataway, NJ, USA). Using a previously
described method (11), these
proteins were expressed in Escherichia coli BL21 (DE3) cells
(Tiandz Inc.), and purified by Ni2+ nitrilotriacetic
acid (for aa1-348; Sigma-Aldrich, St. Louis, MO, USA) or
glutathione beads (for aa349-623; Sigma-Aldrich). An in
vitro pull-down assay was performed using purified GST-fused
ZIPK and His-tagged HsCdc14A with phosphate-buffered saline (PBS),
containing 0.1% Triton X-100 (Sangon Biotech, Shanghai, China) for
4 h at 4°C. The beads were washed three times with PBS containing
1% Triton X-100, and once with PBS alone. The beads were then
boiled in SDS-PAGE sample buffer for 5 min and used for western
blot analysis.
Co-immunoprecipitation
Human embryonic kidney (HEK) 293T cells from the
American Type Culture Collection (Manassas, VA, USA) were grown to
50–60% confluence in Dulbecco’s modified Eagle’s medium (Hyclone,
Logan, UT, USA), supplemented with 10% fetal bovine serum
(Hyclone), at 37°C in an atmosphere containing 5% CO2.
The cells were then co-transfected with green fluorescent protein
(GFP)-HsCdc14A (full length or deletion mutant aa1-348N-terminal or
aa349-623 C-terminal) and FLAG-ZIPK using the standard
CaCl2 method (14),
with a GFP-transfected plasmid as a control. 36 h later, the cells
were collected and lysed in lysis buffer (50 mM HEPES, pH 7.2; 150
mM NaCl; 2 mM ethylene glycol tetraacetic acid; and 0.1% Triton
X-100), together with protease inhibitor mixture (Aprotinin,
Bestatin and Leupeptin Pepstatin A; Sigma-Aldrich). The lysate was
then clarified using centrifugation at 16,000 × g for 10 min at
4°C. The cell lysate was incubated with anti-FLAG antibody
conjugated to agarose beads (Sigma-Aldrich) for 4 h at 4°C, and
then was washed five times using lysis buffer. The proteins were
boiled for 5 min and used for subsequent western blot analysis.
Western blotting analysis
Following SDS-PAGE using a 12% gel, proteins were
transferred onto polyvinylidene difluoride membranes, which had
been obtained from Millipore Corporation (Billerica, MA, USA). The
membranes were incubated with primary antibodies overnight at 4°C
with gentle shaking (Qite Analytical Instrument Co. Ltd., Shanghai,
China). Membranes were then incubated with secondary antibodies at
room temperature for 2 h. Immunoreactive signals were detected
using an enhanced chemiluminescence kit (Pierce Chemical, IL, USA)
and visualized by autoradiography on Kodak BioMAX film (Kodak,
Rochester, NY, USA). The following antibodies were used: Mouse
monoclonal anti-His antibody (1:1,000; Cell Signaling Technology,
Inc., Danvers, MA, USA); rabbit polyclonal anti-GFP antibody
(1:1,000; Cell Signaling Technology, Inc); mouse monoclonal
anti-FLAG antibody (1:2,000; Sigma-Aldrich); and anti-rabbit
(1:2,000) and anti-mouse (1:2,000) horseradish
peroxidase-conjugated secondary antibodies (Cell Signaling
Technology, Inc.).
Cell cycle analysis
A cell cycle analysis was conducted as previously
described (15). Briefly, HEK293T
cells were grown in six-well plates with density ~1×106
cells per well and transfected with the following plasmids: GFP,
GFP-HsCdc14A, GFP-HsCdc14A N-terminus, GFP-HsCdc14A C-terminus,
GFP-ZIPK, GFP-HsCdc14A+ and GFP-ZIPK. 72 h later, cells were
collected and washed with ice cold PBS. Next, they were fixed with
70% ethanol for ≥1 h. The fixed cells were then washed and stained
with propidium iodide (PI; Invitrogen Life Technologies, Carlsbad,
CA, USA) for 1 h at room temperature. Subsequently, the PI-stained
nuclei were analyzed using the BD FACSCalibur™ system (BD
Biosciences, Franklin Lakes, NJ, USA).
Apoptosis assay
Annexin V-fluorescein isothiocyanate (FITC)/PI
staining using an Apoptosis Detection Kit (Invitrogen, Life
Technologies) was used in order to detect apoptotic cells. HEK293T
cells were collected at 72 h post-transfection with the plasmids
listed above. Cells were then trypsinized and collected by
centrifugation at 300 × g for 5 min. Cells were washed once with
PBS, resuspended in 1X binding buffer and stained with Annexin
V-FITC for 15 min. The cell nuclei were then counter-stained with
PI in order to detect necrosis. Flow cytometric analysis was
performed to analyze the percentage of apoptotic cells, using the
BDFACSCalibur™ system.
Statistical analysis
All experiments were performed at least in
triplicate. The results are expressed as the mean ± standard
deviation, and the data were analyzed using Student’s t-test to
detect statistically significant differences among the groups.
P<0.05 was considered to indicate a statistically significant
difference. Statistical analyses was performed using SPSS Version
11 (IBM SPSS, Armonk, NY, USA).
Results
Identification of HsCdc14A as a
ZIPK-interacting protein
Nucleotide sequencing was performed in previous
experiments using full length HsCdc14A cDNA as a bait in order to
screen through a human testis cDNA library. ZIPK was identified as
a potential binding protein. Subsequent assays validated that
HsCdc14A can bind to ZIPK (Fig.
1). To the best of our knowledge, the present study reported
for the first time a potential interaction between ZIPK and
HsCdc14A.
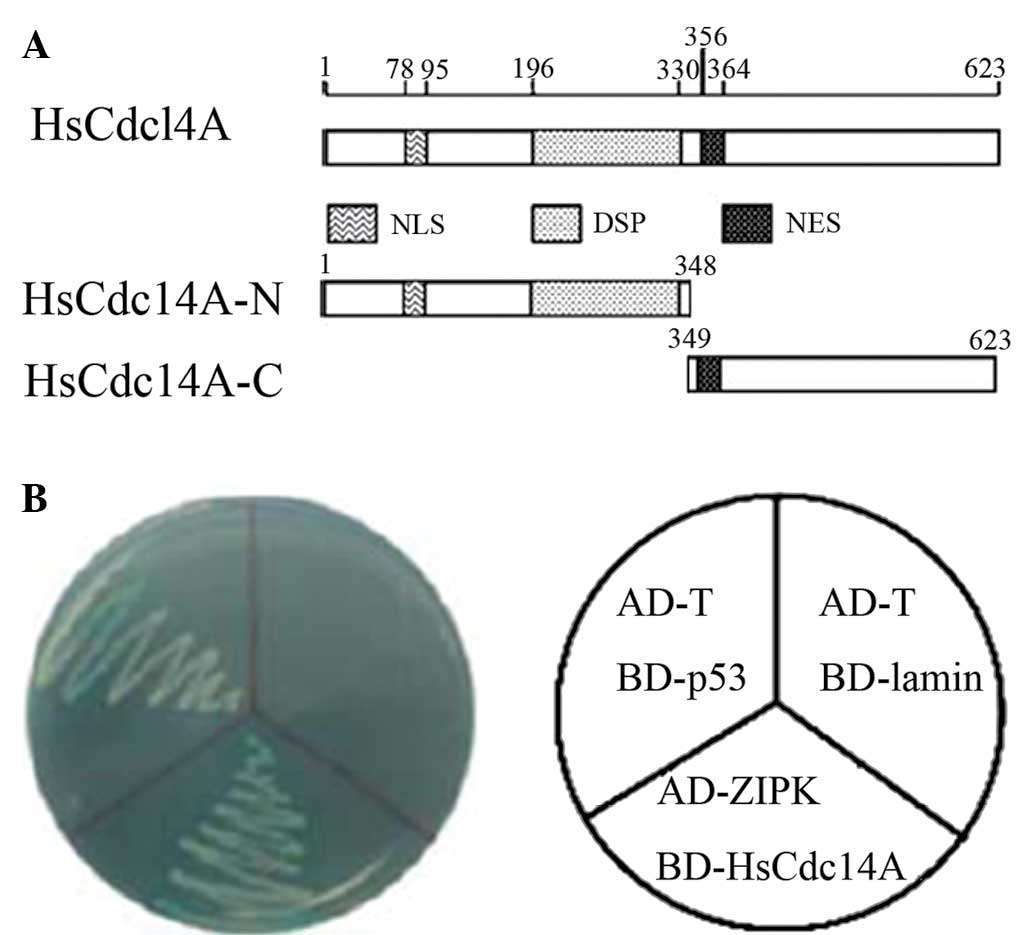 | Figure 1Yeast two-hybrid screening identified
HsCdc14A as a ZIPK-binding protein. Schematic diagrams of (A)
HsCdc14A protein structure, containing the DSP, NLS and NES
domains; and (B) the interaction of ZIPK with HsCdc14A in AH109
yeast cells. BD-p53 and AD-T were co-transformed as the positive
controls, whereas BD-lamin and AD-T were the negative controls.
HsCdc14A, human cell division cycle 14A; ZIPK, zipper-interacting
protein kinase; HsCdc14A-N, N-terminus; HsCdc14A-C, C-terminus;
DPS, dual-specificity phosphatase; NLS, nuclear localization
signal; NES, nuclear export signal. |
ZIPK and HsCdc14A interactions
Subsequent experiments were conducted to verify the
interaction between ZIPK and HsCdc14A. In a GST pull-down assay,
GST-ZIPK was found to pull down HsCdc14A, as compared with GST
alone (Fig. 2A). Various deletion
mutations of HsCdc14A were constructed to map the interaction
fragments of the two proteins. Two deletions of HsCdc14A were
constructed (Fig. 1A). The
N-terminus (aa 1-348) consisted of the nuclear localization signal
(NLS) and dual-specificity phosphatase (DSP) domains, while the
C-terminus (aa 349-623) contained the nuclear export signal (NES)
domain. The results of the subsequent GST pull-down experiment
indicated that ZIPK interacted directly with the N-terminus of
HsCdc14A, but not with the C-terminus of HsCdc14A (Fig. 2A). These results were consistent
with the important function of the DSP domain of HsCdc14A.
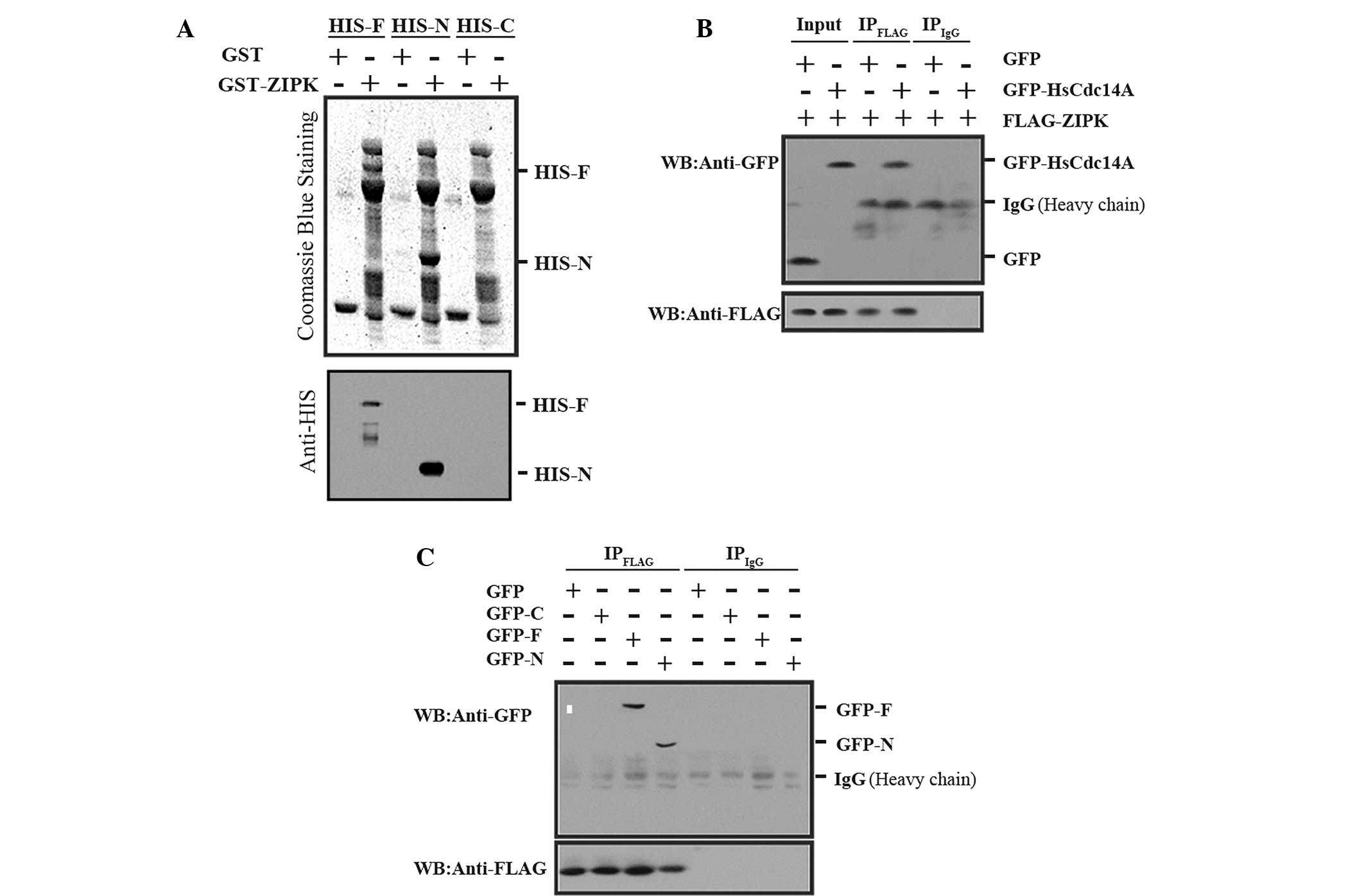 | Figure 2ZIPK interaction with HsCdc14A
N-terminus (aa 1-348). (A) His-HsCdc14A and its N terminus were
pulled down by GST-ZIPK. Three different deletions of HsCdc14A
(His-F, aa 1-623; His-N, aa 1-348; or His-C, aa 349-623) were
separately incubated with GST-ZIPK, followed by GST antibody, and
were separated with SDS-PAGE. The gels were then stained with
Coomassie Blue (upper panel) or analyzed by western blot analysis
using anti-HIS (lower panel). The full length and N-terminus
HsCdc14A bands interacted with ZIPK, whereas the HsCdc14A
C-terminus did not interact with ZIPK (no band). (B) Full length
HsCdc14A was co-immunoprecipitated with ZIPK. The cell lysate
(input) and immunoprecipitate fractions (IPFLAG and
IPIgG, respectively), obtained from the cell lysates,
were immunoprecipitated with anti-FLAG antibodies or non-specific
IgG and subjected to an immunoblotting assay with anti-GFP (upper
panel) or anti-FLAG (lower panel). (C) HsCdc14A N-terminus was
co-immunoprecipitated with ZIPK. The cell lysates (GFP-F, aa 1-623;
GFP-N, aa 1-348; or GFP-C, aa 349-623; co-transfected with
FLAG-tagged ZIPK) were immunoprecipitated with anti-FLAG or
nonspecific IgG, and the immunoprecipitates obtained were analyzed
by western blot analysis using anti-GFP (upper panel) or anti-FLAG
(lower panel). HsCdc14A, human cell division cycle 14A; ZIPK,
zipper-interacting protein kinase; GST, glutathione S-transferase;
-F, full length HsCdc14A; -N, N-terminus; -C, C-terminus; aa, amino
acid; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel
electrophoresis; IgG, immunoglobulin G; GFP, green fluorescent
protein. |
In order to verify the interaction between ZIPK and
HsCdc14A in mammalian cells, HEK 293T cells were transfected with
GFP-tagged HsCdc14A or a control plasmid, along with FLAG-tagged
ZIPK. The cell lysates were immunoprecipitated with anti-FLAG
antibodies. Subsequently, the obtained immunoprecipitates were
analyzed by western blotting using an anti-GFP antibody, in order
to detect the presence of bound HsCdc14A. FLAG-tagged ZIPK was
found to co-precipitate with GFP-tagged HsCdc14A, but not with GFP
alone (Fig. 2B).
The regions responsible for the interaction between
ZIPK and HsCdc14A were also determined. A series of GFP-tagged
HsCdc14A (full length; GFP-HsCdc14A N-terminus or C-terminus) were
co-transfected into HEK 293T cells with FLAG-tagged ZIPK.
Co-immunoprecipitation experiments indicated that ZIPK interacted
strongly with the HsCdc14A N-terminus, containing the NLS and DSP
domains (Fig. 2C).
Co-transfected ZIPK and HsCdc14A plasmids
interfere with mitotic progression and cause apoptosis
Due to the identification of the physical
interaction between ZIPK and HsCdc14A, the present study aimed to
determine the functional relevance of the ZIPK-HsCdc14A interaction
in mitotic progression. HEK 293T cells were separately transfected
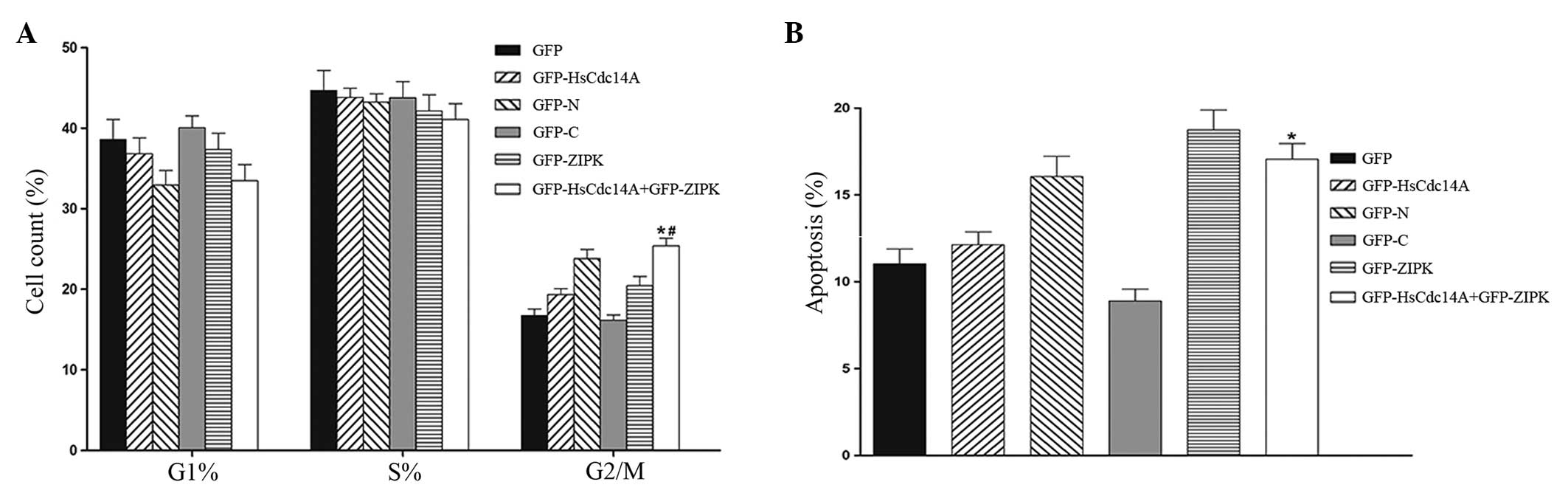
with various plasmids. As shown in Fig. 3A, the cell cycle assay demonstrated
that the G2/M phase cell population was significantly
increased in cells that were co-transfected with ZIPK and HsCdc14A
plasmids, when compared with the cells that were transfected with
HsCdc14A alone. These results were similar to those obtained
following transfection with the HsCdc14A N terminus (the
phosphatase domain and its activity may be inhibited by the
C-terminus). The results indicated that the mitotic progression was
arrested in cells co-transfected with ZIPK and HsCdc14A. In
addition, the percentage of apoptotic cells following transfection
with the various plasmids was determined. A considerable increase
was observed in the number of Annexin V-positive cells upon
co-transfection with ZIPK and HsCdc14A, compared with the cells
transfected with HsCdc14A alone (Fig.
3B). The percentage of apoptotic cells was similar to the cells
transfected with the HsCdc14A N-terminus. These results indicated
that apoptosis was increased in the cells co-transfected with ZIPK
and HsCdc14A. Therefore, the control of HsCdc14A phosphatase
activity by ZIPK is hypothesized to play an important role in
mitotic progression and cell apoptosis.
Discussion
ZIPK was initially identified as a Ser/Thr kinase
that binds activating transcription factor 4 (ATF4). ATF4 is a
member of the activating transcription factor and cyclic adenosine
monophosphate-responsive element-binding protein family of
transcription factors (1). ZIPK
aggregates through its C-terminal leucine zipper structure, thereby
becoming an active enzyme. A previous study identified that the
ectopic expression of ZIPK in NIH-3T3 murine fibroblast cells
induced apoptosis (1). By
contrast, the kinase-inactive ZIPK K42A mutant was not found to
induce apoptosis, indicating that the catalytic activity of ZIPK
stimulates cell apoptosis (1).
Numerous kinases that mediate cell growth are known; however, only
a few protein kinases associated with apoptosis have been
identified, besides ZIPK. Previous studies demonstrated that ZIPK
participates in the regulation and possibly the coordination of
mitosis and cytokinesis, by interacting with the proapoptotic
protein, Par-4, and the CDC5 protein (4,16).
In addition, ZIPK is a centromere-specific histone kinase that may
play a role in the labeling of centromere-specific chromatin for
subsequent mitotic processes (17). Furthermore, previous studies have
indicated that ZIPK, as a regulator of myosin phosphatase, may play
a pivotal role in the regulation of cell motility, reorganization
of actin filaments and control of smooth muscle contraction in
smooth muscle cells (4,6,7).
However, only a few interacting partners of ZIPK have been
identified and the regulatory networks of ZIPK remain to be
elucidated.
The present study demonstrated that ZIPK interacted
with the HsCdc14A protein in vitro, playing an important
role in the regulation of the cell cycle. The interaction was shown
to involve the highly conserved N-terminus of HsCdc14A, indicating
that ZIPK may be involved in the cell cycle regulation.
Cdc14 is a protein phosphatase conserved between
yeast and humans (10). Genetic
analyses have suggested that the yeast Cdc14 plays a pleiotropic
role during the cell cycle, regulating DNA replication and the exit
from mitosis by dephosphorylating cyclin-dependent kinase (Cdk)
targets (18–20). Mammalian cells express two
functional homologs of the yeast Cdc14, known as HsCdc14A and
HsCdc14B (21). These two proteins
remain poorly understood; however, recent evidence has indicated
that they play an isoform-specific role in centrosome
separation/maturation and spindle stability, with the possibility
of additional role in the mitotic exit and cytokinesis (21). The majority of previous studies
have investigated HsCdc14A, which was demonstrated to interact with
interphase centrosomes and regulate the centrosome duplication
cycle (22,23). In addition, HsCdc14A is located at
the central spindle during anaphase, where it appears to be
involved in the spatial regulation of the Aurora B kinase, a key
regulator of chromosome segregation and cytokinesis (24). HsCdc14A may also regulate p53 and
Cdk1/cyclin B; thus, dysregulation of HsCdc14A may play an
important role in carcinogenesis (25). Besides p53, a previous study
revealed that HsCdc14A can dephosphorylate the products of Cdk,
including hCdh1 and cyclin E (26). However, the mechanism through which
the HsCdc14A function is integrated in mitotic regulation, the
substrates involved in chromosome segregation and whether HsCdc14A
plays an active role in mitosis remain unclear.
HsCdc14A contains an NLS motif, a DSP domain and an
NES motif. Furthermore, HsCdc14 exhibits an inhibitory
self-association, in which the C-terminal domain binds and inhibits
the phosphatase domain located at the N-terminus (27). Once the inhibition of C-terminus is
released, the activity of the phosphatase domain, located at the
N-terminus, is increased. The present study demonstrated that ZIPK
interacted with the N-terminus of HsCdc14A, which contains the NLS
and DSP domains. In addition, the effect of co-transfection of HEK
293T cells with ZIPK and HsCdc14A was similar to the effect of the
cells transfected with the HsCdc14A N-terminus; therefore,
ZIPK-mediated phosphorylation may activate the phosphatase activity
of HsCdc14A. Furthermore, the results of the present study
indicated that ZIPK may affect the cell cycle by interacting with
the N-terminus of HsCdc14A. Notably, the effect of ZIPK and
HsCdc14A overexpression on cell apoptosis was similar to the effect
of ZIPK alone, but more evident than the effect of HsCdc14A alone.
Apoptosis was not affected significantly more by the increased
phosphatase activity of HsCdc14A along with ZIPK compared with
cells transfected with HsCdc14A alone. Therefore, these two
proteins may be associated with the same apoptotic pathway. Future
experiments are required to investigate this hypothesis.
In conclusion, the results of the present study
demonstrated that ZIPK may interact with the HsCdc14A protein.
These findings indicated that ZIPK may also be involved in the
regulation of the cell cycle. Further research regarding the
regulation of HsCdc14A by ZIPK is required to provide valuable
insight on the effect of ZIPK on the cell cycle. ZIPK may be a
potential target candidate for the treatment of diseases associated
with cell proliferation.
Acknowledgements
This study was supported by grants from the National
Nature Science Foundation for Young Scientists of China (no.
30900541) and the 985 Project (no. 985III-YFX0302).
References
|
1
|
Kawai T, Matsumoto M, Takeda K, Sanjo H
and Akira S: Zip kinase, a novel serine/threonine kinase which
mediates apoptosis. Mol Cell Biol. 18:1642–1651. 1998.PubMed/NCBI
|
|
2
|
Kogel D, Plottner O, Landsberg G,
Christian S and Scheidtmann KH: Cloning and characterization of
DLK, a novel serine/threonine kinase that is tightly associated
with chromatin and phosphorylates core histones. Oncogene.
17:2645–2654. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Komatsu S and Ikebe M: ZIP kinase is
responsible for the phosphorylation of myosin II and necessary for
cell motility in mammalian fibroblasts. J Cell Biol. 165:243–254.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Engemann H, Heinzel V, Page G, Preuss U
and Scheidtmann KH: DAP-like kinase interacts with the rat homolog
of schizosaccharomyces pombe CDC5 protein, a factor involved in
pre-mRNA splicing and required for G2/M phase transition. Nucleic
Acids Res. 30:1408–1417. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
MacDonald JA, Borman MA, Muranyi A, Somlyo
AV, Hartshorne DJ and Haystead TA: Identification of the endogenous
smooth muscle myosin phosphatase-associated kinase. Proc Natl Acad
Sci USA. 98:2419–2424. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Niiro N and Ikebe M: Zipper-interacting
protein kinase induces Ca(2+)-free smooth muscle contraction via
myosin light chain phosphorylation. J Biol Chem. 276:29567–29574.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sato N, Kawai T, Sugiyama K, et al:
Physical and functional interactions between STAT3 and ZIP kinase.
Int Immunol. 17:1543–1552. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vetterkind S, Illenberger S, Kubicek J, et
al: Binding of par-4 to the actin cytoskeleton is essential for
Par-4/Dlk-mediated apoptosis. Exp Cell Res. 305:392–408. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vetterkind S and Morgan KG: The
pro-apoptotic protein Par-4 facilitates vascular contractility by
cytoskeletal targeting of ZIPK. J Cell Mol Med. 13:887–895. 2009.
View Article : Google Scholar :
|
|
10
|
Li L, Ernsting BR, Wishart MJ, Lohse DL
and Dixon JE: A family of putative tumor suppressors is
structurally and functionally conserved in humans and yeast. J Biol
Chem. 272:29403–29406. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lou Y, Yao J, Zereshki A, et al: NEK2A
interacts with MAD1 and possibly functions as a novel integrator of
the spindle checkpoint signaling. J Biol Chem. 279:20049–20057.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen JS, Hu HY, Zhang S, et al: Brap2
facilitates HsCdc14A Lys-63 linked ubiquitin modification.
Biotechnol Lett. 31:615–621. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gietz RD and Schiestl RH: Applications of
high efficiency lithium acetate transformation of intact yeast
cells using single-stranded nucleic acids as carrier. Yeast.
7:253–63. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng Q, Huang ZH, Tang FX, Huang ZZ and
Che XY: Highly efficient construction of recombinant adenovirus
containing double suicide gene driven by cytomegalovirus promoter
using two-step CaCl2 transformation method. Di Yi Jun Yi Da Xue Xue
Bao. 6:575–577. 2003.(In Chinese).
|
|
15
|
Zhou W, Wang X, Li L, et al: Depletion of
tubulin polymerization promoting protein family member 3 suppresses
HeLa cell proliferation. Mol Cell Biochem. 333:91–98. 2010.
View Article : Google Scholar
|
|
16
|
Preuss U, Bierbaum H, Buchenau P and
Scheidtmann KH: DAP-like kinase, a member of the death-associated
protein kinase family, associates with centrosomes, centromers, and
the contractile ring during mitosis. Eur J Cell Biol. 82:447–459.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Preuss U, Landsberg G and Scheidtmann KH:
Novel mitosis-specific phosphorylation of histone H3 at Thr11
mediated by DlK/ZIP kinase. Nucleic Acids Res. 31:878–885. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hardy CF: Characterization of an essential
Orc2p-associated factor that plays a role in DNA replication. Mol
Cell Biol. 16:1832–1841. 1996.PubMed/NCBI
|
|
19
|
Jaspersen SL, Charles JF, Tinker-Kulberg
RL and Morgan DO: A late mitotic regulatory network controlling
cyclin destruction in saccharomyces cerevisiae. Mol Biol Cell.
9:2803–2817. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stegmeier F and Amon A: Closing mitosis:
the functions of the Cdc14 phosphatase and its regulation. Annu Rev
Genet. 38:203–232. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vazquez-Novelle MD, Esteban V, Bueno A and
Sacristan MP: Functional homology among human and fission yeast
Cdc14 phosphatases. J Biol Chem. 280:29144–29150. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mailand N, Lukas C, Kaiser BK, Jackson PK,
Bartek J and Lukas J: Deregulated human Cdc14A phosphatase disrupts
centrosome separation and chromosome segregation. Nat Cell Biol.
4:317–322. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kaiser BK, Zimmerman ZA, Charbonneau H and
Jackson PK: Disruption of centrosome structure, chromosome
segregation, and cytokinesis by misexpression of human Cdc14A
phosphatase. Mol Biol Cell. 13:2289–2300. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gruneberg U, Neef R, Honda R, Nigg EA and
Barr FA: Relocation of Aurora B from centromeres to the central
spindle at the metaphase to anaphase transition requires MKlp2. J
Cell Biol. 166:167–172. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Paulsen MT, Starks AM, Derheimer FA, et
al: The p53-targeting human phosphatase hCdc14A interacts with the
Cdk1/cyclin B complex and is differentially expressed in human
cancers. Mol Cancer. 5:252006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bembenek J and Yu H: Regulation of the
anaphase-promoting complex by the dual specificity phosphatase
human Cdc14A. J Biol Chem. 276:48237–48242. 2001.PubMed/NCBI
|
|
27
|
Yuan K, Hu H, Guo Z, et al:
Phospho-regulation of HsCdc14A by polo-like kinase 1 is essential
for mitotic progression. J Biol Chem. 282:27414–27423. 2007.
View Article : Google Scholar : PubMed/NCBI
|