Introduction
Advances in understanding of the molecular
mechanisms involved in wound repair have demonstrated a role for
distant stem cells in supporting wound healing (1,2).
Human umbilical cord mesenchymal stem cells (hUMSCs) may represent
an important seed cell for tissue engineering due to their capacity
for self-renewal and multilineage differentiation (3,4).
Stem cells may also represent an effective vehicle for gene
delivery (5,6).
The homeobox (HOX) family of transcription
factors influence bone marrow-derived cell differentiation,
migration and adhesion to injured tissue (7). In multicellular organisms, HOX
genes regulate body morphology formation, and the differentiation
and embryonic development of the axial skeleton, gastrointestinal
tract, urogenital tube, external genital duct, central nervous
system, hematopoietic system, and the limbs and skin (8,9). In
adults, HOX genes are involved in maintaining positional
identity, tissue homeostasis and remodeling in normal tissue and
during wound healing (7,10). HOXA4 gene expression has
been observed in keratinocytes, fibroblasts and mesenchymal stem
cells in embryonic and adult skin, as well as hair follicles
(11). In addition, HOXA4
regulates cell chemotaxis, migration, proliferation and
differentiation during the reconstruction and healing of damaged
tissue (5).
The present study analyzed whether HOXA4
expression by hUMSCs enhanced the differentiation of these cells
into epidermal-like cells in order to promote skin repair. The aim
was to provide a basis for the development of novel combined gene
and stem cell-based therapies for wound repair.
Materials and methods
hUMSC isolation, culture and phenotype
analysis
This study was approved by the institutional review
board of the Second Affiliated Hospital of Nancheng University
(Nanchang, China) in compliance with the Declaration of Helsinki.
Written informed consent was obtained from the patients’ families,
for removal and storage of a fresh human umbilical cord from a
single infant in Hank’s balanced salt solution (Sigma-Aldrich, St
Louis, MO, USA) for 24 h prior to the isolation of hUMSCs as
previously described (12). Cells
were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco
Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal
bovine serum (FBS; GE Healthcare Life Sciences, Logan, UT, USA) and
glucose (4.5 g/l) in 5% CO2 at 37°C for 14 days.
The identity of the isolated hUMSCs was confirmed
using a FACSCalibur Flow Cytometer (BD Biosciences, San Diego, CA,
USA) and fluorescein isothiocyanate (FITC)-conjugated antibodies
against CD105, CD54, CD33 and CD29, and phycoerythrin
(PE)-conjugated antibodies against CD45, CD166, CD34 and CD13 (all
from BD Biosciences). FITC- and PE-conjugated human IgG1 were used
as isotype-matched controls to set gates.
hUMSC infection and differentiation
Cells in the third passage were infected with a
lentiviral vector containing either green fluorescent protein alone
(lenti-GFP) or GFP and HOXA4 (lenti-HOXA4, Invitrogen
Life Technologies, Grand Island, NY, USA) overnight at a
multiplicity of infection (MOI) of 60 plaque-forming units
(PFU)/cell. hUMSCs were incubated in DMEM/F-12 media supplemented
with 0.5 μM dexamethasone (Sigma-Aldrich) and 10 ng/ml epidermal
growth factor (PeproTech, Inc., Rocky Hill, NJ, USA) for 2 weeks.
The efficiency of transduction was determined by analysis of GFP
expression using fluorescence microscopy (TE-2000, Nikon
Corporation, Tokyo, Japan), by calculating the ratio of
GFP-positive cells (green fluorescence) to the total number of
cells.
Flow cytometry
Cells were suspended in DMEM (5×106/ml),
permeabilized in 0.1% Triton X-100 (Sigma-Aldrich), and incubated
with 150 μl mouse anti-human cytokeratin 14 and 18 antibodies
followed by FITC-conjugated anti-mouse antibodies (10 mg/ml; all
from BD Biosciences). Cells were washed twice with
phosphate-buffered saline (PBS), centrifuged at 600 × g for 5 min,
and fixed in 1.5 ml of 4% paraformaldehyde. Control samples were
incubated with PBS in place of a primary antibody or an
isotype-matched control IgG. Cell labeling was evaluated in
1×106 cells using a FACScan (BD Biosciences).
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was isolated using TRIzol®
reagent (Invitrogen Life Technologies), and 1 μg was used for cDNA
synthesis using the Transcriptor Universal cDNA Master (Roche
Diagnostics, Indianapolis, IN, USA). cDNA was obtained by reverse
transcription according to the manufacturer’s instructions (Tiangen
Biotech Co., Ltd., Beijing, China). The PCR reaction was conducted
using 1 μl cDNA, 5 μl of 10× PCR buffer, (Takara Bio, Inc., Otsu,
Japan) 1.5 μl of 2 mM magnesium chloride, 1 μl specific
HOXA4 primers (20 pM per primer), 1 μl of 10 mM dNTP mix
(Takara Bio, Inc.), and 1 μl Taq DNA polymerase (5 U/ml; Takara
Bio, Inc.) in a total volume of 0.5 μl. The HOXA4 primer
sequences were as follows: Forward:
5′-CGCGCTAGCATGACCATGAGCTCGTTTTTG-3′ and reverse:
5′-GCGCGATCGTACTGGTAC TCGAGCAAAAAC-3′. Samples were amplified in a
thermocycler (Applied Biosystems Life Technologies, Foster City,
CA, USA) under the following conditions: 95°C for 2 min; 95°C for
30 sec, 55°C for 54 sec and 72°C for 60 sec for 33 cycles; and 72°C
for 5 min. The PCR products were separated with an agarose gel and
stained with ethidium bromide (Sigma-Aldrich).
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde,
permeabilized and blocked with 0.4% Triton X-100 and 5% bovine
serum albumin (Sigma-Aldrich) in PBS, and incubated with
human-cytokeratin 14 and 18 antibodies (BD Biosciences). Following
incubation with biotinylated goat anti-rabbit IgG, staining was
visualized using an avidin biotin complex (ABC)-peroxidase kit
(Sigma-Aldrich), 3,3-diaminobenzidine tetrahydrochloride and
counterstained with hematoxylin. For the negative control, the
primary antibody was omitted. The slides were observed under an
Olympus microscope (BX51, Olympus Corporation, Tokyo, Japan), in
order to calculate the ratio of positive cells (brown fluorescence)
to the total number of cells.
In vivo model of full-thickness skin
defects
Collagen was prepared from the tails of
Sprague-Dawley rats as previously described (13). All animal procedures used approved
protocols in accordance with the regulations of the Animal Care and
Experiment Committee of the Nanchang University School of Medicine.
Forty-five nude mice were purchased from Shanghai Experimental
Animal Laboratory (Shanghai, China). Following intraperitoneal
administration of ketamine (10 mg/100 g; Sigma-Aldrich),
full-thickness skin defects (1.5×1.5 cm2) were made on
the backs of all nude mice, which were randomly divided into the
following groups, with 15 mice in each group: Collagen membrane
with 1×106 lenti-HOXA4 hUMSCs; collagen membrane
with 1×106 lenti-GFP hUMSCs and collagen membrane alone.
Cells were seeded evenly on the collagen membrane, which was then
directly applied to the wound as previously described (14). At day 21, the mice were sacrificed
by cervical dislocation following anesthesia with IP ketamine (100
mg/kg) and xylazine (5 mg/kg; Sigma-Aldrich), and wound tissue
samples were harvested. One section of tissue was fixed with 10%
formalin, cut into 3-μm sections, stained with hematoxylin and
eosin, and observed with a light microscope (BX51, Olympus
Corporation). The remaining piece was processed for western blot
analysis.
Western blot analysis
Total protein was extracted using a triplex lysis
buffer (Roche Diagnostics, Basel, Switzerland). Total protein (20
μg) was separated on a 10% SDS polyacrylamide gel and transferred
to a polyvinylidene fluoride membrane (EMD Millipore, Billerca, MA,
USA). The membrane was blocked with non-fat dry milk and incubated
with mouse monoclonal anti-HOXA4 antibody (1:100) or mouse
monoclonal anti-actin antibody (1:500) (Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) at 4°C overnight, followed by incubation
with monoclonal rabbit anti-mouse horseradish peroxidase
(HRP)-conjugated secondary antibody (1:5,000; Santa Cruz
Biotechnology, Inc.) at room temperature for 1–2 h. Color was
developed using an enhanced chemiluminescence kit (Thermo Fisher,
Scientific, Waltham, MA, USA). The relative HOXA4 protein
expression was determined as the gray value of HOXA4 band/gray
value of actin band using a gel imaging analysis system (Syngene,
Frederick, MD, USA).
Statistical analysis
Continuous variables are presented as the mean ±
standard deviation. For comparisons of three treatment groups,
one-way analysis of variance was undertaken. When a significant
difference between groups was apparent, multiple comparisons were
performed using the Bonferroni procedure with type I error
adjustment. All statistical assessments were two-sided and
P<0.05 was considered to indicate a statistically significant
difference. SAS software package, version 9.2 (SAS Institute, Inc.,
Cary, NC, USA) was used for statistical analysis.
Results
Identity of the isolated hUMSCs
The percentage of cells positive for the markers
investigated was as follows: CD13, 1.31±0.27%; CD34, 0.19±0.04%;
CD45, 2.71±0.56%; CD33, 2.34±0.64%; CD29, 33.35±0.89%; CD54,
67.51±4.58%; CD105, 74.33±6.08%; and CD166, 81.99±6.56%. Thus, the
isolated cells were confirmed as hUMSCs.
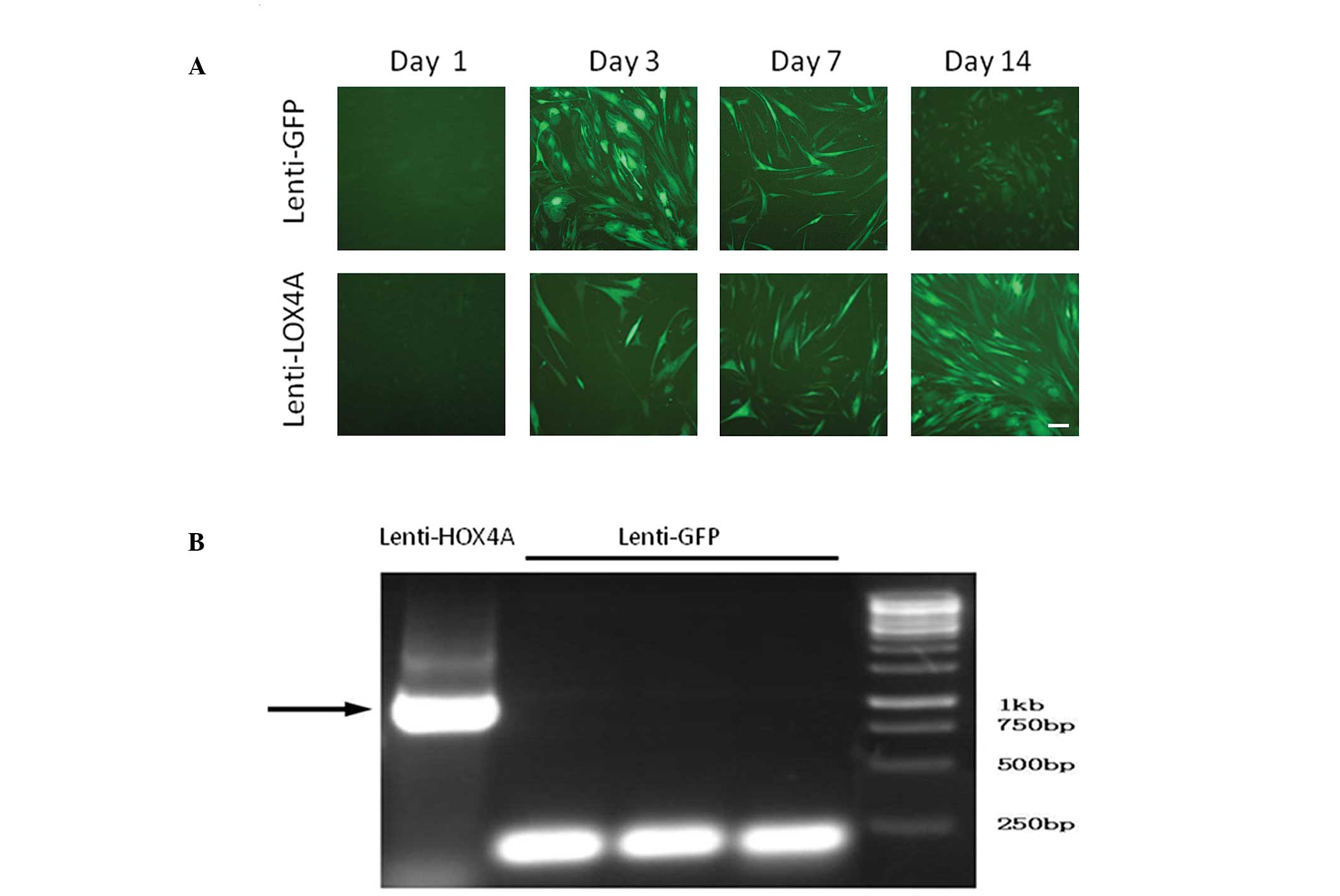
HOXA4 gene transduction of hUMSCs
The efficiency of lenti-HOXA4 gene
transduction was determined in each group simultaneously by
evaluating the percentages of GFP-positive hUMSCs at days 3, 7 and
14, which were 90.3, 81.6 and 70.4%, respectively (Fig. 1A). HOXA4 gene expression was
analyzed using RT-PCR following transduction and culture in a
normal medium. At day 7, hUMSCs transduced with lenti-HOXA4
expressed higher levels of HOXA4 than those cells infected
with lenti-GFP (Fig. 1B).
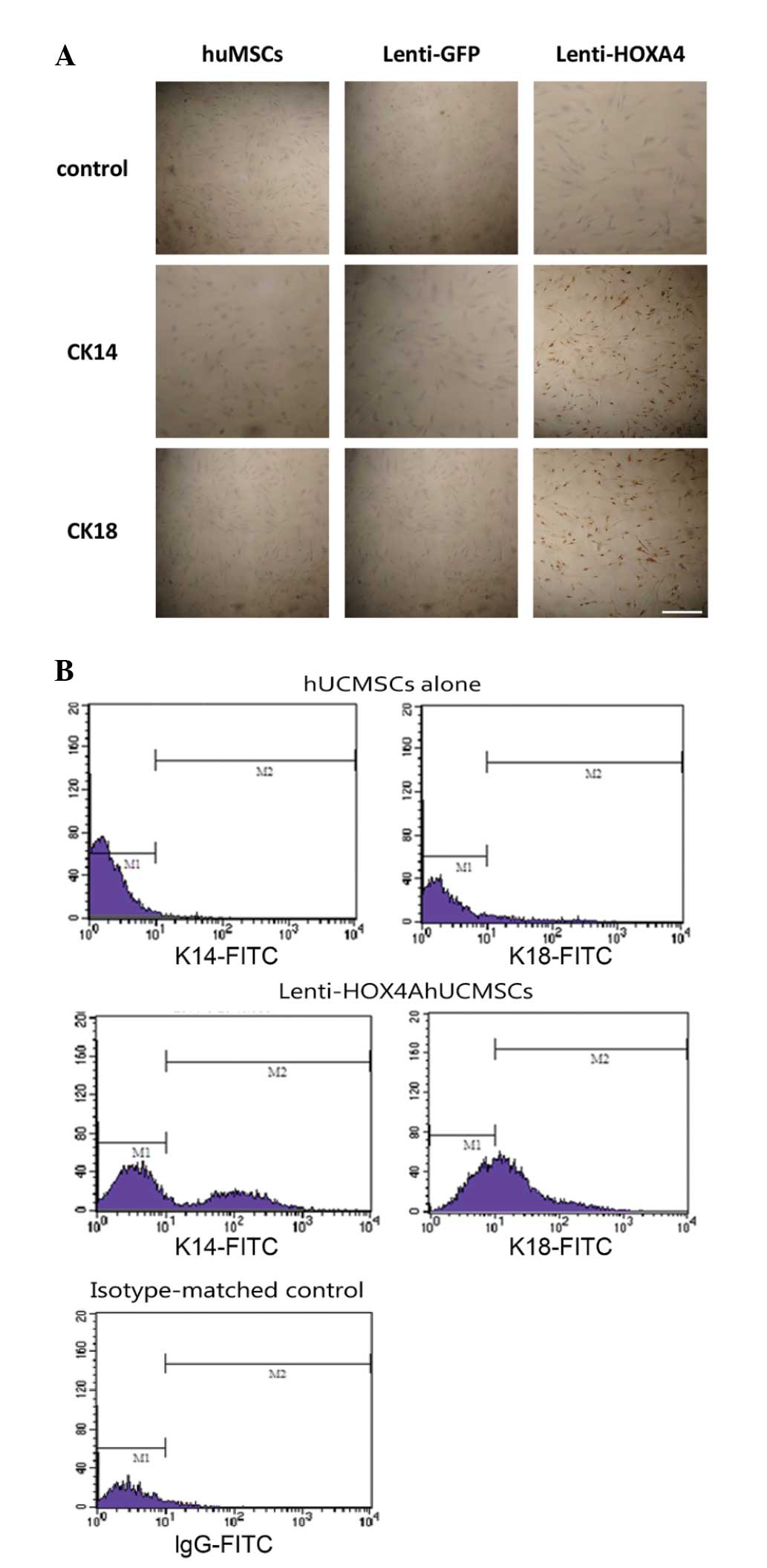
Increased epidermal cell marker
expression in lenti-HOXA4 hUMSCs
At 14 days post-lenti-HOXA4 infection,
expression of cytokeratin 14 and 18 was increased compared with
that in the control groups (Fig.
2A). The proportion of CK14-positive cells in the hUMSCs,
lenti-GFP hUMSCs and lenti-HOXA4 hUMSCs groups was
1.22±0.13, 1.31±0.24 and 32.98±5.12%, respectively. The proportion
of CK18-positive cells was 1.22±0.13, 1.3±0.14 and 45.13±4.5% in
the hUMSCs, lenti-GFP hUMSCs and lenti-HOXA4 hUMSCs,
respectively. These results were confirmed using flow cytometric
analysis. In hUMSCs, CK14 and CK18 expression was observed in 1.58
and 5.17% of the cells, respectively; whereas these markers were
detected in 32.84 and 41.14%, respectively, of cells transduced
with lenti-HOXA4 (Fig.
2B).
In vivo epidermal regeneration with
HOXA4-expressing hUMSCs
Mice with full-thickness skin defects were treated
with collagen membrane alone, or evenly seeded with lenti-GFP
hUMSCs or lenti-HOXA4 hUMSCs, and histological analysis was
undertaken at day 21 post-injury (Fig.
3), at which time macroscopic evidence of wound healing was
also evident. In the collagen membrane control group, the epidermis
did not heal, or scar tissue was formed (Fig. 3A; arrow). Mice treated with
lenti-HOXA4 hUMSCs had a thicker epidermis with more
organized cell layers compared with that of the lenti-GFP hUMSC
group (Fig. 3B and C,
respectively). Furthermore, western blot analysis of the tissues
indicated persistent HOXA4 expression in the wounds of mice treated
with lenti-HOXA4 hUMSCs (Fig.
4A, lane 1), which was significantly higher than that in the
hUMSCs and lenti-GFP hUMSC groups (P<0.001, Fig. 4B). Expression of cytokeratins 14
and 18 was also detected in mice treated with lenti-GFP hUMSCs and
lenti-HOX4A (Fig. 4C, lane
2); however, the expression of each of these cytokeratins was
significantly higher in the lenti-HOXA4 hUMSC group than in
the lenti-GFP hUMSC group (P≤0.003 and P≤0.001, respectively;
Fig. 4C and 4D).
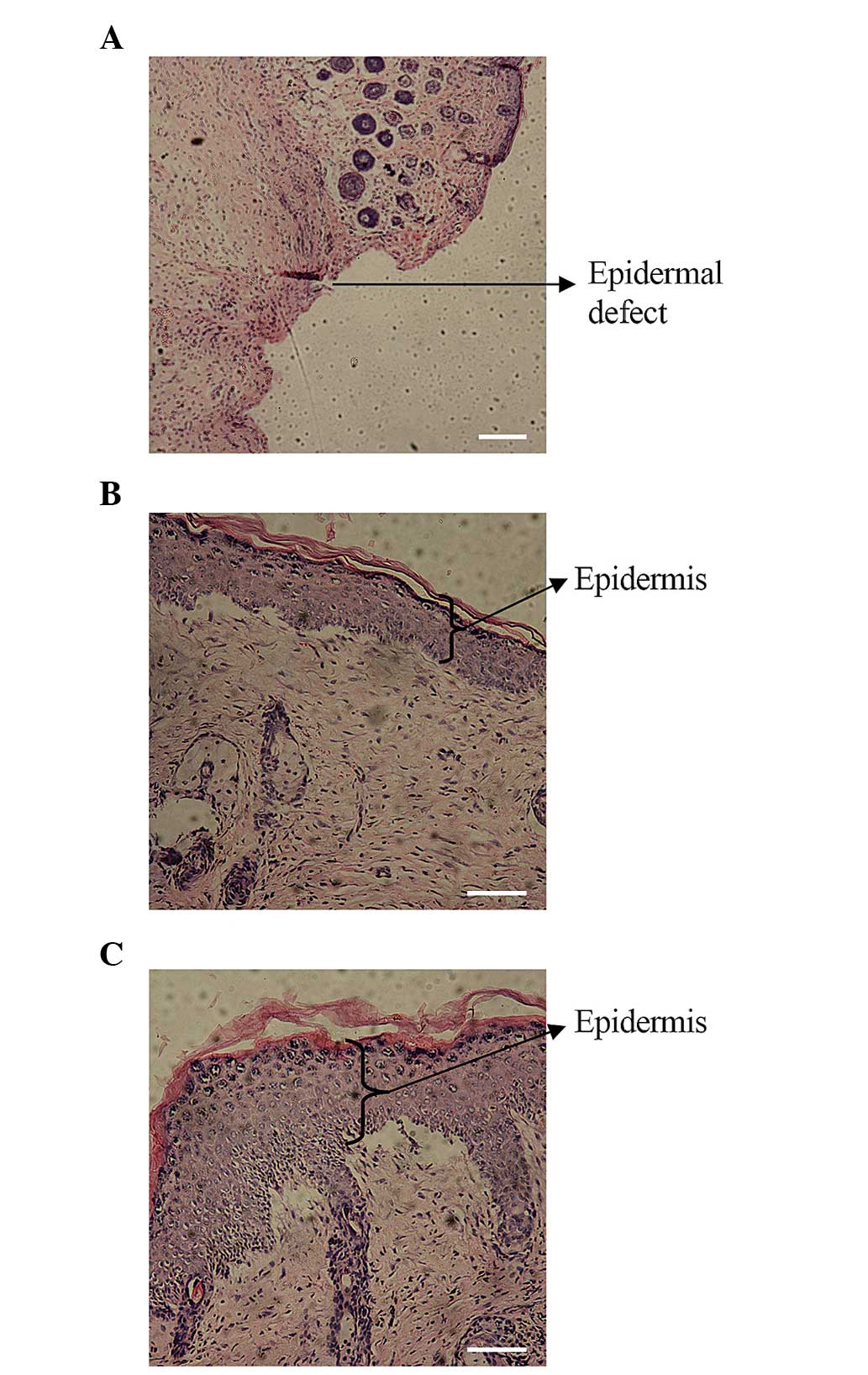 | Figure 3In vivo epidermal regeneration
following xenografts with HOXA4-expressing hUMSCs.
Full-thickness skin defects were treated with (A) a collagen
membrane alone, or a collagen membrane evenly seeded with either
(B) lenti-GFP hUMSCs or (C) lenti-HOXA4 hUMSCs. At day 21,
histological analysis of the wounds was undertaken following
hematoxylin and eosin staining. In the collagen membrane control
group, the skin was not fully healed, and new epidermis was not
formed (arrow; magnification, ×40; scale bar, 200 μm). In the
lenti-GFP hUMSC and lenti-HOXA4 hUMSC groups, the epidermis
(arrows) was formed (magnification, ×100; scale bar, 100 μm).
However, the epidermis was thicker and the cell layers more
organized in the wounds treated with HOXA4-expressing hUMSCs
than in the control group. Lenti-HOXA4, lentivirus
expressing homeobox A4; hUMSCs, human umbilical cord mesenchymal
stem cells; lenti-GFP, lentivirus expressing green fluorescent
protein. |
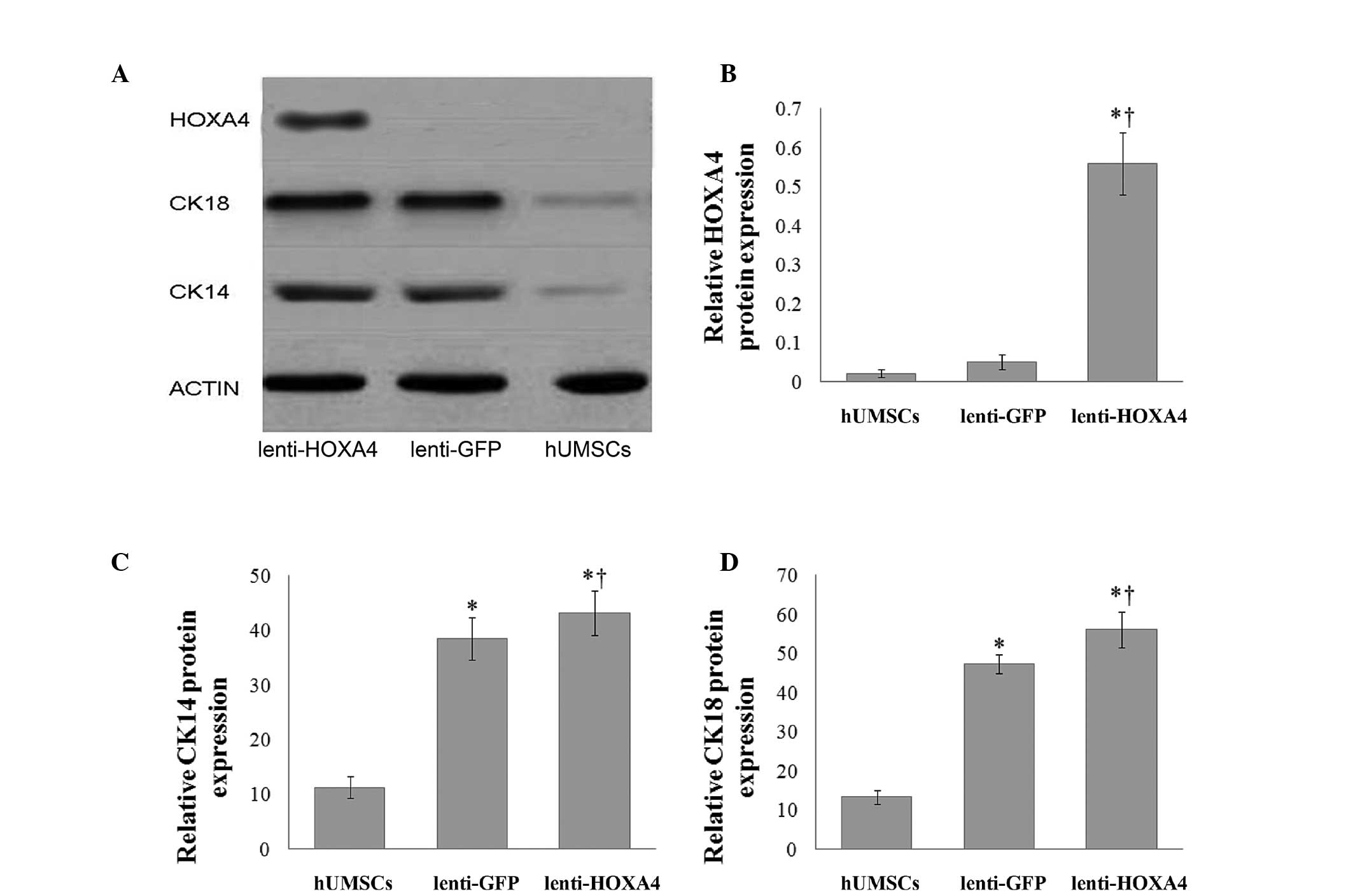 | Figure 4HOXA4, CK14 and CK18 protein
expression during epidermal regeneration following xenografts with
HOXA4-expressing hUMSCs in vivo. (A) Representative
western blot of HOXA4, CK14 and CK18 protein expression.
Quantitative analysis of (B) HOXA4, (C) CK14 and (D) CK18 protein
expression in the hUMSCs, lenti-GFP hUMSCs, and lenti-HOXA4
hU MSCs groups (n = 15 per group). *P<0.05, compared
with the hUMSC control group and †P<0.05, compared
with the lenti-GFP hUMSC group. HOXA4, homeobox A4; hUMSCs, human
umbilical cord mesenchymal stem cells; lenti-GFP, lentivirus
expressing green fluorescent protein; lenti-HOXA4,
lentivirus expressing HOXA4; CK, cytokeratin. |
Discussion
The present study sought to determine whether
HOXA4 expression in hUMSCs enhanced their differentiation
into epidermal-like cells and therefore facilitated the process of
skin repair. In lenti-HOXA4 hUMSCs, increased cytokeratin 14
and 18 expression was observed along with enhanced repair of
full-thickness skin defects.
Since embryonic MSCs are able to differentiate into
epidermal cells in vitro and in vivo (15), the application of these cells in
acute and chronic wound therapy has been investigated. Studies have
shown that following transplantation via tail vein injection,
hUMSCs migrate to the skin and differentiate into epithelial cells
(16,17). In addition, an artificial skin,
consisting of epidermal stem cells and fibroblasts on a collagen
lattice, was shown to promote wound healing in mice (18). Besides their regenerative capacity,
the use of MSCs as vehicles for gene delivery has been explored
(5,6). Improved wound healing was reported
following the administration of MSCs expressing various growth
factors and cytokines (19–21).
In the present study, persistent expression of HOXA4 in
hUMSCs was observed at 21 days following transduction, suggesting
that these cells are also capable of carrying foreign genes, thus
potentially enhancing cell therapy.
The HOXA4 gene is expressed in skin, amongst
other tissues (22,23) and is known to regulate cell
differentiation during the reconstruction and healing of damaged
tissue (5). We therefore
hypothesized that its expression by hUMSCs may enhance their
capacity to differentiate into epidermal cells. Following
incubation with epidermal-inducing agents, HOXA4-expressing
hUMSCs differentiated into epidermal-like cells and were
demonstrated to express cytokeratins 14 and 18. Further studies may
thus directly assess the effects of HOXA4 expression on
hUMSC differentiation into keratinocytes. Furthermore, recent data
have indicated that Mospd1 gene expression may regulate
mesenchymal to epithelial transition (13,18);
therefore, future studies may be conducted in order to assess its
value in promoting differentiation.
In the current study, wounds treated with
lenti-HOXA4 hUMSCs appeared to have a thicker epidermis on
day 21 following injury. Thus, lenti-HOXA4 hUMSCs may
improve wound healing and reduce scar formation. However, the
mechanism underlying these effects remains unknown. The expression
of HOXA4 may, for example, increase the expression of Cyclin
D1 and Bcl-2, or it may reduce the expression of P21WAF/CIPI,
thereby promoting cell proliferation and inhibiting apoptosis
(24). This group intends to
investigate these possible mechanisms in future studies.
The present study is limited in that the
re-epithelialized epidermis was not evaluated for the presence of
hUMSC-derived keratinocytes. In addition, the extent of wound
healing was only assessed on day 21; therefore, differences at
earlier time points may have been missed. Furthermore, changes in
the epidermal thickness were not quantified. Finally, although
HOXA4 mRNA expression was determined following transduction
of the hUMSCs, its protein expression levels were not assessed.
In conclusion, lenti-HOXA4 hUMSCs enhanced
wound repair and may represent a novel therapeutic approach for
burns and chronic wounds. Further studies are required to elucidate
the molecular mechanisms governing this promotion of wound
healing.
Acknowledgements
The authors would like to thank Professor Yucheng
Dai (Institution of Hematology, the Second Affiliated Hospital of
Nanchang University) for his assistance. This study was supported
by the National Natural Science Foundation of China (grant no.
30760261), the Natural Science Foundation of Jiangxi Province
(grant no. 0540084) and the Project of Cultivating Academic and
Technical Leaders for Major Disciplines in Jiangxi province (grant
no. 2014BCB22009).
References
|
1
|
Ballas CB, Zielske SP and Gerson SL: Adult
bone marrow stem cells for cell and gene therapies: implications
for greater use. J Cell Biochem. 38(Suppl): 20–28. 2002. View Article : Google Scholar
|
|
2
|
Fathke C, Wilson L, Hutter J, Kapoor V,
Smith A, Hocking A and Isik F: Contribution of bone marrow-derived
cells to skin: collagen deposition and wound repair. Stem Cells.
22:812–822. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chamberlain G, Fox J, Ashton B and
Middleton J: Concise review: mesenchymal stem cells: their
phenotype, differentiation capacity, immunological features, and
potential for homing. Stem Cells. 25:2739–2749. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Branski LK, Gauglitz GG, Herndon DN and
Jeschke MG: A review of gene and stem cell therapy in cutaneous
wound healing. Burns. 35:171–180. 2009. View Article : Google Scholar
|
|
6
|
Gauglitz GG and Jeschke MG: Combined gene
and stem cell therapy for cutaneous wound healing. Mol Pharm.
8:1471–1479. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mahdipour E and Mace KA: Hox transcription
factor regulation of adult bone-marrow-derived cell behaviour
during tissue repair and regeneration. Expert Opin Biol Ther.
11:1079–1090. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Akiyama M, Smith LT and Holbrook KA:
Growth factor and growth factor receptor localization in the hair
follicle bulge and associated tissue in human fetus. J Invest
Dermatol. 106:391–396. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Favier B and Dollé P: Developmental
functions of mammalian Hox genes. Mol Hum Reprod. 3:115–131. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang KC, Helms JA and Chang HY:
Regeneration, repair and remembering identity: the three Rs of Hox
gene expression. Trends Cell Biol. 19:268–275. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stelnicki EJ, Kömüves LG, Kwong AO, Holmes
D, Klein P, Rozenfeld S, Lawrence HJ, Adzick NS, Harrison M and
Largman C: HOX homeobox genes exhibit spatial and temporal changes
in expression during human skin development. J Invest Dermatol.
110:110–115. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hsieh JY, Wang HW, Chang SJ, Liao KH, Lee
IH, Lin WS, Wu CH, Lin WY and Cheng SM: Mesenchymal stem cells from
human umbilical cord express preferentially secreted factors
related to neuroprotection, neurogenesis, and angiogenesis. PLoS
One. 8:e726042013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu K, Dai Y and Yuan J: An experimental
study on the preparation of artificial dermis. J Xian Jiaotong Univ
(Medical Sciences). 27:117–119. 1312006.(In Chinese).
|
|
14
|
Hu K, Dai Y, Hu Q, Li J, Yuan J, Li J and
Wu Q: An experimental study on the repair of full skin loss of nude
mice with composite graft of epidermal stem cells. Burns.
32:416–422. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu M, Yang L, Liu S, Li H, Hui N, Wang F
and Liu H: Differentiation potential of human embryonic mesenchymal
stem cells for skin-related tissue. Br J Dermatol. 155:282–291.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dai Y, Li J, Li J, Dai G, Mu H, Wu Q, Hu K
and Cao Q: Skin epithelial cells in mice from umbilical cord blood
mesenchymal stem cells. Burns. 33:418–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gang EJ, Jeong JA, Hong SH, Hwang SH, Kim
SW, Yang IH, Ahn C, Han H and Kim H: Skeletal myogenic
differentiation of mesenchymal stem cells isolated from human
umbilical cord blood. Stem Cells. 22:617–624. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thaler R, Rumpler M, Spitzer S, Klaushofer
K and Varga F: Mospd1, a new player in mesenchymal versus epidermal
cell differentiation. J Cell Physiol. 226:2505–2515. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deodato B, Arsic N, Zentilin L, Galeano M,
Santoro D, Torre V, Altavilla D, Valdembri D, Bussolino F,
Squadrito F and Giacca M: Recombinant AAV vector encoding human
VEGF165 enhances wound healing. Gene Ther. 9:777–785. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ha XQ, Lü TD, Hui L and Dong F: Effects of
mesenchymal stem cells transfected with human hepatocyte growth
factor gene on healing of burn wounds. Chin J Traumatol.
13:349–355. 2010.PubMed/NCBI
|
|
21
|
Liechty KW, Nesbit M, Herlyn M, Radu A,
Adzick NS and Crombleholme TM: Adenoviral-mediated overexpression
of platelet-derived growth factor-B corrects ischemic impaired
wound healing. J Invest Dermatol. 113:375–383. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cillo C, Cantile M, Faiella A and
Boncinelli E: Homeobox genes in normal and malignant cells. J Cell
Physiol. 188:161–169. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ford HL: Homeobox genes: a link between
development, cell cycle, and cancer? Cell Biol Int. 22:397–400.
1998. View Article : Google Scholar
|
|
24
|
Li J, Li Y, Ma S, Gao Y, Zuo Y and Hu J:
Enhancement of bone formation by BMP-7 transduced MSCs on
biomimetic nano-hydroxyapatite/polyamide composite scaffolds in
repair of mandibular defects. J Biomed Mater Res A. 95:973–981.
2010. View Article : Google Scholar : PubMed/NCBI
|