Introduction
Liver regeneration is important in the recovery from
injury induced by surgery, trauma, poisoning, infection, necrosis
or liver transplantation (1).
Consequently, research investigating the improvement of the
regeneration ability of the liver, is of great significance. During
regeneration, quiescent mature hepatocytes reenter the cell cycle
in order to proliferate and divide, thus leading to hepatic
regeneration without the involvement of stem cells. Although the
exact mechanisms have not been fully characterized, a study
demonstrated that liver regeneration primarily comprises cell
proliferation, lipid metabolism, various growth factors, and a
number of cytokines and their signaling pathways (2).
Bile acid, which is synthesized from cholesterol, is
the chief components of bile. The primary functions of bile are to
digest the fat soluble molecules in food and to aid in the
intestinal absorption of lipids in vivo. Recent studies have
shown that bile acid acts as a signaling molecule by activating
signaling pathways, and that it participates in the process of
liver regeneration (3,4). A number of transport proteins for
bile acid have been identified in the liver and are known to be
regulated by nuclear receptors (5). Nuclear receptors (NRs) are
ligand-activated transcription factors that are members of a super
family, consisting of 48 proteins (6). The farnesoid X receptor (FXR), a
member of the sub-cluster of metabolic NRs, was originally isolated
from a rat liver cDNA library and cloned in 1995 (7). FXR is predominantly expressed in the
liver, kidney, intestine and adrenal glands, and is involved in
regulating the metabolism of bile acid and cholesterol (8,9).
Furthermore, the interaction of bile acid with FXR is essential for
glucose metabolism, liver inflammation and liver regeneration
(10–12).
In the current study, bile acid, glucose and FXR
were administrated in vivo and in vitro, and the
effects of these molecules on liver regeneration and lipid
metabolism were compared. The mechanisms underlying these effects
were also explored.
Materials and methods
Animals
Male Sprague-Dawley rats (weight, 250–300 g) were
obtained from the Experimental Animal Center of Jiamusi University
(Jiamusi, China) and housed in a temperature- and light-controlled
room at 19-22°C with a 12 h light/dark cycle. The animals had free
access to food and water. The animal study protocol was reviewed
and approved by the Institutional Animal Care and Use Committee of
Jiamusi University.
Rat model of hepatectomy
Rats were fasted for 12 h and assigned to one of the
following three groups: Control group (rats were fed with normal
diet), bile acid group (rats were fed with normal diet plus 0.2%
bile acid) and glucose group (rats were fed with normal diet plus
10% glucose). There were 36 rats in each group. At day 7, rats were
subjected to 50 or 70% hepatectomy and the weight of removed liver
tissues was recorded as mR. At 24, 48 or 72 h following
hepatectomy, the rats were sacrificed by cervical dislocation
following anesthesia induction, and liver tissues and blood were
collected immediately. Total liver mass was calculated based on the
ratio of removed liver weight and volume, and recorded as mT. That
is, for rats in which 50% of the was liver removed, the weight of
the removed portion was divided by 0.5 to obtain the total volume,
and for those in which 70% was removed, it was divided by 0.7. The
liver weight of sacrificed rats was recorded as mS and the hepatic
regeneration rate was calculated as [mS-(mT-mR)]/mT × 100.
Immunohistochemistry
Fresh liver samples were immediately fixed in
neutral formalin and embedded in paraffin. Paraffin-embedded liver
samples were cut into 5-μm sections deparaffinized in
xylene, rehydrated in serial dilutions of ethanol, placed in
antigen retrieval solution and microwaved at low power for 10 min.
The activity of endogenous peroxidase was blocked by incubation
with 3% H2O2 (Sinopharm Chemical Reagent,
Shanghai, China). A mouse monoclonal antibody against proliferating
cell nuclear antigen (PCNA; 1:50; sc-252820; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) was added and incubated
at 4°C overnight. Sections were then incubated with a horseradish
peroxidase (HRP)-conjugated goat anti-mouse antibody (1:200; A0216;
Beyotime Institute of Biotechnology, Haimen, China) for 30 min at
37°C. Positive signals were visualized by 3,3′-Diaminobenzidine
(Solarbio, Beijing, China) and sections were then counterstained
with hematoxylin. Images of the sections were captured using a
microscope (x400; BX51, Olympus, Tokyo, Japan). A brown or yellow
color was regarded as a positive reaction.
Isolation of hepatocytes and experimental
setup
Hepatocyte isolation was performed as previously
described (13). Briefly,
following administration of anesthesia with 10% chloral hydrate
(Sinopharm Chemical Reagent), a cannula was introduced into the
portal vein and D-Hank’s solution (Solarbio) was perfused in order
to remove the blood. Collagenase (0.05%; Invitrogen, Carlsbad, CA,
USA) was then perfused to hydrolyse the collagen molecules. The
liver was removed from the capsule, cut into sections and passed
through a 200 mesh sieve. The viability of isolated hepatocytes was
>95%, as assessed by trypan blue (Beyotime Institute of
Biotechnology) exclusion. Primary hepatocytes were cultured in
Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY,
USA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan,
UT, USA) at 37°C in a humidified 5% CO2 atmosphere. The
medium was replaced every 2 days, and cells were passaged when they
reached 80–90% confluence. The experiments were initiated following
the third passage, with cells being divided into nine groups: The
control group (hepatocytes were cultured in normal medium); the
bile acid group (hepatocytes were cultured in normal medium with 40
μmol/l bile acid; Sigma-Aldrich, St. Louis, MO, USA); the
glucose group (hepatocytes were cultured in normal medium with 25
mmol/l glucose); the FXR agonist group (hepatocytes were cultured
in normal medium with 10 μmol/l FXR agonist, GW4064;
Sigma-Aldrich); the FXR antagonist group (hepatocytes were cultured
in normal medium with 100 μmol/l FXR antagonist,
Guggulsterones; Sigma-Aldrich); the bile acid and FXR agonist group
(hepatocytes were cultured in normal medium with 40 μmol/l
bile acid and 10 μmol/l FXR agonist, GW4064); the bile acid
and FXR antagonist group (hepatocytes were cultured in normal
medium with 40 μmol/l bile acid and 100 μmol/l FXR
antagonist, Guggulsterones); the glucose and FXR agonist group
(hepatocytes were cultured in normal medium with 25 mmol/l glucose
and 10 μmol/l FXR agonist, GW4064); and the glucose and FXR
antagonist group (hepatocytes were cultured in normal medium with
25 mmol/l glucose and 100 μmol/l FXR antagonist,
Guggulsterones). Cells were incubated at 37°C with 5%
CO2 and saturated humidity conditions in a culture box
(Heal Force, Shanghai. China) and collected at 72 h.
Analysis of total bile acid (TBA),
triglyceride (TG), total cholesterol (TC), high density lipoprotein
(HDL), low density lipoprotein (LDL) and free fatty acid (FFA)
The levels of TBA in liver and serum were measured
with the TBA assay kit (Nanjing Jiancheng Bioengineering Institute,
China) according to manufacturer’s instructions. The levels of TG,
TC, HDL, LDL and FFA in hepatocytes were assessed using an ELISA
kit (Rat TG ELISA kit, Ximei, Shanghai, China; Rat TC ELISA kit,
Rat HDL ELISA kit, Hyperheal, Shanghai, China; Rat LDL ELISA kit,
J&L Biological, Shanghai, China; Rat FFA ELISA kit, Lianshuo
Biological, Shanghai, China, respectively). Optical density was
measured using a microplate reader (BioTek Instruments, Winooski,
VT, USA).
Western blotting
Total liver protein was extracted with a
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, China) and total cellular protein was extracted with
NP-40 buffer (Beyotime Institute of Biotechnology). Equal
quantities of total protein (40 μg) were loaded and
separated using 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis, transferred to polyvinylidene fluoride membranes
(EMD Millipore, Billerica, MA, USA) and blocked in 5% fat-free milk
for 1 h. Membranes were incubated with primary antibody at 4°C
overnight. The following primary antibodies were used: A rabbit
polyclonal antibody to FXR (1:100; sc-13063; Santa Cruz
Biotechnology, Inc.), a rabbit polyclonal antibody to Caveolin 1
[1:1,000 (animal) or 1:2,000 (cell); ab2910; Abcam, Cambridge, UK],
a rabbit polyclonal antibody to ASBT (1:1,000; bs-4189R; Bioss,
Beijing, China), a rabbit polyclonal antibody to BSEP (1:1,000;
ab99088), a rabbit monoclonal antibody to NTCP (1:1,000; ab133670)
(Abcam) a goat polyclonal antibody to SHP (1:100; sc-15283) and a
rabbit polyclonal antibody to CYP7A1 (1:100; sc-25536) (Santa Cruz
Biotechnology, Inc.). Membranes were then washed and incubated with
goat anti-rabbit (A0208) or donkey anti-goat (A0108) HRP-conjugated
secondary antibodies (1:5,000, Beyotime Institute of Biotechnology)
at 37°C for 45 min. Immunoreactive bands were visualized by
enhanced chemiluminescence solution (Qihai Biotech, Shanghai,
China) according to the manufacturer’s instructions.
Statistical analysis
Data are presented as the mean ± standard deviation.
Two-tailed Student’s t test was used to assess statistical
significance. Data were analyzed using GraphPad Prism 5.0 software
(GraphPad Software, Inc., San Diego, CA, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Hepatic regeneration rate and expression
of PCNA
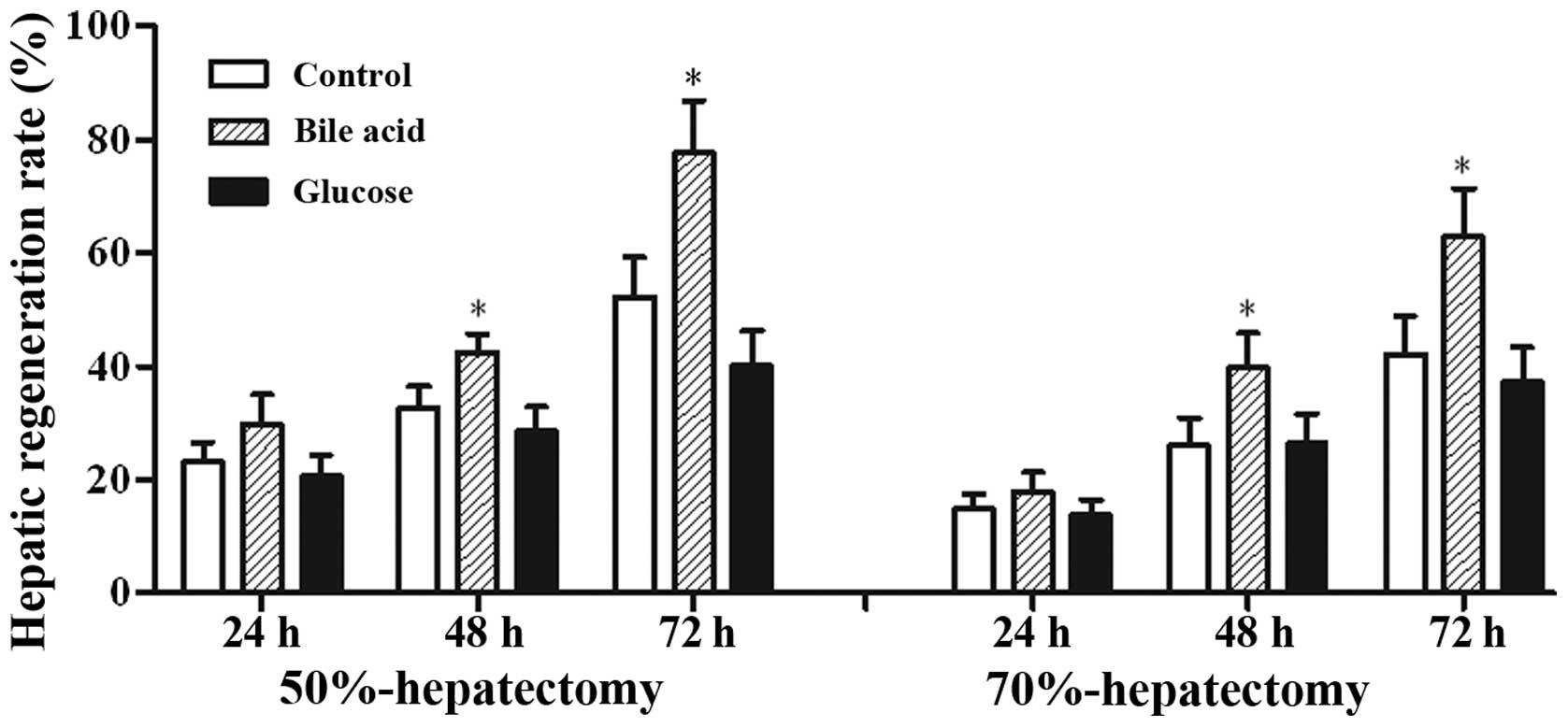
The restituted liver mass in the bile acid group was
markedly increased, with a peak at 72 h, and was higher than that
in the control group. By contrast, the hepatic regeneration rate of
the rats fed with glucose was reduced compared with the control
group (Fig. 1). The calculated
regeneration rate of rats that survived 70% liver resection was
less than that in the 50% resection group (survival, n=6 per
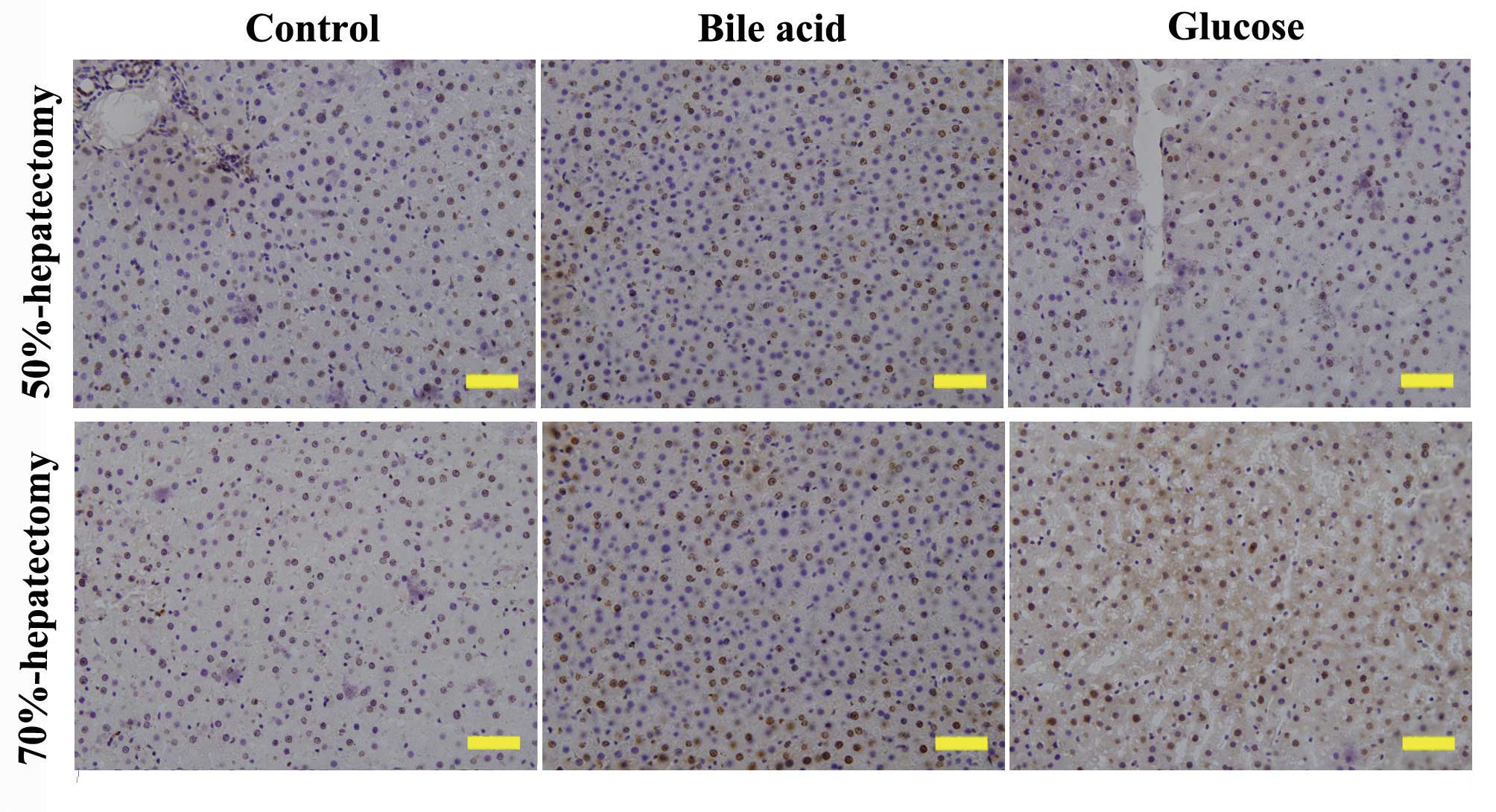
group). PCNA is a protein marker of DNA synthesis that is involved
in the initiation of cell proliferation. The expression of PCNA is
correlated with the S-phase of the cell cycle. At 72 h following
hepatectomy, the expression of PCNA was highest in the bile acid
group (Fig. 2), which was in
accordance with the results of the hepatic regeneration rate.
Expression of FXR and Caveolin-1 in
liver
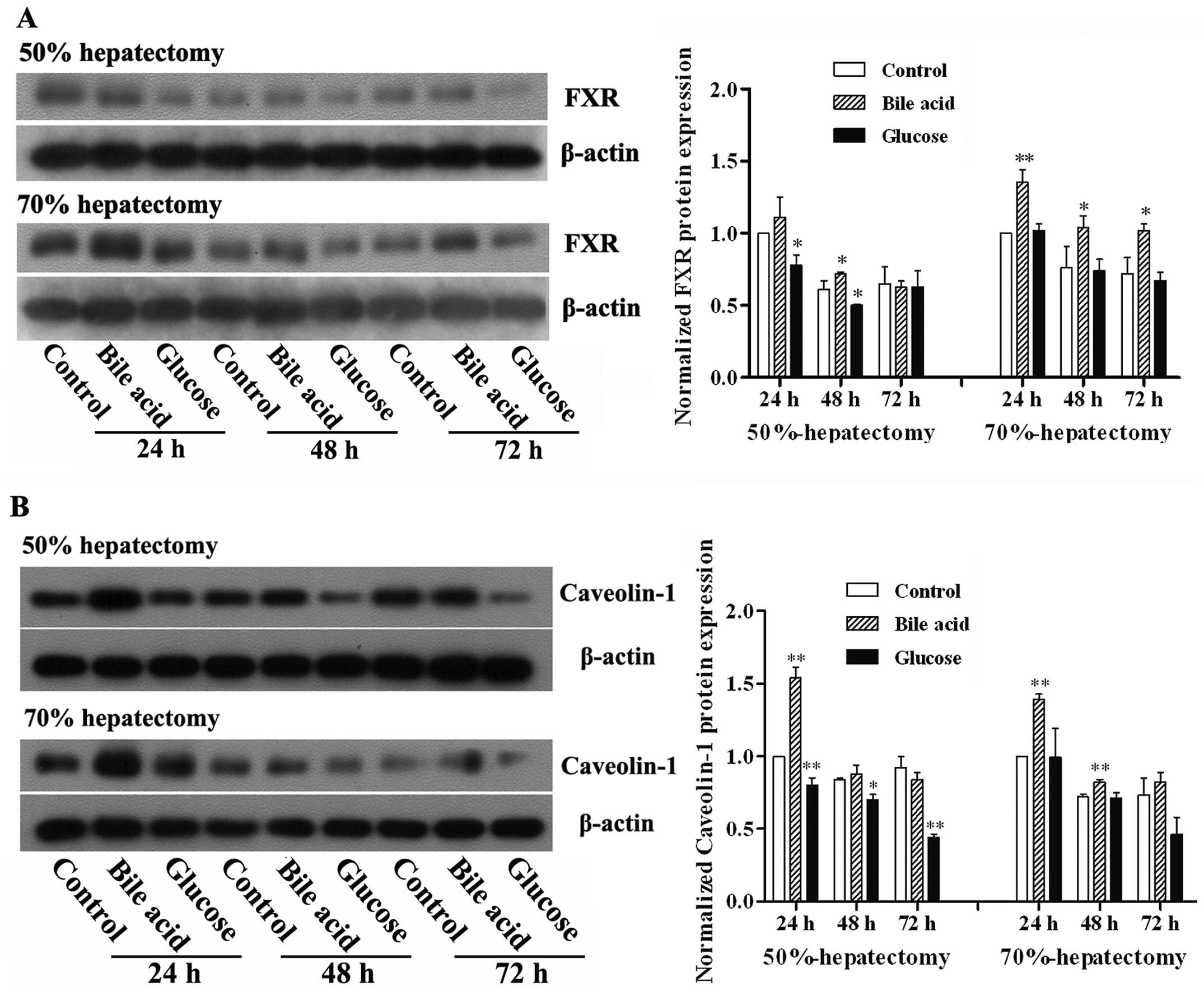
FXR is known to be an important receptor for bile
acid and its activation affects lipid and glucose metabolism
(14). Caveolin-1 is involved in
the regulation of intracellular homeostasis, such as lipid
metabolism, cell activation and cell proliferation (15). Based on the results of the initial
experiments, which demonstrated the facilitation of hepatic
regeneration by bile acid, the effect of bile acid on the
expression of FXR and Caveolin-1 was investigated. As shown in
Fig. 3, the protein expression of
FXR and Caveolin-1 was elevated in response to administration of
bile acid. By contrast, treatment with glucose decreased the
expression of FXR and Caveolin-1.
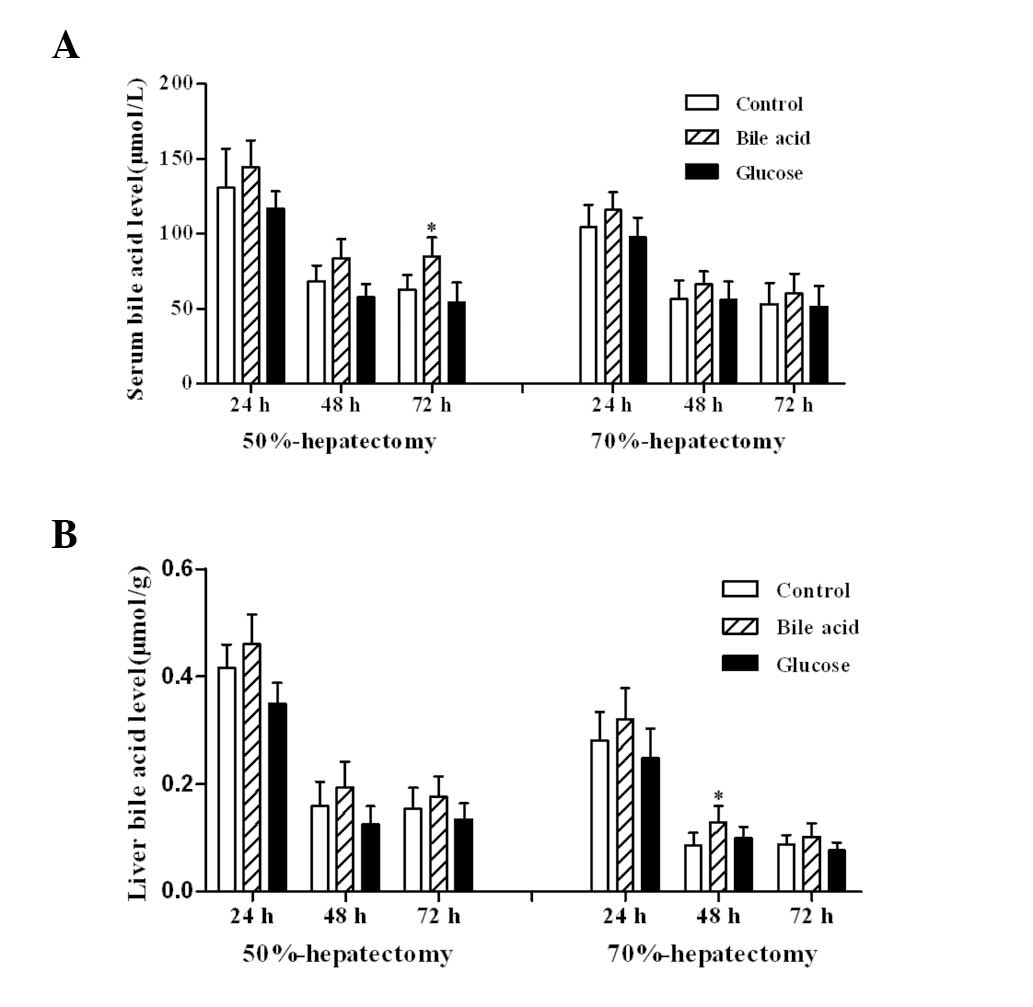
Bile acid levels following
hepatectomy
Serum and liver bile acid levels were similar to
those measured in a previous study (16). Oral administration of 0.2% bile
acid for 7 days significantly increased the concentration of serum
and liver bile acid, whereas the concentration decreased in the
glucose-fed group (Fig. 4).
Changes in levels of lipid
metabolism-related factors in hepatocytes in vitro
In order to investigate the effects of bile acid and
FXR on lipid metabolism in hepatocytes, TG, TC, HDL, LDL and FFA
levels in liver cells were measured (Table I). Levels of TG, LDL and FFA were
reduced in the bile acid and FXR agonist groups, whilst they were
increased in the glucose and FXR antagonist groups. By contrast,
levels of TC and HDL were increased in the bile acid and FXR
agonist groups, and decreased in the glucose and FXR antagonist
groups. These differences were statistically significant
(P<0.05).
 | Table IChanges of lipid metabolism related
factors in hepatocytes in vitro. |
Table I
Changes of lipid metabolism related
factors in hepatocytes in vitro.
| Group | TG (μg) | TC (mmol) | HDL
(μmol) | LDL
(μmol) | FFA
(μmol) |
|---|
| Control | 36.59±0.70 | 0.66±0.03 | 1.15±0.02 | 0.90±0.01 | 1.67±0.03 |
| Bile acid | 20.85±0.35b | 0.81±0.05a | 2.58±0.07b | 0.70±0.01b | 1.21±0.02b |
| Glucose | 42.58±1.87a | 0.45±0.05b | 0.96±0.10a | 0.92±0.02 | 1.62±0.18 |
| FXR agonist | 17.50±0.64b | 0.90±0.00b | 2.77±0.10b | 0.73±0.02b | 1.24±0.05b |
| FXR antagonist | 53.37±2.32b | 0.34±0.07b | 0.83±0.04b | 1.09±0.00b | 2.03±0.01b |
| Bile acid+FXR
agonist | 16.03±0.59b | 0.94±0.03b | 2.93±0.05b | 0.67±0.02b | 1.12±0.04b |
| Bile acid+FXR
antagonist | 48.29±2.51b | 0.43±0.05b | 0.96±0.07a | 1.04±0.03b | 1.92±0.07b |
| Glucose+FXR
agonist | 25.13±1.13b | 0.69±0.07 | 1.75±0.10b | 0.73±0.02b | 1.23±0.04b |
| Glucose+FXR
antagonist | 58.45±1.88b | 0.20±0.00b | 0.69±0.03b | 1.12±0.00b | 2.12±0.02b |
Expression of FXR signaling-related
proteins in hepatocytes
In order to verify the mechanism by which bile acid
and FXR affect lipid metabolism, the expression of FXR signaling-
and lipid metabolism-associated proteins was determined. As shown
in Fig. 5, the expression of bile
salt export pump (BSEP), Caveolin-1 and small heterodimer partner
(SHP) were elevated in response to bile acid and FXR agonist,
whereas that of apical sodium-dependent bile acid transporter
(ASBT), Na+/taurocholate cotransporting polypeptide and
cholesterol 7α-hydroxylase (NTCP) and CYP7A1 were downregulated.
The effects of glucose and the FXR antagonist on FXR signaling and
lipid metabolism proteins were the opposite of those of bile acid
and the FXR agonist.
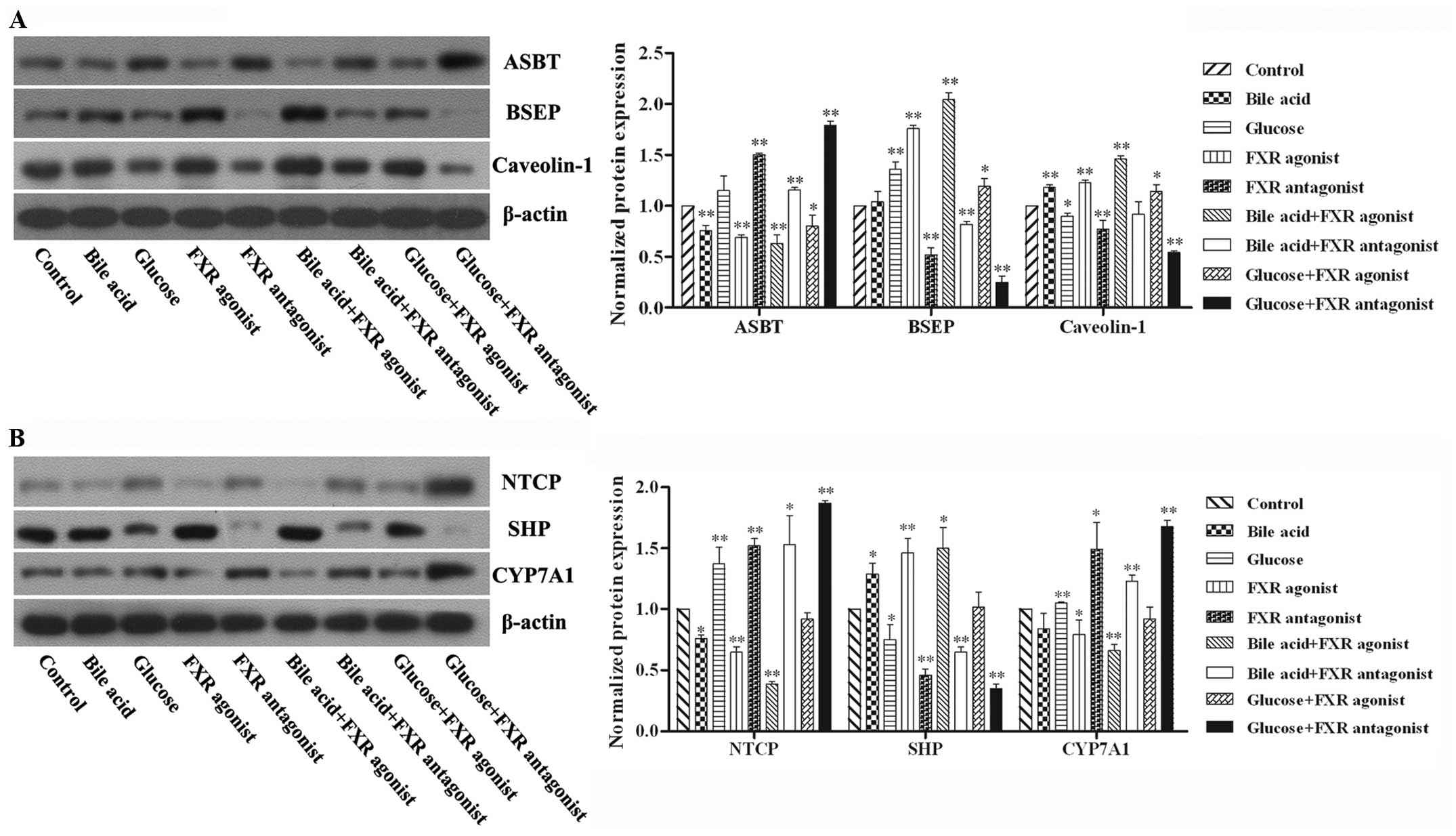 | Figure 5FXR signaling- and lipid
metabolism-associated proteins in hepatocytes. Hepatocytes isolated
from the rats were treated with bile acid, glucose, FXR agonist or
antagonist for 72 h. Expression of (A) ASBT, BSEP, Caveolin-1
protein and (B) NTCP, SHP, CYP7A1 in the hepatocytes were detected
using western blotting. Normalized expression was analyzed by
gradation. Data are presented as the mean ± standard deviation,
n=6, *P<0.05 and **P<0.01 compared with
control. FXR, farnesoid X receptor; ASBT, apical sodium-dependent
bile acid transporter; BSEP, bile salt export pump; NTCP,
Na+/taurocholate cotransporting polypeptide and
cholesterol 7α-hydroxylase; SHP, small heterodimer partner. |
Discussion
Liver regeneration is crucial for patients who
undergo partial hepatectomy or liver transplantation. It is
therefore important to investigate methods of improving the
capacity of the liver to regenerate in response to damage. A number
of factors are involved in liver regeneration, including a variety
of cytokines and growth factors. Previous studies have shown that
bile acid and FXR are required for early liver regeneration
(17,18). In vitro studies have
demonstrated that physiological concentrations of bile acid promote
hepatocyte proliferation (19).
However, excess bile acid cause degeneration and necrosis of liver
cells (19,20). In one study, fatalities occurred in
mice that had been fed with 1% bile acid and subjected to 70%
hepatectomy, suggesting that bile acid not only failed to promote
liver regeneration, but was likely to be cytotoxic, perhaps as a
result of the higher dose to that employed in the present study
(21). Furthermore, the mRNA
expression of FXR in 2/3 hepatectomy rats was significantly
increased, and the hepatic regeneration rate of FXR-knockout mice
subjected to 70% hepatectomy, was shown to be inhibited (11). In addition, FXR alleviated
age-related proliferation defects by activating Fork head Box m1b
transcription in regenerating mouse livers (22), and was also shown to regulate liver
repair following CCl4-induced toxic injury (23). Borude et al (24) demonstrated that hepatocyte-specific
deletion of FXR delayed, but did not completely inhibit, liver
regeneration following partial hepatectomy, by delaying cyclin D1
activation. Another study suggested that hepatic-FXR and
intestinal-FXR participate in the promotion of liver regeneration
and repair in mice (25). All
studies have shown that bile acid and FXR are important in the
process of liver regeneration. Our study showed that 0.2% bile acid
significantly increased the liver growth of rats that had undergone
hepatectomy and that this result was reversed in the glucose group,
as indicated by the level of expression of PCNA. PCNA is a marker
for DNA synthesis that acts as a scaffold for DNA-related enzymes
by encircling dsDNA and is commonly used as an indicator of cell
proliferation (26,27). In addition, 0.2% bile acid
increased the serum and liver bile acid levels, demonstrating that
bile acid is associated with improved liver regeneration following
major hepatectomy. The protein levels of FXR and Caveolin-1 were
found to be elevated in the bile acid group and reduced in the
glucose group. Caveolin-1 is a structural protein of caveolae,
which is a subtype of cholesterol-enriched lipid microdomains that
are usually observed as vesicles pinching off from the plasma
membrane (28). Caveolin-1 has
previously been reported to regulate lipid metabolism, apoptosis
and endocytosis in cells (29,30).
In the present study, administration of bile acid upregulated the
expression of caveolin-1 and the hepatic regeneration rate, which
was consistent with previous research.
This study provided evidence that bile acid and FXR
are involved in liver regeneration. TG, TC, HDL, LDL and FFA levels
in liver cells were then compared in vitro. Primary cultured
hepatocytes from rats were treated with bile acid, glucose, FXR
agonist or FXR antagonist, and changes in lipid metabolism factors
were measured. The results indicated that bile acid and FXR
agonists reduced the levels of TG, LDL and FFA, and increased the
levels of TC and HDL. In the subsequent study, examining the
expression of FXR signaling-related proteins in hepatocytes, the
mechanisms underlying the effects of bile acid and FXR on lipid
metabolism and liver regeneration were investigated. Activated FXR
stimulated expression of SHP, which integrated with the liver
receptor homolog and inhibited transcription of CYP7A1 (31,32).
Bile acid is known to be a regulator of cysteine sulfinic acid
decarboxylase via mechanisms shared in part with CYP7A1, and may
affect cholesterol via CYP7A1 through the downregulation of the
hepatic FXR/SHP pathway (33,34).
Bile acid levels were shown to be increased with an increased
expression of BSEP and the expression of NTCP and ASBT are also
known to be involved in the regulation of bile acid metabolism
(35,36,37).
In the present study, the expression of BSEP, Caveolin-1 and SHP
significantly increased, while that of ASBT, NTCP and CYP7A1
decreased, in accordance with previous studies.
In conclusion, the current study demonstrated that
bile acid and FXR are involved in the regulation of liver
regeneration, and may affect the lipid metabolism and
glycometabolism of the liver. In view of these properties, it is
possible that bile acid regulates energy metabolism through FXR
signaling pathways and that physiological concentrations of bile
acid promote liver regeneration.
Acknowledgments
This study was supported by a grant from the
National Natural Science Foundation of China (grant no.
81141047).
References
|
1
|
Fausto N, Campbell JS and Riehle KJ: Liver
regeneration. Hepatology. 43(2 Suppl 1): S45–S53. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mohammed FF and Khokha R: Thinking outside
the cell: proteases regulate hepatocyte division. Trends Cell Biol.
15:555–563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gerbino A, Ranieri M, Lupo S, et al:
Ca2+-dependent K+ efflux regulates
deoxycholate-induced apoptosis of BHK-21 and Caco-2 cells.
Gastroenterology. 137:955–964. e951–e952. 2009. View Article : Google Scholar
|
|
4
|
Drudi Metalli V, Mancino MG, Mancino A, et
al: Bile salts regulate proliferation and apoptosis of liver cells
by modulating the IGF1 system. Dig Liver Dis. 39:654–662. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Staudinger JL, Woody S, Sun M and Cui W:
Nuclear-receptor-mediated regulation of drug- and
bile-acid-transporter proteins in gut and liver. Drug Metab Rev.
45:48–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carlberg C and Seuter S: Dynamics of
nuclear receptor target gene regulation. Chromosoma. 119:479–484.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Forman BM, Goode E, Chen J, et al:
Identification of a nuclear receptor that is activated by farnesol
metabolites. Cell. 81:687–693. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang YD, Chen WD, Moore DD and Huang W:
FXR: a metabolic regulator and cell protector. Cell Res.
18:1087–1095. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gadaleta RM, van Mil SW, Oldenburg B,
Siersema PD, Klomp LW and van Erpecum KJ: Bile acids and their
nuclear receptor FXR: Relevance for hepatobiliary and
gastrointestinal disease. Biochim Biophys Acta. 1801:683–692. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peterson DF, Coote JH, Gilbey MP and
Futuro-Neto HA: Differential pattern of sympathetic outflow during
upper airway stimulation with smoke. Am J Physiol. 245:R433–R437.
1983.PubMed/NCBI
|
|
11
|
Xing X, Burgermeister E, Geisler F, et al:
Hematopoietically expressed homeobox is a target gene of farnesoid
X receptor in chenodeoxycholic acid-induced liver hypertrophy.
Hepatology. 49:979–988. 2009. View Article : Google Scholar
|
|
12
|
Lefebvre P, Cariou B, Lien F, Kuipers F
and Staels B: Role of bile acids and bile acid receptors in
metabolic regulation. Physiol Rev. 89:147–191. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
El-Shenawy NS: Effects of insecticides
fenitrothion, endosulfan and abamectin on antioxidant parameters of
isolated rat hepatocytes. Toxicol In Vitro. 24:1148–1157. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matsubara T, Li F and Gonzalez FJ: FXR
signaling in the enterohepatic system. Mol Cell Endocrinol.
368:17–29. 2013. View Article : Google Scholar
|
|
15
|
Fernández MA, Albor C, Ingelmo-Torres M,
et al: Caveolin-1 is essential for liver regeneration. Science.
313:1628–1632. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hoekstra LT, van Lienden KP, Schaap FG,
Chamuleau RA, Bennink RJ and van Gulik TM: Can plasma bile salt,
triglycerides, and apoA-V levels predict liver regeneration? World
J Surg. 36:2901–2908. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Csanaky IL, Aleksunes LM, Tanaka Y and
Klaassen CD: Role of hepatic transporters in prevention of bile
acid toxicity after partial hepatectomy in mice. Am J Physiol
Gastrointest Liver Physiol. 297:G419–G433. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Otao R, Beppu T, Isiko T, et al: External
biliary drainage and liver regeneration after major hepatectomy. Br
J Surg. 99:1569–1574. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Perez MJ and Briz O: Bile-acid-induced
cell injury and protection. World J Gastroenterol. 15:1677–1689.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kren BT, Rodrigues CM, Setchell KD and
Steer CJ: Modulation of steady-state messenger RNA levels in the
regenerating rat liver with bile acid feeding. Liver Transpl.
7:321–334. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Huang X, Meng Z, et al:
Significance and mechanism of CYP7a1 gene regulation during the
acute phase of liver regeneration. Mol Endocrinol. 23:137–145.
2009. View Article : Google Scholar :
|
|
22
|
Chen WD, Wang YD, Zhang L, et al:
Farnesoid X receptor alleviates age-related proliferation defects
in regenerating mouse livers by activating forkhead box m1b
transcription. Hepatology. 51:953–962. 2010.
|
|
23
|
Meng Z, Wang Y, Wang L, et al: FXR
regulates liver repair after CCl4-induced toxic injury. Mol
Endocrinol. 24:886–897. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Borude P, Edwards G, Walesky C, et al:
Hepatocyte-specific deletion of farnesoid X receptor delays but
does not inhibit liver regeneration after partial hepatectomy in
mice. Hepatology. 56:2344–2352. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang L, Wang YD, Chen WD, et al:
Promotion of liver regeneration/repair by farnesoid X receptor in
both liver and intestine in mice. Hepatology. 56:2336–2343. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dionne I, Brown NJ, Woodgate R and Bell
SD: On the mechanism of loading the PCNA sliding clamp by RFC. Mol
Microbiol. 68:216–222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hlinkova V, Xing G, Bauer J, et al:
Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring
assembly and opening. Acta Crystallogr D Biol Crystallogr.
64:941–949. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Woudenberg J, Rembacz KP, van den Heuvel
FA, et al: Caveolin-1 is enriched in the peroxisomal membrane of
rat hepatocytes. Hepatology. 51:1744–1753. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fernández-Rojo MA, Restall C, Ferguson C,
et al: Caveolin-1 orchestrates the balance between glucose and
lipid-dependent energy metabolism: implications for liver
regeneration. Hepatology. 55:1574–1584. 2012. View Article : Google Scholar
|
|
30
|
Meyer C, Liu Y, Kaul A, Peipe I and Dooley
S: Caveolin-1 abrogates TGF-beta mediated hepatocyte apoptosis.
Cell Death Dis. 4:e4662013. View Article : Google Scholar
|
|
31
|
Li G, Thomas AM, Hart SN, Zhong X, Wu D
and Guo GL: Farnesoid X receptor activation mediates head-to-tail
chromatin looping in the Nr0b2 gene encoding small heterodimer
partner. Mol Endocrinol. 24:1404–1412. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Anakk S, Watanabe M, Ochsner SA, McKenna
NJ, Finegold MJ and Moore DD: Combined deletion of FXR and SHP in
mice induces Cyp17a1 and results in juvenile onset cholestasis. J
Clin Invest. 121:86–95. 2011. View
Article : Google Scholar :
|
|
33
|
Kerr TA, Matsumoto Y, Matsumoto H, et al:
Cysteine sulfinic acid decarboxylase regulation: A role for
farnesoid X receptor and small heterodimer partner in murine
hepatic taurine metabolism. Hepatol Res. 44:E218–E228. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matsui S, Yamane T, Takita T, Oishi Y and
Kobayashi-Hattori K: The hypocholesterolemic activity of Momordica
charantia fruit is mediated by the altered cholesterol- and bile
acid-regulating gene expression in rat liver. Nutr Res. 33:580–585.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu QN, Xie HM, Zhang D, Liu J and Lu YF:
Hepatic bile acids and bile acid-related gene expression in
pregnant and lactating rats. Peer J. 1:e1432013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Miura T, Kimura N, Yamada T, et al:
Sustained repression and translocation of Ntcp and expression of
Mrp4 for cholestasis after rat 90% partial hepatectomy. J Hepatol.
55:407–414. 2011. View Article : Google Scholar
|
|
37
|
Hoang MH, Houng SJ, Jun HJ, et al: Barley
intake induces bile acid excretion by reduced expression of
intestinal ASBT and NPC1L1 in C57BL/6 J mice. J Agric Food Chem.
59:6798–6805. 2011. View Article : Google Scholar : PubMed/NCBI
|