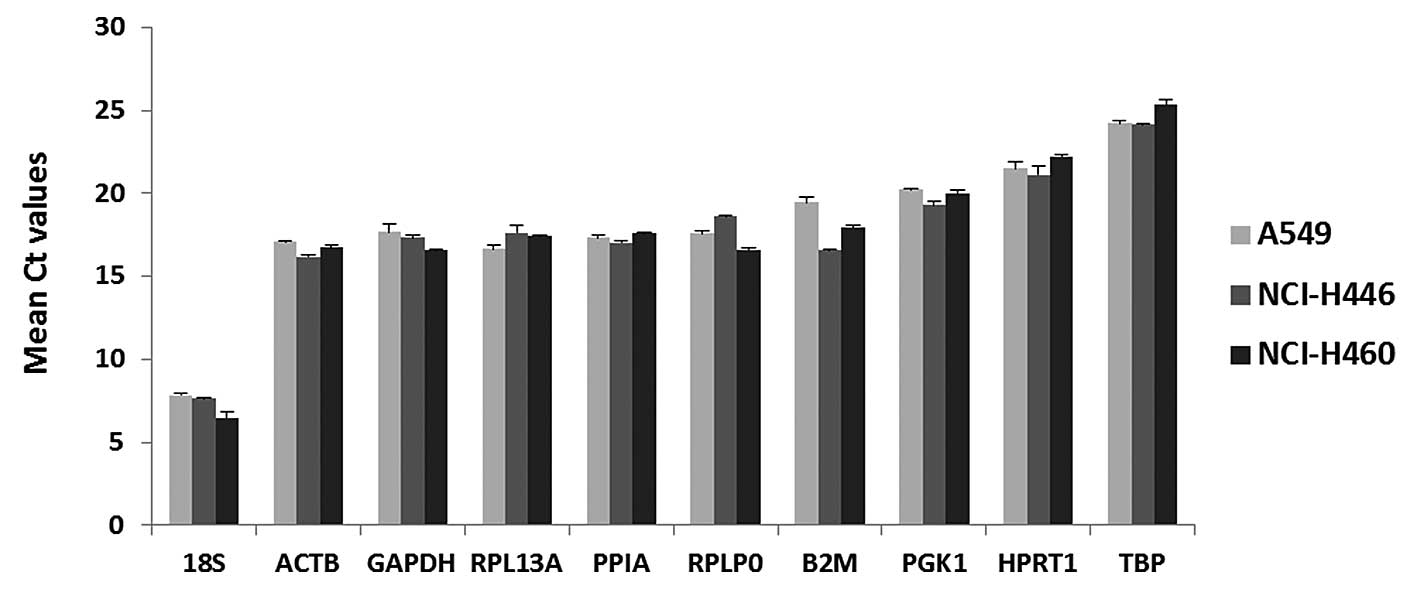
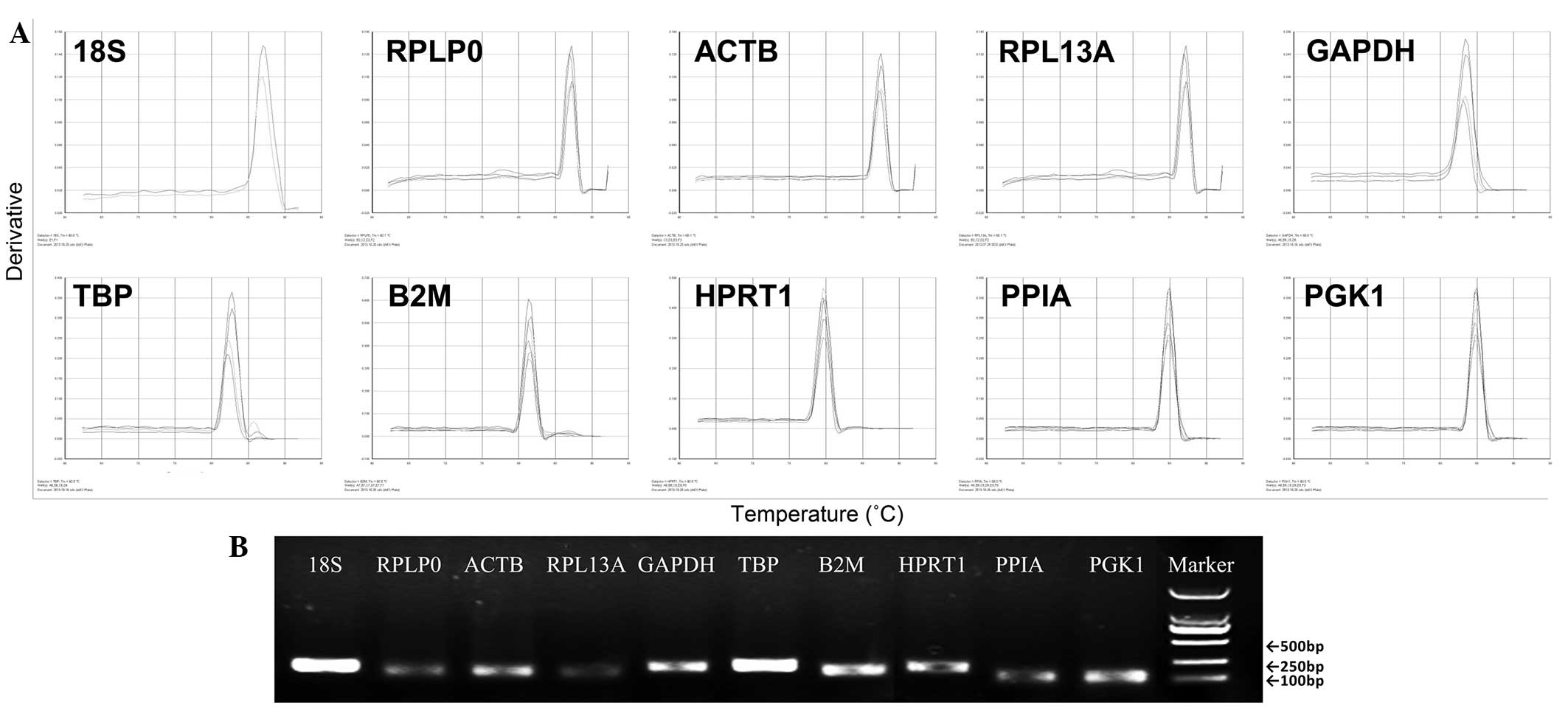
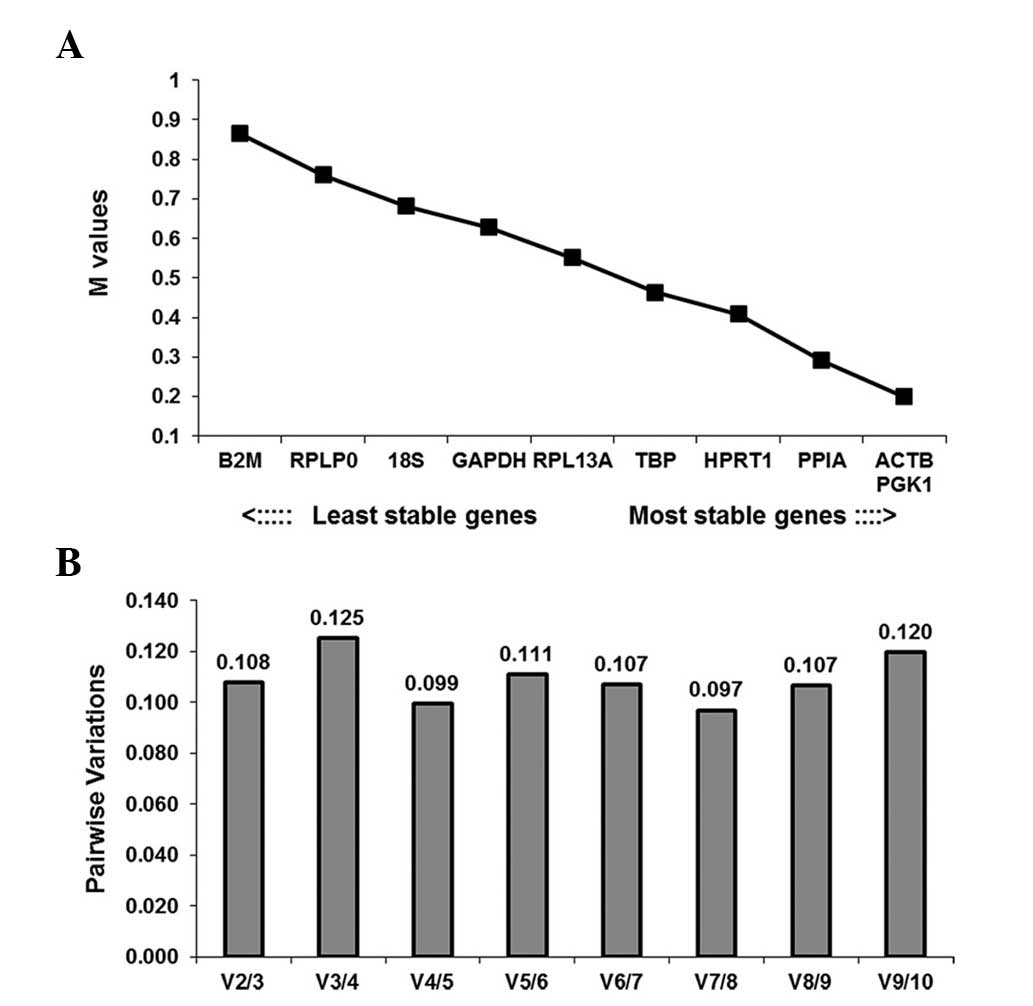
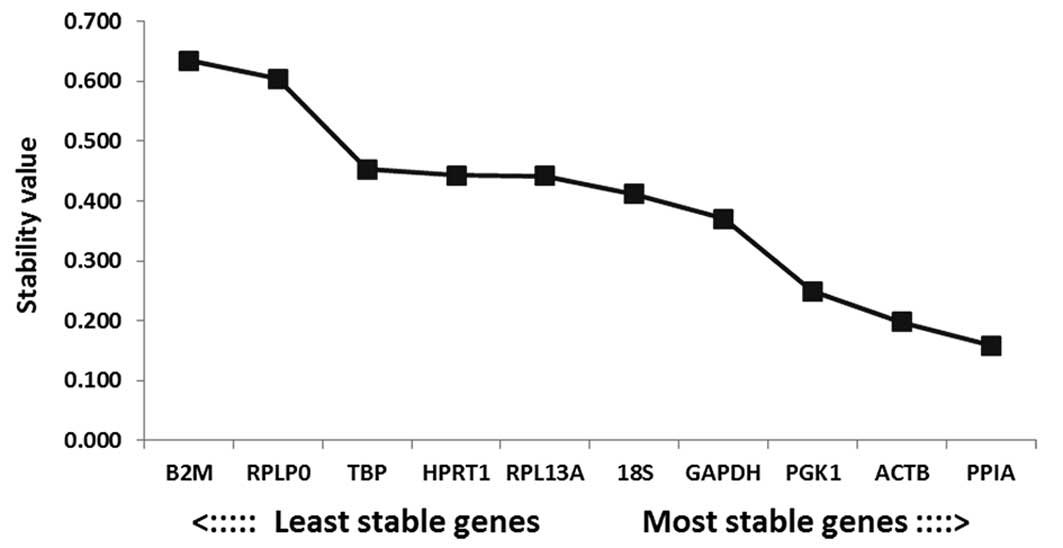
|
1
|
Koutsokera A, Kiagia M, Saif MW, Souliotis
K and Syrigos KN: Nutrition habits, physical activity and lung
cancer: an authoritative review. Clin Lung Cancer. 14:342–350.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perroud MW Jr, Honma HN, Barbeiro AS, et
al: Mature autologous dendritic cell vaccines in advanced non-small
cell lung cancer: a phase I pilot study. J Exp Clin Cancer Res.
30:652011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Watanabe T, Miura T, Degawa Y, et al:
Comparison of lung cancer cell lines representing four
histopathological subtypes with gene expression profiling using
quantitative real-time PCR. Cancer Cell Int. 10:22010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guenin S, Mauriat M, Pelloux J, Van
Wuytswinkel O, Bellini C and Gutierrez L: Normalization of qRT-PCR
data: the necessity of adopting a systematic, experimental
conditions-specific, validation of references. J Exp Bot.
60:487–493. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dheda K, Huggett JF, Bustin SA, Johnson
MA, Rook G and Zumla A: Validation of housekeeping genes for
normalizing RNA expression in real-time PCR. Biotechniques.
37:112–114. 2004.PubMed/NCBI
|
|
7
|
Hendriks-Balk MC, Michel MC and Alewijnse
AE: Pitfalls in the normalization of real-time polymerase chain
reaction data. Basic Res Cardiol. 102:195–197. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eisenberg E and Levanon EY: Human
housekeeping genes, revisited. Trends Genet. 29:569–574. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arenas-Hernandez M and Vega-Sanchez R:
Housekeeping gene expression stability in reproductive tissues
after mitogen stimulation. BMC Res Notes. 6:2852013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huggett J, Dheda K, Bustin S and Zumla A:
Real-time RT-PCR normalisation; strategies and considerations.
Genes Immun. 6:279–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barber RD, Harmer DW, Coleman RA and Clark
BJ: GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression
in a panel of 72 human tissues. Physiol Genomics. 21:389–395. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bas A, Forsberg G, Hammarstrom S and
Hammarstrom ML: Utility of the housekeeping genes 18S rRNA,
beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for
normalization in real-time quantitative reverse
transcriptase-polymerase chain reaction analysis of gene expression
in human T lymphocytes. Scand J Immun. 59:566–573. 2004. View Article : Google Scholar
|
|
13
|
Selvey S, Thompson EW, Matthaei K, Lea RA,
Irving MG and Griffiths LR: Beta-actin - an unsuitable internal
control for RT-PCR. Mol Cell Probes. 15:307–311. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Andersen CL, Jensen JL and Orntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: a model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Can Res. 64:5245–5250. 2004. View Article : Google Scholar
|
|
15
|
Rodriguez-Mulero S and Montanya E:
Selection of a suitable internal control gene for expression
studies in pancreatic islet grafts. Transplantation. 80:650–652.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sorby LA, Andersen SN, Bukholm IR and
Jacobsen MB: Evaluation of suitable reference genes for
normalization of real-time reverse transcription PCR analysis in
colon cancer. J Exp Clinical Cancer Res. 29:1442010. View Article : Google Scholar
|
|
17
|
Chua SL, See Too WC, Khoo BY and Few LL:
UBC and YWHAZ as suitable reference genes for accurate
normalisation of gene expression using MCF7, HCT116 and HepG2 cell
lines. Cytotechnology. 63:645–654. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim B, Lee HJ, Choi HY, et al: Clinical
validity of the lung cancer biomarkers identified by bioinformatics
analysis of public expression data. Cancer Res. 67:7431–7438. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schlotter YM, Veenhof EZ, Brinkhof B, et
al: A GeNorm algorithm-based selection of reference genes for
quantitative real-time PCR in skin biopsies of healthy dogs and
dogs with atopic dermatitis. Vet Immunol Immunopathol. 129:115–118.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Ishikawa T, Michiue T, Zhu BL,
Guan DW and Maeda H: Stability of endogenous reference genes in
postmortem human brains for normalization of quantitative real-time
PCR data: comprehensive evaluation using geNorm, NormFinder and
BestKeeper. Int J Legal Med. 126:943–952. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper - Excel-based tool using pair-wise correlations.
Biotechno Lett. 26:509–515. 2004. View Article : Google Scholar
|
|
22
|
de Oliveira LA, Breton MC, Bastolla FM, et
al: Reference genes for the normalization of gene expression in
eucalyptus species. Plant Cell Physiol. 53:405–422. 2012.
View Article : Google Scholar
|
|
23
|
Bunn PA Jr: Worldwide overview of the
current status of lung cancer diagnosis and treatment. Arch
Pathology Lab Med. 136:1478–1481. 2012. View Article : Google Scholar
|
|
24
|
Tsuboi M, Ohira T, Saji H, et al: The
present status of postoperative adjuvant chemotherapy for
completely resected non-small cell lung cancer. Ann Thoracic
Cardiovasc Surg. 13:73–77. 2007.
|
|
25
|
Liu J, Lee W, Jiang Z, et al: Genome and
transcriptome sequencing of lung cancers reveal diverse mutational
and splicing events. Genome Res. 22:2315–2327. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sokilde R, Kaczkowski B, Podolska A, et
al: Global microRNA analysis of the NCI-60 cancer cell panel. Mol
Cancer The. 10:375–384. 2011. View Article : Google Scholar
|
|
27
|
Che CL, Zhang YM, Zhang HH, et al: DNA
microarray reveals different pathways responding to paclitaxel and
docetaxel in non-small cell lung cancer cell line. Int J Clin Exp
Path. 6:1538–1548. 2013.
|
|
28
|
Gazdar AF, Girard L, Lockwood WW, Lam WL
and Minna JD: Lung cancer cell lines as tools for biomedical
discovery and research. J Natl Cancer Inst. 102:1310–1321. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gazdar AF, Gao B and Minna JD: Lung cancer
cell lines: Useless artifacts or invaluable tools for medical
science? Lung Cancer. 68:309–318. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
van Eijk R, Licht J, Schrumpf M, et al:
Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle
aspirates from non-small-cell lung cancer using allele-specific
qPCR. PloS One. 6:e177912011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saviozzi S, Cordero F, Lo Iacono M,
Novello S, Scagliotti GV and Calogero RA: Selection of suitable
reference genes for accurate normalization of gene expression
profile studies in non-small cell lung cancer. BMC Cancer.
6:2002006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ho-Pun-Cheung A, Bascoul-Mollevi C,
Assenat E, et al: Validation of an appropriate reference gene for
normalization of reverse transcription-quantitative polymerase
chain reaction data from rectal cancer biopsies. Anal Biochem.
388:348–350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ohl F, Jung M, Xu C, et al: Gene
expression studies in prostate cancer tissue: which reference gene
should be selected for normalization? J Mol Medicine. 83:1014–1024.
2005. View Article : Google Scholar
|
|
34
|
Jacob F, Guertler R, Naim S, et al:
Careful selection of reference genes is required for reliable
performance of RT-qPCR in human normal and cancer cell lines. PloS
One. 8:e591802013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mafra V, Kubo KS, Alves-Ferreira M, et al:
Reference genes for accurate transcript normalization in citrus
genotypes under different experimental conditions. PloS One.
7:e312632012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mallona I, Lischewski S, Weiss J, Hause B
and Egea-Cortines M: Validation of reference genes for quantitative
real-time PCR during leaf and flower development in Petunia
hybrida. BMC Plant Biol. 10:42010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu DW, Chen ST and Liu HP: Choice of
endogenous control for gene expression in nonsmall cell lung
cancer. Eur Respir J. 26:1002–1008. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gresner P, Gromadzinska J and Wasowicz W:
Reference genes for gene expression studies on non-small cell lung
cancer. Acta Biochim Pol. 56:307–316. 2009.PubMed/NCBI
|