Introduction
The cluster of differentiation (CD)95 antigen, also
termed apoptosis antigen (APO)-1 or Fas, is a cell surface protein
with a molecular weight of 200,000 Da, which differs to the
molecular weight of the tumor necrosis factor (TNF) receptor
(1). Anti-CD95 induces cell-death
and its activity is indistinguishable from the cytolytic activity
of TNF (1). A previous study
observed that anti-CD95 induces apoptosis in vivo. Nanogram
quantities of anti-CD95 completely inhibited the proliferation of
cells containing APO-1 in vitro, characteristic of the
process of programmed cell death or apoptosis (2). Complementary DNA (cDNA) encoding the
Fas cell surface antigen were isolated from a cDNA library of human
T-cell lymphoma KT-3 cells by Itoh et al (3) The nucleotide sequence of the cDNA
revealed that the molecule coding for the Fas antigen determinant
is a 319 amino acid polypeptide with a single transmembrane domain.
The extracellular domain is rich in cysteine residues and is
similar to that of human TNF receptors, the human nerve growth
factor receptor and the CD40 human B cell antigen (3,4).
Apoptosis is the predominant form of eukaryotic cell
death and occurs during tissue replacement, organ development,
metamorphosis, tissue atrophy and tumor regression (4–6).
Apoptosis is induced by a diverse range of agents, including
glucocorticoids, cytostatic drugs, cytolytic cytokines, including
TNF and lymph toxin, and in target cells of various killer cells,
including cytotoxic T lymphocytes (4–6). The
most prominent morphological features of apoptosis are chromatin
condensation and membrane blebbing (4–6). In
cells undergoing apoptosis, an endonuclease is induced, which
cleaves the genomic DNA into polynucleosomal fragments and is
observed on agarose gels as a ‘DNA ladder’ (4). Apoptosis and the Fas system are
important in the process of converting liver cirrhosis into
hepatocellular carcinoma. Downregulation in the expression of Fas
and upregulation in the expression of Fas ligand in hepatocytes,
and elevation of serum levels of Fas are important in tumor evasion
from immune surveillance and in hepatic carcinogenesis (5,6). The
present study investigated the relative expression of CD95 in liver
cancer cells to determine whether there is a link between CD95 and
liver cancer.
Materials and methods
Reagents
The rabbit polyclonal immunoglobulin G (IgG)
anti-CD95 antibody (N-18; cat. no. sc-714), mouse monoclonal
IgG1 anti-caspase-8 antibody (1.1.40; cat. no.
sc-81656), rabbit polyclonal IgG anti-caspase-3 antibody (H-277;
cat. no. sc-7148), goat polyclonal IgG anti-poly(ADP-ribose)
polymerase 1 (PARP1) antibody (A-20; cat. no. sc-1562) and mouse
monoclonal IgG1 anti-β-actin antibody (C4; cat. no.
sc-47778) were purchased from Santa Cruz Biotechnology, Inc. (Santa
Cruz, CA, USA). The radioimmunoprecipitation buffer and enhanced
bicinchoninic acid assay kit were purchased from Beyotime Institute
of Biotechnology (Jiangsu, China). Polyvinylidene difluoride (PVDF)
membranes were purchased from EMD Millipore (Billerica, MA, USA).
The LipoFiter™ Liposomal Transfection reagent was purchased from
HanBio (Shanghai, China). The SP test kit and diaminobenzidine
(DAB) colorization test kit were purchased from Beijing Zhongshan
Golden Bridge Biotechnology, Co., Ltd. (Beijing, China). The
AnnexinV-fluorescein isothiocyanate (FITC)/propidium iodide (PI)
apoptosis detection kit was purchased from KeyGen Biotech, Co.,
Ltd. (Nanjing, China).
Human liver cancer tissues
Frozen tumor samples and the corresponding normal
liver tissues were obtained from 40 patients with gastric cancer,
between 2006 and 2012 at Guangxi Medical University (Nanning,
Guangxi, China) and paraffinembedded samples were obtained from 66
patients with gastric cancer, between 2006 and 2012 at Guangxi
Medical University, Guilin Medical University, Guilin, Guangxi,
China and Jianghan University, Wuhan, Hubei, China. Written
informed consent was obtained from all patients. These samples were
used following approval of the Institutional Review Board. The
study was approved by the ethics committee of the School of
Medicine, Jianghan University (Wuhan, China).
Cell lines
The human HepG2 liver cancer cell line and the
SGC7901 cell line were obtained from the Cell Center of Basic
Medicine, Chinese Academy of Medical Sciences (Beijing, China).
CD95 expression plasmid construction
The CD95 expression plasmid was donated by Dr
JunFeng Zhang (Institute of Genetics and Developmental Biology,
Chinese Academy of Sciences, Beijing, China). The total RNA was
extracted from the SGC7901 cell line using TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) and the first
strand cDNA was synthesized by reverse transcription using a
ReverAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific,
Waltham, MA, USA) at 42°C, according to the manufacturer’s
instructions. The entire CD95 coding region was amplified using the
following primers: CD95, forward 5′-cactcgagCTTTCACTTCGGAGGATTGC-3′
(the added XhoI site is lowercase) and reverse
5′-gtgaattctGACCAAGCTTTGGATTTCATTTC-3′ (the added EcoRI site
is underlined). The primers were designed using Primer 5.0 (Primer
Biosoft International, Palo Alto, CA, USA) and were synthesized by
the Sangon Biotech Co., Ltd. (Shanghai, China). Polymerase chain
reactions were performed using a Ready-To-Use PCR kit (Tag DNA
polymerase; no. SK2082, Sangon Biotech Co., Ltd) as follows: 94°C
for 4 min, 30 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C
for 70 sec with a final extension at 72°C for 10 min. The resulting
fragment was digested using XhoI and EcoRI (Sangon
Biotech Co., Ltd) and then subcloned into the pEgreen fluorescent
protein (GFP)-N1, resulting in the N-terminal fusion to GFP. The
resulting construct, pCMV IE: CD95-enhanced GFP (CD95-vector), was
sequenced to confirm the in-frame fusion of CD95 and GFP and was
used for transient expression in the HepG2 cells.
Cell culture
All the cells were maintained in RPMI-1640 medium
containing 10 % fetal bovine serum (FBS; Invitrogen Life
Technologies) at 37°C in a 5 % CO2 atmosphere.
Cell transfection
All the transfections were performed using
LipoFiter™ Liposomal Transfection reagent, according to the
manufacturer’s instructions. The HepG2 cells were plated
(6×106 cells) in 100 mm dishes and incubated overnight
prior to replacing with fresh RPMI-1640 medium supplemented with
10% FBS. The cells were transfected with 4.6 μg of either
CD95-vector or vector-control using 4.8 μl LipoFiter™ Liposomal
Transfection reagent. The media was replaced 6 h after transfection
and the cells were incubated at 37°C for 48 h. pEGFP-N1 was used as
the vector-control. These transfected cells were used for
subsequent experiments.
Immunohistochemistry
Immunohistochemical staining was performed on 5
μm-thick tumor sections using a ‘two-step’ method. The tissue
slides were de-paraffinized with 100% xylene for 10 min and
rehydrated gradually in an alcohol series. The endogenous
peroxidase activity was inhibited by incubation in a 3% hydrogen
peroxide/methanol buffer for 10 min. Antigen retrieval was
performed by immersing the slides in 0.5 mol/l ethylenediamine
tetraacetic acid buffer (pH 8.0) for 10 min, followed by boiling in
a waterbath for 25 min. The slides were rinsed in
phosphate-buffered saline (PBS) and subsequently incubated with
polyclonal anti-CD95 antibody (1:200) overnight at 4°C in a
humidified chamber. Following incubation, the slides were washed
three times with PBS containing 0.05% Tween-20 for 2 min each time.
The slides were then incubated with 100 μl horseradish peroxidase
polymer-anti-Mouse/Rabbit immunoglobulin G covering the tissue
section in a moist chamber for 15 min. The slides were then washed
three times, as previously, and the tissue was incubated for 5 min
with 100 μl DAB chromogen. Following development of the appropriate
color, the slides were washed gently under tap water for ~1–2 min,
prior to counterstaining with 100 μl Mayer’s hematoxylin (Sangon
Biotech Co., Ltd) covering the tissue completely for ~20 sec. The
slides were rinsed thoroughly with tap water for ~1–2 min and
subsequently incubated in PBS for 20 sec until the color turned
blue. The slides were then rinsed with distilled water, followed by
tap water.
The levels of CD95 staining were scored as follows:
0, no staining or staining observed in <10% tumor cells; 1+,
faint/barely perceptible staining detected in ≥10% tumor cells;
2+/3+, moderate/strong staining, respectively, observed in ≥10%
tumor cells. A score of 0/1+ was considered negative and a score of
2+/3+ was considered positive. The immunostained slides were
evaluated independently by two pathologists in a blinded-manner. In
the majority of cases, the results of the evaluation the two
pathologists were identical; discrepancies were resolved by
re-examination and consensus.
Western blot analysis
For western blot analysis, the cells were washed
with cold PBS and lysed in a lysis buffer containing 50 mM Tris-HCl
(pH 8.0), 150 mM NaCl, 0.25 mM EDTA (pH 8.0), 0.1% SDS, 1% Triton
X-100 and 50 mM NaF, supplemented with MS-SAFE™ Protease and
Phosphatase Inhibitor Cocktail (1:100; Sigma-Aldrich, St. Louis,
MO, USA) and phosphatase inhibitors (Sigma-Aldrich). The protein
concentrations were determined using an Enhanced Bicinchoninic Acid
Protein Assay kit (Beyotime Institute of Biotechnology). The cell
lysates were mixed with loading buffer (Beyotime Institute of
Biotechnology), separated using 12% SDS-PAGE gels and transferred
onto a PVDF membrane (EMD Millipore, Billerica, MA, USA). The
membranes were subsequently probed with various primary antibodies,
appropriate secondary antibodies [goat anti-mouse IgG-horseradish
peroxidase (HRP; sc-2005), goat anti-rabbit IgG-HRP (sc-2004) and
donkey anti-goat IgG-HRP (sc-2020)] and visualized using enhanced
chemiluminescence detection reagents (DNR Bio-Imaging Systems,
Ltd., Jerusalem, Israel). The density of the protein bands were
assessed using Totallab analysis software, version 2.01 (Nonlinear
USA, Inc., Durham, NC, USA).
Apoptotic assay
The cells were stained using an Annexin V-FITC
apoptosis detection kit (Nanjing KeyGen Biotech, Co., Ltd.,
Nanjing, China) according to the manufacturer’s instructions, to
detect early apoptotic cells (Annexin V+PI- events) and necrotic or
late apoptotic cells (Annexin V+PI+ events) by flow cytometry.
Briefly, the HepG2 cells were transfected with either the
vector-CD95 or vector-control for 48 h. The cells were then
collected and resuspended in the culture medium at a density of
1×106 cells/ml, stained with 5 μL Annexin V-FITC and 5
μL PI in 300 μL binding buffer containing 10 mM HEPES, (pH 7.4),
140 mM NaOH and 2.5 mM CaCl2 according to the
manufacturer’s instructions for 15 min at room temperature in the
dark. Quantification of the apoptotic cells was assessed using a
FACScan flow cytometer (Beckman Coulter, Brea, CA, USA).
Statistical analysis
Statistical analysis was performed using SPSS 12.0
software (SPSS, Inc., Chicago, IL, USA). The data are expressed as
the mean ± standard deviation of three replicates and were compared
using Student’s t-test and analysis of variance. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were performed at least three times to ensure
reproducibility of the results.
Results
Association between the expression of
CD95 and clinicopathological features in liver cancer
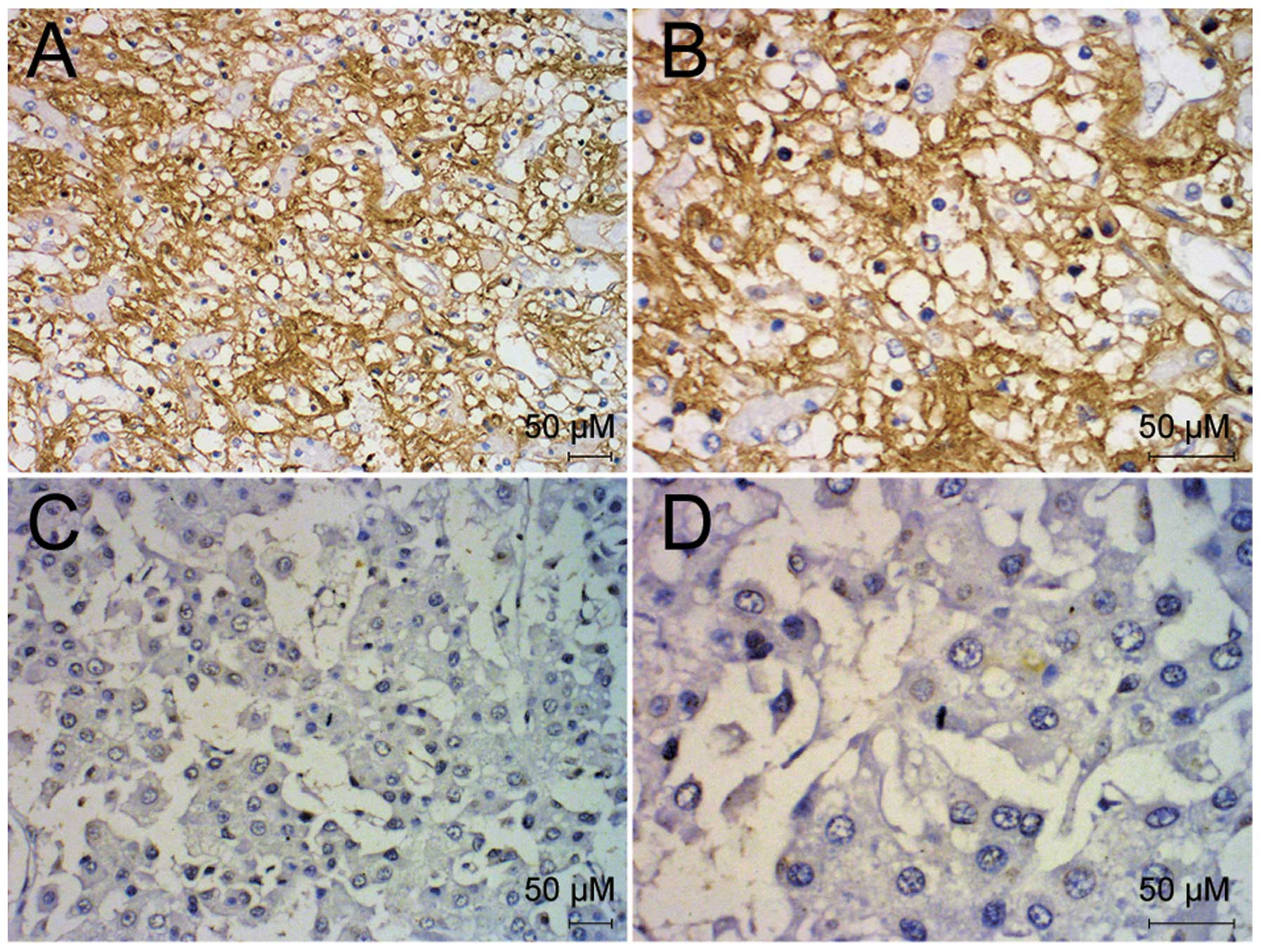
The expression of CD95 was examined in the 66 liver
cancer samples using immunohistochemistry. The expression of CD95
was detected predominantly in the cytoplasm of the liver cancer
cells and at the plasma membrane, however, no expression was
detected in the nuclei (Fig. 1).
Positive expression of CD95 was detected in 17 of the liver cancer
samples. The expression of CD95 correlated with histological
differentiation, liver cirrhosis, lymph node metastasis and distant
metastasis (P<0.05), however, no correlations with gender, age,
quantity of tumor nodules or T stage were observed (P>0.05;
Table I).
 | Table IAssociation between the expression of
CD95 and the clinicopathological features of liver cancer. |
Table I
Association between the expression of
CD95 and the clinicopathological features of liver cancer.
| Expression of
CD95 | |
|---|
|
| |
|---|
| Factor | Positive (n) | Negative (n) | P-value |
|---|
| Gender |
| Male | 10 | 31 | 0.74 |
| Female | 7 | 18 | |
| Age (years) |
| ≤50 | 5 | 16 | 0.80 |
| >50 | 12 | 33 | |
| Histological
differentiation |
| High | 12 | 13 | |
| Medial | 2 | 8 | 0.002 |
| Low | 3 | 28 | |
| Tumor nodules |
| Single | 6 | 20 | 0.68 |
| Multiple | 11 | 29 | |
| Liver cirrhosis |
| Positive | 8 | 37 | 0.029 |
| Negative | 9 | 12 | |
| T stage |
| T1 | 5 | 7 | |
| T2 | 6 | 11 | 0.064 |
| T3 | 4 | 21 | |
| T4 | 2 | 10 | |
| N stage |
| N0 | 13 | 19 | 0.016 |
| N1–3 | 4 | 30 | |
| M stage |
| M0 | 16 | 32 | 0.047 |
| M1 | 1 | 17 | |
Expression of CD95 is associated with the
expression levels of caspase-8, caspase-3 and PARP1
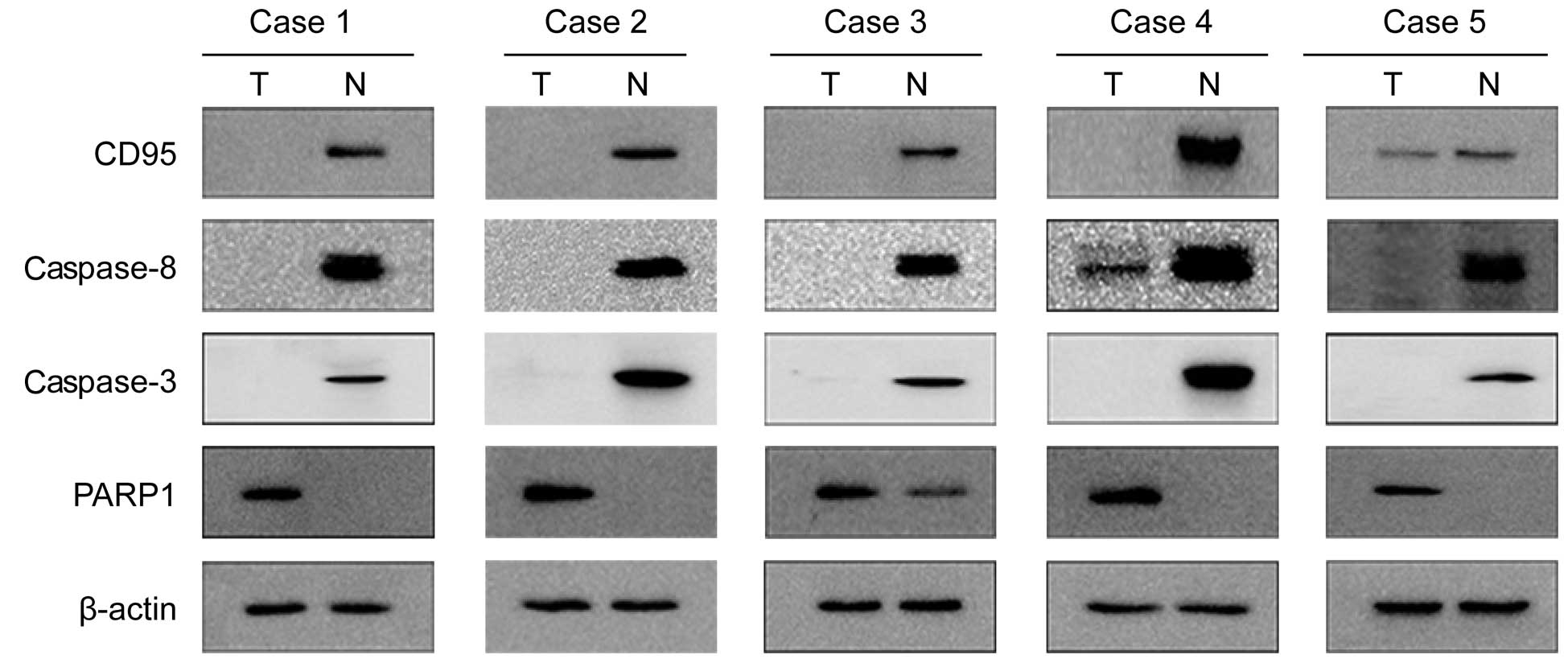
The protein expression levels of CD95, caspase-8,
caspase-3 and PARP1 in the tumor tissues and the corresponding
normal tissues of 40 liver cancer samples were assessed by western
blot analysis. The positive expression of CD95, caspase-8 and
caspase-3 was detected in 14 (35%), 14 (35%) and 13 (32.5%) of the
40 tumor specimens, respectively. These levels were lower compared
with those in the normal tissues, in which the positive expression
of CD95, caspase-8 and caspase-3 was observed in 32 (80%), 33 (75%)
and 36 (90%) specimens, respectively (Fig. 2). The expression of PARP1 was
detected in 28 (70%) tumor specimens, compared with 16 (40 %) in
the normal liver tissue specimens (Fig. 2). The expression of CD95 correlated
with the expression of caspase-8, caspase-3 and PARP1 in the tumor
specimens (Table II,
P<0.01).
 | Table IIExpression of CD95 is associated with
the expression of caspase-8, caspase-3 and PARP1. |
Table II
Expression of CD95 is associated with
the expression of caspase-8, caspase-3 and PARP1.
| Expression of
CD95 | |
|---|
|
| |
|---|
| Factor | Positive (n) | Negative (n) | κ-value/P-value |
|---|
| Caspase-8 |
| Positive | 12 | 2 | κ=0.78 |
| Negative | 2 | 24 | P<0.01 |
| Caspase-3 |
| Positive | 10 | 3 | κ=0.609 |
| Negative | 4 | 23 | P<0.01 |
| PARP1 |
| Positive | 3 | 25 | P<0.01 |
| Negative | 11 | 1 | |
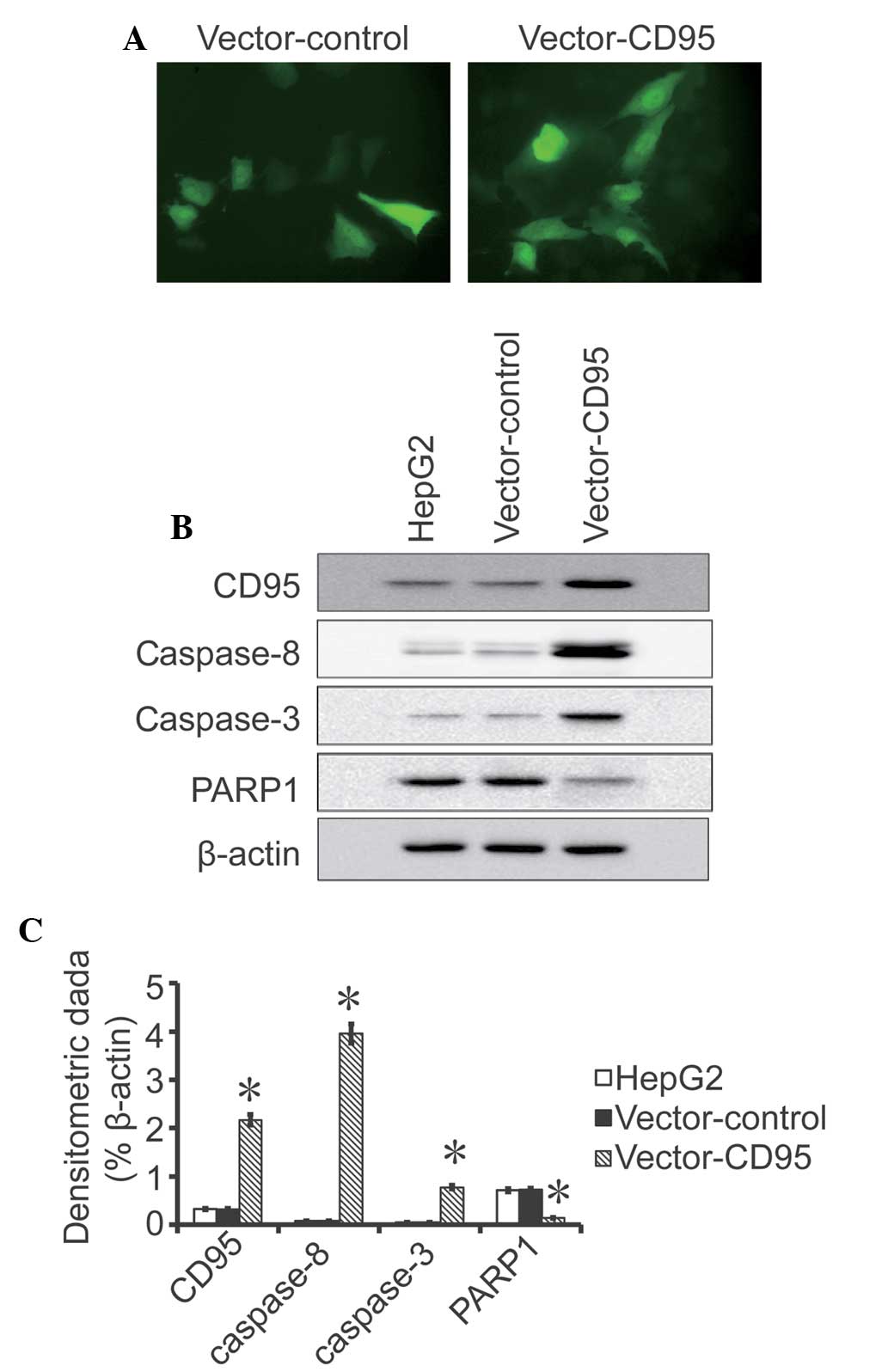
Cell transfection
A positive GFP signal was detected in the High95
cells and in the control cells 24 h after transfection, which
indicated that the transfection was successful (Fig. 3). Western blot analysis revealed
that the expression of CD95 was higher in the High95 cells compared
with the HepG2 cells and the control cells (P<0.05; Fig. 3). No difference was observed
between the HepG2 cells and the control cells.
Expression levels of CD95, caspase-8,
caspase-3 and PARP1
Western blot analysis indicated that the expression
levels of CD95, caspase-8 and caspase-3 were higher in the High95
cells compared with the HepG2 cells and control cells (P<0.05;
Fig. 4). The western blot analysis
results also revealed that the expression of PARP1 was lower in the
High95 cells compared with the HepG2 cells and control
cells(P<0.05; Fig. 4). No
difference was observed in the expression levels of CD95,
caspase-8, caspase-3 and PARP between the HepG2 cells and control
cells.
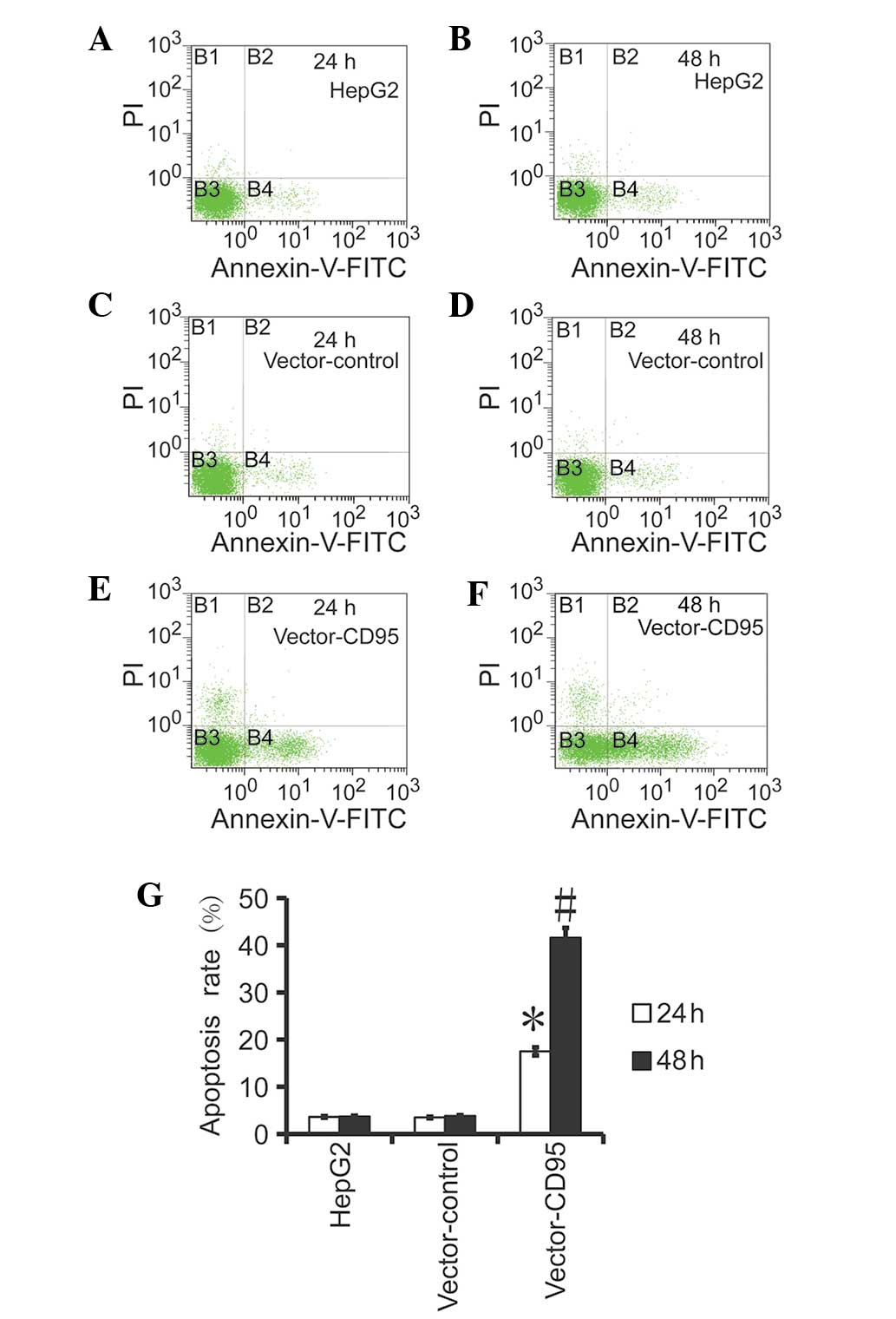
CD95 promotes apoptosis in the HepG2
cells
The results of the flow cytometry revealed that the
level of apoptosis in the High95 cells was higher compared with the
HepG2 cells and the control cells (Fig. 5). Furthermore, the
apoptotic rate was higher at 48 h compared with 24 h, which
indicated that CD95 mediated apoptosis in the HepG2 cells.
Discussion
CD95 is widely expressed in normal and diseased
tissues and has been implicated in tumor progression in several
types of cancer (5). Reduced
expression levels of CD95 have been observed in a number of tumor
types(7–12). The present study demonstrated that
the expression of CD95 was lower in liver cancer tissues compared
with normal liver tissues and correlated with histological
differentiation, liver cirrhosis, lymph node metastasis and distant
metastasis (P<0.05). Conversely, no correlations were observed
with gender, age, quantity of tumor nodules or T stage, indicating
that the expression of CD95 was associated with liver cancer.
In the present study, western blot analysis revealed
that the expression of CD95 correlated with the expression levels
of caspase-8, caspase-3 and PARP1. Caspase-8 is important in the
CD95-mediated activation of the mitogen-activated protein kinases
(MAPKs). A mechanism has been proposed, in which the catalytic
activity and substrate specificity of caspase-8 are determined by
the conformation and cleavage status of procaspase-8 (13). Procaspase-8 processing is required
for the CD95-induced activation of MAPKs and conditions impairing
MAPK activation are accompanied by reduced procaspase-8 processing
(14). The activation of
procaspase-8 is hypothesized to occur through an ‘induced
proximity’ mechanism, involving the dimerization of procaspase-8
molecules, which facilitates activation through subsequent
self-processing. This is in contrast to the executioner caspases,
caspase-3 and caspase-7, which are constitutively dimeric and
inactive due to the ‘strain’ caused by their short interdomain
linker region on the active site and only become active on
proteolytic cleavage (13). CD95
may rely exclusively on the activation of caspase-8 and the
mitochondrial activation of caspase-3, which can process more
procaspase-8 and, thus, propagate the amplification of the
apoptotic signal (15). The
present study revealed that the expression of caspase-8 was lower
in the liver cancer tissues compared with the normal liver
tissues.
Normal cells contain only a small quantity of
caspases, in the form of inactive zymogens, and activated caspases
are transformed to proteases via the catalytic activity of enzymes,
which are capable of cleaving a number of substrate proteins
resulting in apoptosis (16,17).
Caspase-3 is activated by a series of cascade reactions until DNase
is activated, which belongs to the Mg2+-dependent
endonucleases and acts as an apoptotic factor (16). As caspase-3 is an effector caspase
in apoptotic pathways, previous studies have hypothesized that a
loss in the expression of caspase-3 may be important in the
carcinogenesis of hepatocellular carcinoma (17). The present study revealed that the
expression of caspase-3 was lower in the liver cancer tissues
compared with the normal liver tissues.
PARP1 is a nuclear enzyme, which catalyzes PARP in
target proteins in response to DNA damage and is considered to be
important in DNA repair/recombination, cell death, cell
proliferation and for stabilization of the genome (18). In several types of cancer, the
expression of PARP1 is high and, therefore, inhibiting the
expression of PARP1 may improve outcomes in patients with cancer
(19–22). The present study demonstrated that
the expression of PARP1 was higher in the liver cancer tissues
compared with the normal liver tissues. In addition, the expression
of PARP1 decreased as the expression of CD95 increased. The results
demonstrated that inhibition of the expression of PARP1 may improve
outcomes in patients with liver cancer.
Acknowledgements
This study was supported by the National Natural
Science foundation of Guangxi (no. 0848014), the National Natural
Science Foundation of China (no. 30870981), the Jianghan University
Doctor Foundation (no. 1010-08110001) and the Science Foundation of
Health Office of Hubei Province (no. NX200727).
References
|
1
|
Yonehara S, Ishii A and Yonehara M: A
cell-killing monoclonal antibody (anti-Fas) to a cell surface
antigen co-downregulated with the receptor of tumor necrosis
factor. J Exp Med. 169:1747–1756. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Trauth BC, Klas C, Peters AM, et al:
Monoclonal antibody-mediated tumor regression by induction of
apoptosis. Science. 245:301–305. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Itoh N, Yonehara S, Ishii A, et al: The
polypeptide encoded by the cDNA for human cell surface antigen Fas
can mediate apoptosis. Cell. 66:233–243. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oehm A, Behrmann I, Falk W, et al:
Purification and molecular cloning of the APO-1 cell surface
antigen, a member of the tumor necrosis factor/nerve growth factor
receptor superfamily. Sequence identity with the Fas antigen. J
Biol Chem. 267:10709–10715. 1992.PubMed/NCBI
|
|
5
|
Hammam O, Mahmoud O, Zahran M, et al: The
role of fas/fas ligand system in the pathogenesis of liver
cirrhosis and hepatocellular carcinoma. Hepat Mon. 12:e61322012.
View Article : Google Scholar
|
|
6
|
El Bassiouny AE, El-Bassiouni NE, Nosseir
MM, et al: Circulating and hepatic Fas expression in HCV-induced
chronic liver disease and hepatocellular carcinoma. Medscape J Med.
10:1302008.PubMed/NCBI
|
|
7
|
Lee YB, Kyung Kim E, Park HJ, et al:
Expression of Fas and Fas ligand in primary cutaneous squamous cell
carcinoma in association with grade of tumor differentiation. Int J
Dermatol. 52:1092–1097. 2013. View Article : Google Scholar
|
|
8
|
Sjostrom-Mattson J, Von Boguslawski K,
Bengtsson NO, Mjaaland I, Salmenkivi K and Blomqvist C: The
expression of p53, bcl-2, bax, fas and fasL in the primary tumor
and lymph node metastases of breast cancer. Acta Oncol.
48:1137–1143. 2009. View Article : Google Scholar
|
|
9
|
Bebenek M, Dus D and Kozlak J: Fas
expression in primary breast cancer is related to neoplastic
infiltration of perilymphatic fat. Adv Med Sci. 53:49–53. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang D, Torres CM, Bardhan K, Zimmerman M,
McGaha TL and Liu K: Decitabine and vorinostat cooperate to
sensitize colon carcinoma cells to Fas ligand-induced apoptosis in
vitro and tumor suppression in vivo. J Immunol. 188:4441–4449.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu Y, Bae S, Kim H, et al: The anti-tumor
activity of vitamin C via the increase of Fas (CD95) and MHC I
expression on human stomach cancer cell line, SNU1. Immune Netw.
11:210–215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Volkmann M, Schiff JH, Hajjar Y, et al:
Loss of CD95 expression is linked to most but not all p53 mutants
in European hepatocellular carcinoma. J Mol Med (Berl). 79:594–600.
2001. View Article : Google Scholar
|
|
13
|
Hughes MA, Harper N, Butterworth M, Cain
K, Cohen GM and MacFarlane M: Reconstitution of the death-inducing
signaling complex reveals a substrate switch that determines
CD95-mediated death or survival. Mol Cell. 35:265–279. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kober AM, Legewie S, Pforr C, et al:
Caspase-8 activity has an essential role in CD95/Fas-mediated MAPK
activation. Cell Death Dis. 2:e2122011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bajt ML, Vonderfecht SL and Jaeschke H:
Differential protection with inhibitors of caspase-8 and caspase-3
in murine models of tumor necrosis factor and Fas receptor-mediated
hepatocellular apoptosis. Toxicol Appl Pharmacol. 175:243–252.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao LJ, Zhao S, Zhao EH, et al:
Clinicopathological and prognostic significance of Ki-67, caspase-3
and p53 expression in gastric carcinomas. Oncol Lett. 6:1277–1284.
2013.PubMed/NCBI
|
|
17
|
Sun BH, Zhang J, Wang BJ, et al: Analysis
of in vivo patterns of caspase 3 gene expression in primary
hepatocellular carcinoma and its relationship to p21(WAF1)
expression and hepatic apoptosis. World J Gastroenterol. 6:356–360.
2000.
|
|
18
|
Lindahl T, Satoh MS, Poirier GG and
Klungland A: Post-translational modification of poly(ADP-ribose)
polymerase induced by DNA strand breaks. Trends Biochem Sci.
20:405–411. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bi FF, Li D and Yang Q: Hypomethylation of
ETS transcription factor binding sites and upregulation of PARP1
expression in endometrial cancer. Biomed Res Int. 2013:9462682013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu W, Kong Z, Duan X, et al: Inhibition of
PARP1 by small interfering RNA enhances docetaxel activity against
human prostate cancer PC3 cells. Biochem Biophys Res Commun.
442:127–132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He W, Liu T, Shan Y, Zhu K and Li Y: PARP1
polymorphisms increase the risk of gastric cancer in a Chinese
population. Mol Diagn Ther. 16:35–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim J, Pyun JA, Cho SW, Lee K and Kwack K:
Lymph node metastasis of gastric cancer is associated with the
interaction between poly (ADP-ribose) polymerase 1 and matrix
metallopeptidase 2. DNA Cell Biol. 30:1011–1017. 2011. View Article : Google Scholar : PubMed/NCBI
|