Introduction
Approximately 12–15% of couples of reproductive age
worldwide are sterile and approximately half of these cases are due
to male factors (1). In at least
one-third of male infertility cases, the cause is elusive (2) and is referred to as idiopathic
infertility. Idiopathic male infertility predominantly manifests as
oligozoospermia or azoopermia. When more than 30% of the
spermatozoa have DNA damage, the fecundity of an individual
decreases significantly in vivo. Thus, screening sperm for
DNA damage is necessary in male patients with idiopathic
infertility and patients pursuing assisted reproduction (3).
MicroRNAs (miRNAs/miRs) are non-coding RNA molecules
approximately 22 nucleotides long, which regulate
post-transcriptional gene expression by binding to the 3′
untranslated region (UTR) of their target mRNA (4,5).
miRNAs affect a series of physiological activities involved in cell
proliferation through their combination with target genes, causing
mRNA degradation or translation inhibition. Recent studies in the
field of reproduction indicate that miRNA is an important regulator
of spermatogenesis. A study on the Dicer-dependent pathway
demonstrated that miRNA loss has pernicious effects on male
fertility (6). A further study
indicated that Dicer-deleted primordial germ cells and
spermatogonia exhibit proliferation disorders, and spermatogenesis
is suppressed during the early stage of proliferation and early
differentiation in Dicer-deleted testis (7). These studies suggested a role for
certain miRNAs in reproduction. For instance, miR-383 is a negative
regulator of cell proliferation, miR-383-pRb pathways are
associated with spermatogenesis (8), and miRNA-372 and miR-373 are
oncogenes in testicular germ cell tumors (9).
miR-145, located at 5q32–33, is an anti-oncogene,
which targets several tumor-associated genes that are involved in
tumor growth, metastasis and tumor angiogenesis. miR-145 regulates
the expression of the pluripotency factors OCT4, sex-determining
region Y Box (SOX)2 and -9 as well as KLF4, which also have
oncogenic features (10). Sachdeva
et al (11) reported that
miR-145 inhibits the expression of proto-oncogene c-Myc via p53.
Another study by Chiyomaru et al (12) illustrated that miR-145 expression
is markedly restrained in tumor organization, wherein FSCN1 is
overexpressed.
The transcription factor SOX9 is located at
17q24.3-q25.1 and is expressed in the heart, brain, kidneys,
prostate, testicles and other organs in human adults. The SOX9 gene
is expressed in the brain and testis, as well as during the resting
or reproductive stages of the perichondrium of the fetus. SOX9
mutations are associated with sex reversal and SOX9 affects the
development of bones and testicles through expression in
mesoblastomas.
According to a report on miRNA chips, miR-145 is
downregulated among altered miRNA in the testicular tissues of
patients with non-obstructive azoospermia (NOA) (13). Among all species, miR-145 contains
a unique seed sequence which is conserved in Xenopus, zebrafish and
various mammals (14,15). In living organisms, Homo
sapiens (hsa)-miR-145 is abundant in the uterus, ovaries,
testes, prostate and heart, which are all mesoderm-derived tissues
(16). A previous study also
revealed that SOX9 expression differs between normal and infertile
samples based on DNA arrays, and SOX9 is required for Sertoli cell
maturation and normal spermatogenesis (17). However, the association between
SOX9 and miR-145 in reproductive cells has remained to be
elucidated, and the function of hsa-miR-145 in human testes
requires further study. In the present study, the testicular
embryonic carcinoma cell line NTERA-2 (NT2) and a normal testis
cell line, Hs 1.tes (18,19) were used to investigate the function
of hsa-miR-145 during spermatogenesis by identifying its target
genes.
Materials and methods
Database analysis
Through miRBase (http://www.mirbase.org/) (20), it was confirmed that the sequence
of hsa-miR-145 was 16-GUCCAGUUUUCCCAGGAAUCCCU-38. A search using
TargetScan (http://www.targetscan.org/), miRBase and pictar
(http://pictar.mdc-berlin.de/) revealed
that the putative 3′UTR of SOX9 did not complement miR-145
(21) (Table I).”
 | Table ITarget validation of the SOX9 3′UTRs
by TargetScan, miRBase and Pictar. |
Table I
Target validation of the SOX9 3′UTRs
by TargetScan, miRBase and Pictar.
| Gene | Target sites | Position |
|---|
| SOX9 | 5′ …
UUUUUGUUGAAAACAAACUGGAA
3′.…. UCCCUAAGGACCCUUUUGACCUG | 1402–1409 |
Construction of miR-145 expression
plasmid
A total of two single-stranded DNA sequences were
designed based on the hsa-miR-145 sequence in the miRBase database.
The hsa-miR-145 expression recombinant and a control plasmid were
constructed by the cloning of annealed oligonucleotides of
hsa-miR-145 (sense, 5′-GATCCCCGTCCAGTTTTCCCAGGAATCCCTTTTTTT
GTCGACA-3′ and antisense, 5′-AGCTTGTCGACAAAAAA
AGGGATTCCTGGGAAAACTGGACGGG-3′), or control (sense,
5′-GATCCCCTTCTCCGAACGTGTCACGTATTA TTTTTTGTCGACA-3′ and antisense,
5′-AGCTTGTCGACAA AAAATAATACGTGACACGTTCGGAGAAGGG-3′) into pGenesil-1
plasmid. The recombinants were transformed into Escherichia
coli DH5α (TransGen, Beijing, China) competent cells, which
were cultured in lysogeny broth (10 g/l tryptone, 5 g/l yeast
extract, 10 g/l NaCl; Beyotime Institute of Biotechnology,
Shanghai, China) at 37°C. Restriction enzymes SalI and
PstI were used in the initial survey and DNA sequencing was
conducted to identify the correct plasmid (22). DNA sequencing was completed by
Sangon Biotech Co., Ltd (ABI3730xl; Shanghai, China).
Cell culture
NT-2 [American Type Culture Collection (ATCC),
Manassas, VA, USA; CRL-1973™] and Hs 1.tes (ATCC; CRL-7002™) were
grown in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Logan,
UT, USA) with high glucose, supplemented with 10% fetal bovine
serum (FBS; Gibco-BRL, Gaithersburg, MD, USA) and 1% antibiotics
(100 U/ml penicillin and 100 mg/ml streptomycin). The cells were
cultured at 37°C with 5% CO2.
Recombinant plasmid transfection
Transfection of NT-2 and Hs 1.tes cells was
performed using Lipofectamine 2000 reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions. When the cell density in the six orifice plates
reached 80%, the cells were transfected with 5 ng
pGenesil-1-miR-145 or 5 ng pGenesil-1-miR-NC in each orifice. The
plasmids with pGenesil-1-miR-145 or pGenesil-1-miR-NC were
extracted from E. coli using an Endofree plasmid minikit
(Omega, Tarzana, CA, USA). Epifluorescence imaging of cells was
carried out on an inverted epifluorescence microscope (Nikon Ti-E
fluorescent microscope; Nikon, Tokyo, Japan). At 48 h after
transfection, the cells were collected for RNA and protein
extraction to be used for further analyses.
RNA isolation
Total RNA was isolated from the collected NT-2 or Hs
1.tes cells using a Total RNA kit II (Omega) according to the
manufacturer’s instructions. Spectrophotometry (Eppendorf
BioSpectrometer® basic; Eppendorf, Hamburg, Ger many)
was used for RNA quantification. Optical density (OD) was measured
at 260 and 280 nm. The OD260/OD280 ratio was used to estimate RNA
purity, and the RNA with OD ratios of 1.8–2.0 were used for
subsequent studies (23).
miRNA detection
Reverse transcription quantitative polymerase chain
reaction (RT-qPCR) analysis of miR-145 was performed in analogy
with methods used in previous studies (24,25).
PCR was performed on cDNA generated from 2 μg total RNA
using the protocol of the All-in-One™ miRNA RT-qPCR Detection kit
(GeneCopoeia, Rockville, MD, USA). A 25-μl reaction mixture
containing 2 μg total RNA, 1 μl 2.5 U/μl
polymerase A, 1 μl RTase mix and 5 μl reaction buffer
was prepared according to the manufacturer’s instructions. The
cycle parameters for the RT reaction were 37°C for 60 min and 85°C
for 5 min. Subsequently, 0.4 μl RT product was PCR-amplified
in 20 μl qPCR reaction mixture containing 10 μl 2X
All-in-One qPCR mix (GeneCopoeia), 0.2 μM All-in-One qPCR
Primer hsa-miR-145 (GeneCopoeia) and the hsnRNA U6 control
(GeneCopoeia). The amplification parameters for RT and qPCR were
set according to the manufacturer’s instructions of the All-in-One
miRNA RT-qPCR Detection kit using a Slan-96S real-time PCR system
(Shanghai Hongshi Medical Technology Co., Ltd, Shanghai, China).
The amplification parameters were as follows: 95°C for 3 min,
followed by 40 cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C
for 16 sec. hsnRNA U6 was used as a control to normalize the
expression levels of each miRNA. The relative expression levels
were obtained using the 2−ΔΔCT method, with the relative
gene expression=2−(∆Ct sample-∆Ct control). All
experiments were performed in triplicate.
RT-qPCR analysis
To determine the expression levels of SOX9
(NC_018928.1) in Hs 1.tes cells, RT-qPCR was performed using the
All-in-One First-Strand cDNA Synthesis kit and All-in-One qPCR Mix
(GeneCopoeia), followed by RT-qPCR using SYBR Green (GeneCopoeia).
GAPDH (NM_002046.4) was used as the control. A 25-μl
reaction mixture containing 1 μg total RNA, 10 μM
random primer, 1 U/μl RNase inhibitor, 8 U/μl M-MLV
RTase, 1 mM dNTP and 5 μl reaction buffer was prepared
according to the manufacturer’s instructions. The cycling
parameters for the RT reaction were 37°C for 60 min and 85°C for 5
min. Subsequently, a reaction mixture containing 10 μl 2X
All-in-One qPCR Mix and the appropriate primers was added to 2
μl cDNA template to a final reaction volume of 20 μl.
The amplification parameters were 95°C for 5 min, followed by 30
cycles of 95°C for 10 sec, 62.8°C for 20 sec and 72°C for 16 sec.
PCR was performed using a Slan-96S real-time PCR system. Data were
analyzed using the 2−ΔΔCT method. The primers (Sangon
Biotech Co., Ltd) used for RT-qPCR were as follows: SOX9 forward,
5′-TGGTCTTTAACTCTGACCGTTACCT-3′ and reverse,
5′-TATTCCGGATCTTAATCAGAGAAAGTG-3′; GAPDH forward,
5′-ACGGATTTGGTCGTATTGGGC-3′ and reverse,
5′-CTCGCTCCTGGAAGATGGTGAT-3′
The expression levels of OCT4 (GenBank ID,
NM_002701.4), SOX2 (NM_003106.3), c-Myc (NM_002467.4), KLF4
(NM_004235.4) and FSCN1 (NM_003088.3) in NT-2 cells were determined
using an All-in-One First-Strand cDNA synthesis kit and an
All-in-One qPCR mix (GeneCopoeia). The RT reaction and RT-qPCR were
operated as described above. The melting temperature for OCT4,
SOX2, c-Myc and KLF4 was 60°C and the melting temperature for FSCN1
was 65°C. GAPDH was used as a control and data were analyzed using
the 2−ΔΔCT method. The primers (Sangon Biotech Co., Ltd)
used for RT-qPCR were as follows: OCT4 forward,
5′-CTGGGTTGATCCTCGGACCT-3′ and reverse, 5′-CACAGAACTCATACGGCGGG-3′;
SOX2 forward, 5′-CCCAGCAGACTTCACATGT-3′ and reverse,
5′-CCTCCCATTTCCCTCGTTTT-3′; c-Myc forward,
5′-CGTCTCCACACATCAGCACAA-3′ and reverse,
5′-TCTTGGCAGCAGGATAGTCCTT-3′; KLF4 forward,
5′-CAGCTCCCCAGCAGGACTACC-3′ and reverse,
5′-CATCTGAGCGGGCGAATTTC-3′; FSCN1 forward,
5′-CTGGCTACACGCTGGAGTTC-3′ and reverse, 5′-CTGAGTCCCCTGCTGTCTCC-3′.
All experiments were performed in triplicate.
Western blot analysis
GAPDH was used as the internal control. The cell
lysates were used for protein extraction. Infected cells were
washed three times with ice-cold phosphate-buffered saline
(Beyotime Institute of Biotechnology) and the cells were
resuspended in 100 μl radioimmunoprecipitation assay buffer
(Beyotime Institute of Biotechnology). Cells were incubated on ice
for 30 min, centrifuged at 15,000 × g and the supernatant was
collected. Following measuring the protein concentration using an
ELISA, the equivalent denatured protein samples were mixed with
loading buffer. The samples were resolved using 10% SDS-PAGE and
transferred onto a polyvinylidene fluoride membrane via the wet
transfer method. Following blocking for 1 h in 5% nonfat dried milk
(Yili Industrial Group Co., Ltd, Inner Mongolia, China) in
Tris-buffered saline and Tween 20 (TBST; Amresco, Solon, OH, USA)
at room temperature, the blots were incubated overnight with rabbit
polyclonal anti-SOX9 (BS1597; Bioworld Technology, Nanjing, China;
diluted 1:1,000) or mouse monoclonal anti-GAPDH (200306; Zen
Bioscience, Chengdu, China; diluted 1:5,000) antibodies at 4°C for
15. Following washing three times in TBST, the blots were incubated
with horseradish peroxidase (HRP)-conjugated secondary antibodies
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA; diluted
1:2,000) at room temperature for 1 h. The blots were visualized
using Immobilon Western chemiluminescent HRP substrate (Millipore,
Billerica, MA, USA) following the manufacturer’s instructions. A
Kodak 440CF imaging system (Kodak, Tokyo, Japan) was used to
visualize the blots.
Cell proliferation assay
Growth inhibition of NT-2 cells was determined via
MTT cell viability assays using MTT Cell Proliferation kit
(Solarbio, Beijing, China). The NT-2 cells were cultured in 96-well
plates at an initial number of 1×104 cells/well in DMEM
supplemented with 10% FBS. Following transfecting the recombinant
plasmids for 48 h at 37°C, 10 μl MTT reagent was added and allowed
to react for 4 h in an incubator. A total of 110 μl formazan
reagent was added, which was allowed to react for 10 min. Substrate
cleavage was monitored at 490 nm via ELISA. Experiments were
performed in triplicate.
Cell apoptosis assay
Cell apoptosis was detected using Annexin
V-allophycocyanin (APC)/7-aminoactinomycin D (7-AAD) double
staining and flow cytometric analysis. NT-2 cells were cultured in
six-well plates. The NT-2 cells were transfected with
pGenesil-1-miR-145 and pGenesil-1-miR-NC. After 48 h, the cells
were collected and manipulated according to the instructions of the
Annexin V-APC/7-AAD apoptosis detection kit (KeyGen Biotech Co.,
Ltd, Nanjing, China). A total of 500 μl binding buffer was
added to make a cell suspension. 5 μl Annexin V-APC was
added, followed by 5 μl 7-AAD. The mixture was allowed to
react in the dark for 15 min at room temperature. After 1 h, the
mixture was analyzed via flow cytometry (BD FACSCalibur; BD
Biosciences, San Jose, CA, USA).
Statistical analysis
The data were analyzed using SPSS 19.0 software
(IBM, Armonk, N Y, USA). A t-test was used for statistical
comparisons between groups, whereas a one-way analysis of variance
was used for comparisons among multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Identification of miR-145 expression
vector and control construct
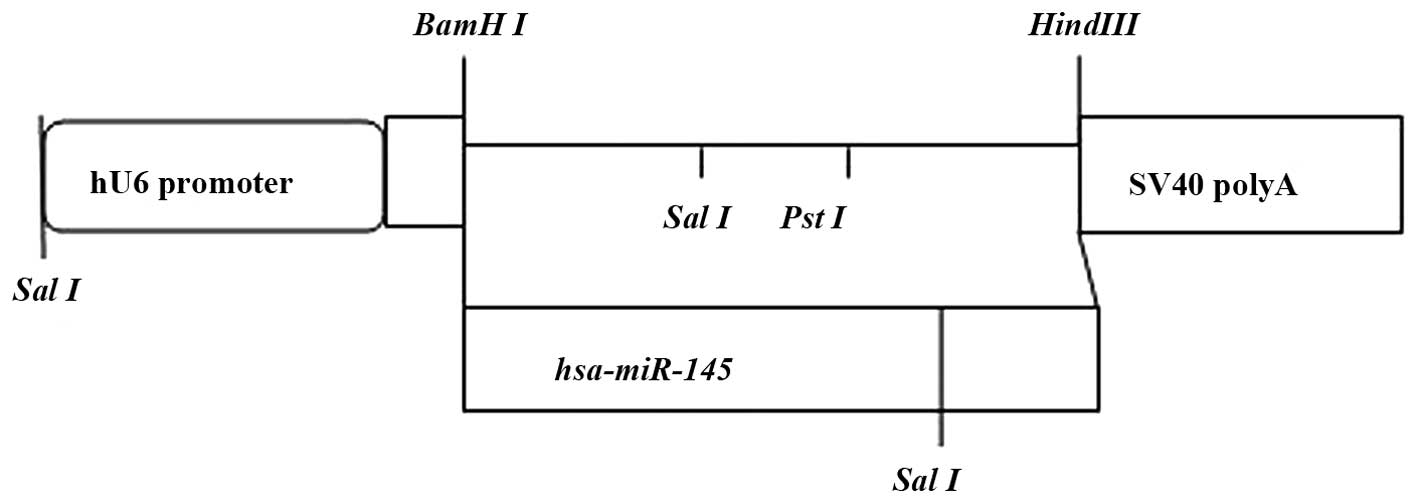
Plasmid pGenesil-1 is a eukaryotic expression vector
containing a U6 promoter, an enhanced green fluorescent protein
(EGFP) gene and an anti-kanamycin gene. Multiple cloning sites were
located behind U6, including the restriction enzyme sites
BamHI, HindIII, SalI, and PstI.
SalI and PstI are between BamHI and
HindIII. Double digestion was used with restriction enzymes
BamHI and HindIII to obtain a linear fragment, which
was inserted into SalI to verify the recombinant plasmids (Fig. 1). The correct plasmid was
identified via electrophoresis. SalI digestion produced a
350-bp fragment, whereas PstI digestion did not. DNA
sequencing confirmed that the plasmids were reconstructed
successfully. The samples were retested through sequencing by
Sangon Biotech (Shanghai, China).
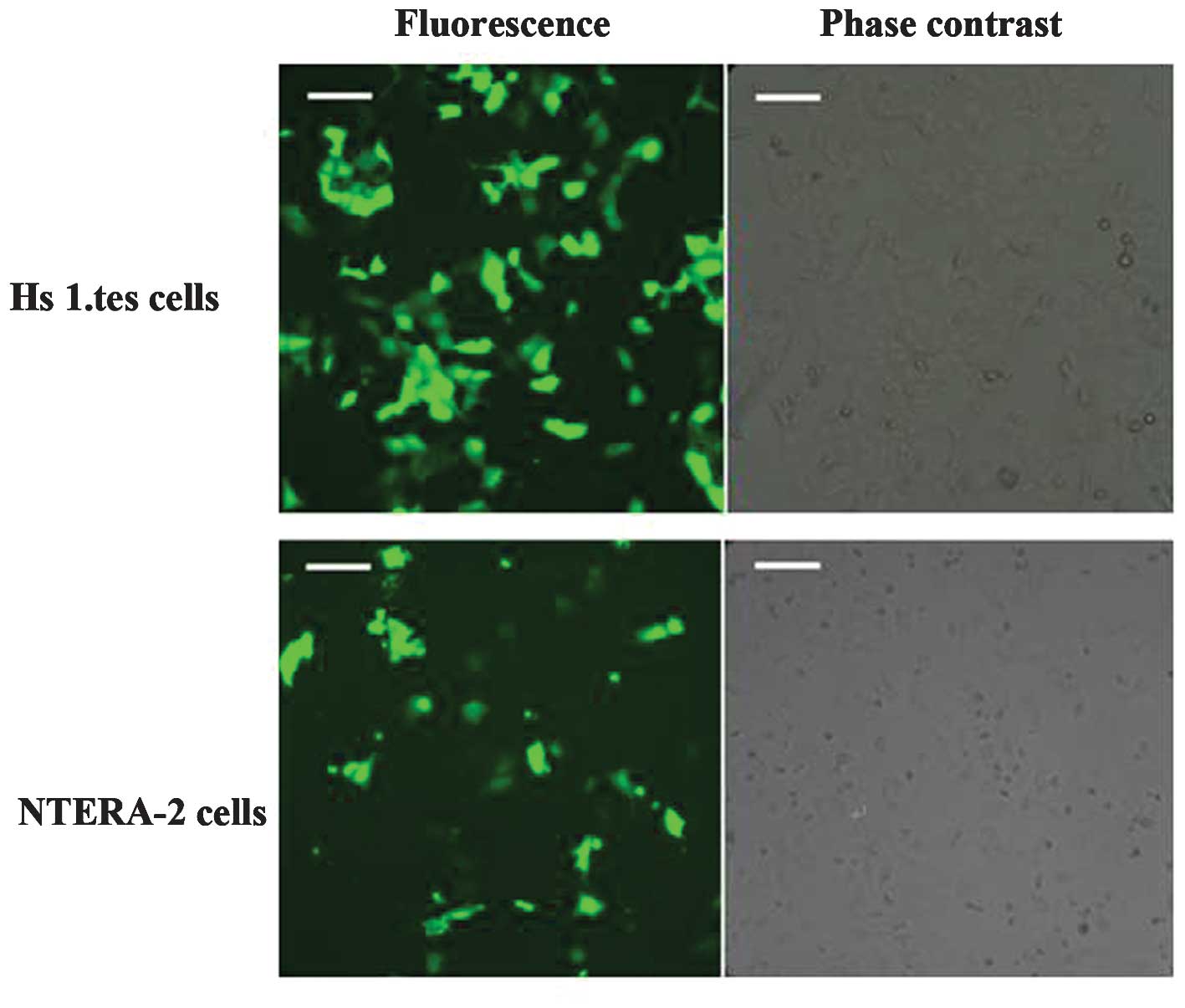
Transfection
The pGenesil-1-miR-145 and pGenesil-1-control
constructs with enhanced green florescence protein were transfected
into the NT-2 cell line and the Hs 1.tes cell line, respectively.
The constructs exhibited green fluorescence under inverted
fluorescence microscopy. Fluorescence was observed under
fluorescence microscopy in cells containing pGenesil-1-miR-145 and
pGenesil-1-miR-NC within 24 h after transfection. No fluorescent
signal was observed in the blank control group (Fig. 2).
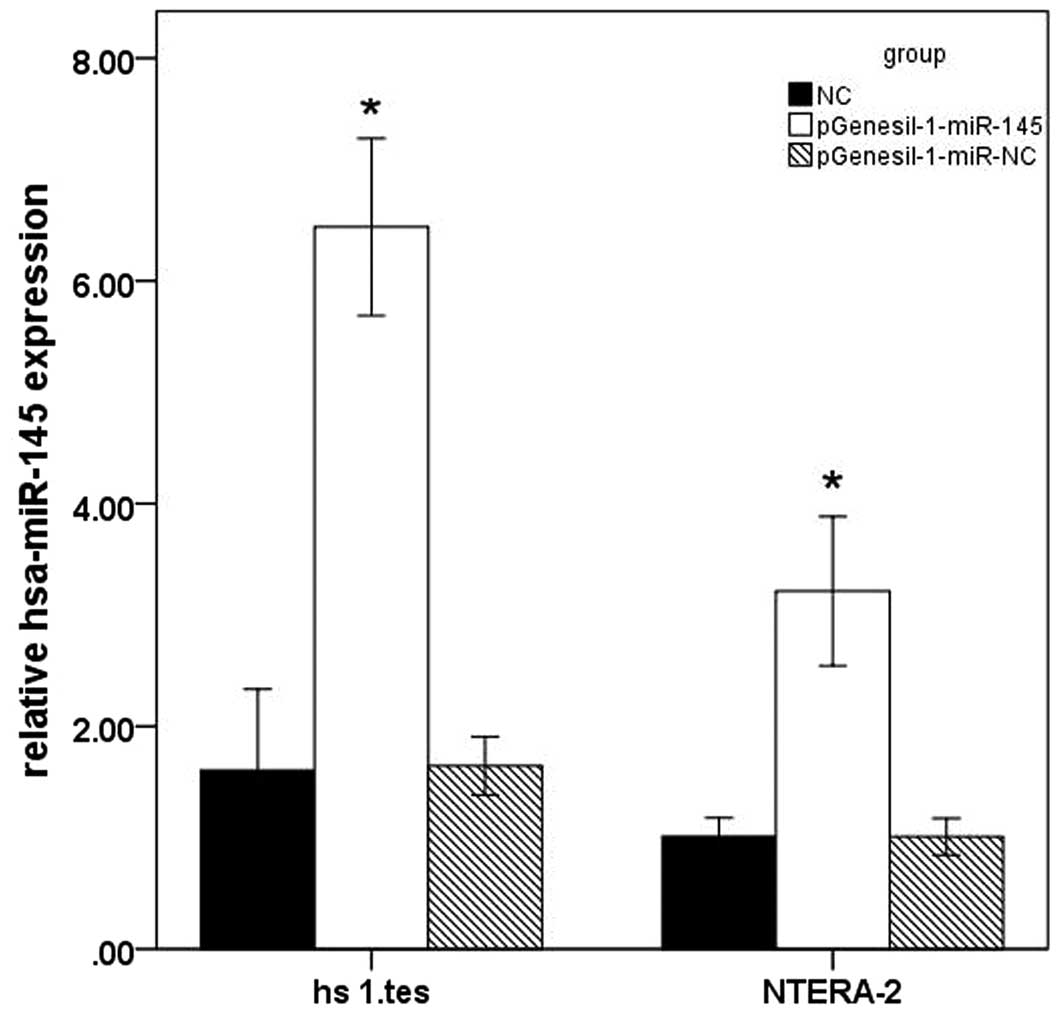
miR-145 overexpression in
pGenesil-1-miR-145-transfected cells
The pGenesil-1-miR-145-transfected groups and
pGenesil-1-miR-NC-transfected groups were analyzed at 48 h after
transfection. miR-145 expression of the NT-2 and Hs 1.tes cells in
the control and the transfected groups were analyzed via RT-qPCR
(Fig. 3). The mRNA levels were
normalized to the internal control U6. Statistical analysis
revealed that the miR-145 expression levels in the NT-2 and Hs
1.tes cells in pGenesil-1-miR-145 groups were higher than those in
the control and the pGenesil-1-miR-NC groups (P<0.05).
pGenesil-1-miR-NC had no effect on miR-145 expression in NT-2 and
Hs 1.tes cells (P>0.05, vs. control).
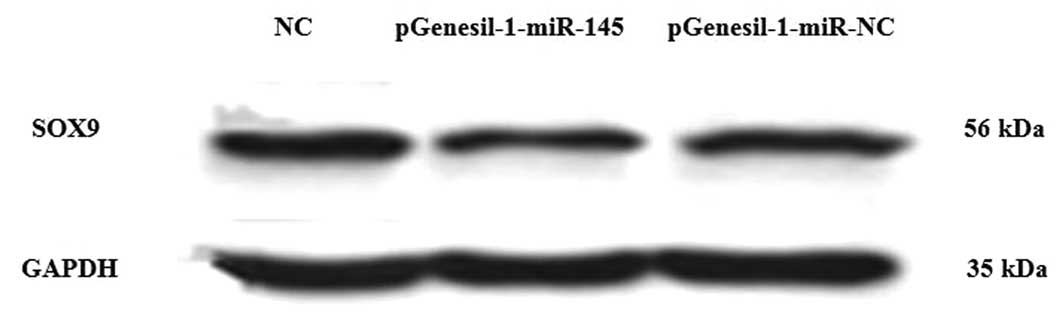
miR-145 inhibits SOX9 expression in Hs
1.tes cells at the mRNA and protein level
To demonstrate that miR-145 acts as an inhibitor of
SOX9 protein expression, pGenesil-1-miR-145 and pGenesil-1-miR-NC
were transfected into Hs 1.tes cells. Western blot analysis
revealed that SOX9 protein levels were markedly lower in the cells
with miR-145 overexpression than those in the negative control
(Fig. 4). The relative protein
expression of SOX9 in Hs 1.tes cells was as follows: NC group,
0.88±0.02; pGenesil-1-miR-145 group, 0.59±0.01; and
pGenesil-1-miR-NC group, 0.82±0.03. RT-qPCR analysis demonstrated
that the SOX9 mRNA levels were decreased in the cells with miR-145
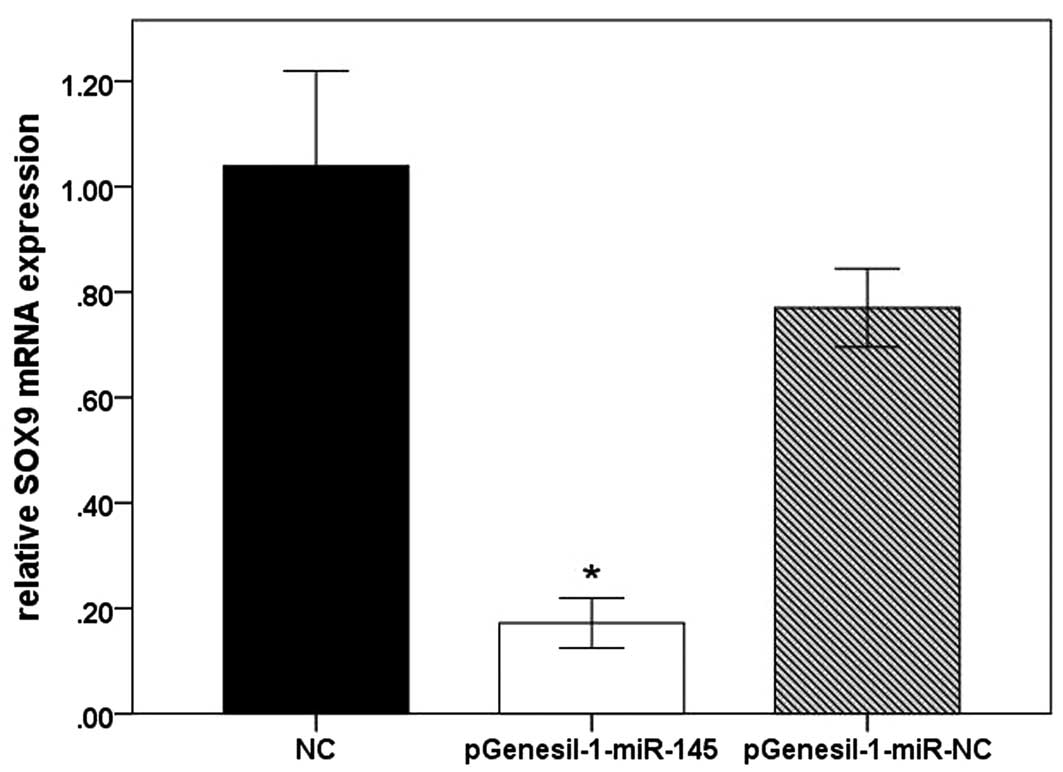
overexpression (Fig. 5). The
relative mRNA expression of SOX9 in Hs 1.tes cells was as follows:
NC group, 1.03±0.73; pGenesil-1-miR-145 group, 0.17±0.02; and
pGenesil-1-miR-NC group, 0.77±0.03. SOX9 had no marked change in
the mRNA and protein levels in the pGenesil-1-miR-NC-transfected
cells. Thus, the results demonstrated that miR-145 inhibited the
protein and mRNA expression of SOX9 in Hs 1.tes cells.
miR-145 inhibits the expression of OCT4,
SOX2, c-Myc, KLF4, and FSCN1 at the mRNA level in NT-2 cells
To investigate the effect of miR-145 in germ cell
neoplasms, it was determined whether miR-145 inhibited the
expression of endogenous OCT4, SOX2, KLF4, c-Myc and FSCN1 in NT-2
cells. miR-145 was upregulated and the mRNA levels of these genes
among three groups were detected. RT-qPCR revealed that the mRNA
levels of OCT4, SOX2, c-Myc and FSCN1, but not KLF4, were decreased
in the pGenesil-1-miR-145 group (Table II). However, no significant
differences were detected in the expression of these genes in
pGenesil-1-miR-NC-transfected cells.
 | Table IIExpression levels of OCT4, SOX2,
c-Myc, KLF4 and FSCN1 mRNA. |
Table II
Expression levels of OCT4, SOX2,
c-Myc, KLF4 and FSCN1 mRNA.
| Gene/group | NC |
pGenesil-1-miR-145 |
pGenesil-1-miR-NC |
|---|
| OCT4 | 1.00±0.14 | 0.60±0.15a | 0.80±0.05 |
| SOX2 | 1.11±0.29 | 0.22±0.05a | 0.84±0.10 |
| c-Myc | 1.00±0.07 | 0.43±0.14a | 0.89±0.14 |
| KLF4 | 1.02±0.24 | 0.78±0.15 | 0.97±0.27 |
| FSCN1 | 1.01±0.21 | 0.15±0.08a | 0.99±0.17 |
Inhibition of proliferation by miR-145 in
NT-2 cells
To analyze the biological effect of upregulated
miR-145 expression, the growth of NT-2 cells treated with
pGenesil-1-miR-145 was investigated using an MTT assay (Table III). At 48 h after transfection,
it was observed that pGenesil-1-miR-145 significantly reduced the
growth of Hs 1.tes and NT-2 cells. By contrast, the growth in the
control group exhibited no significant changes.
 | Table IIINTERA-2 cell proliferation inhibition
by transfection with recombinants for 48 h. |
Table III
NTERA-2 cell proliferation inhibition
by transfection with recombinants for 48 h.
| Group | Absorbance (A) | Inhibition
ratio |
|---|
| NC | 0.22±0.07 | – |
|
pGenesil-1-miR-145 | 0.12±0.03a | 45.45% |
|
pGenesil-1-miR-NC | 0.21±0.09 | 4.45% |
Promotion of NT-2 cell apoptosis by
miR-145
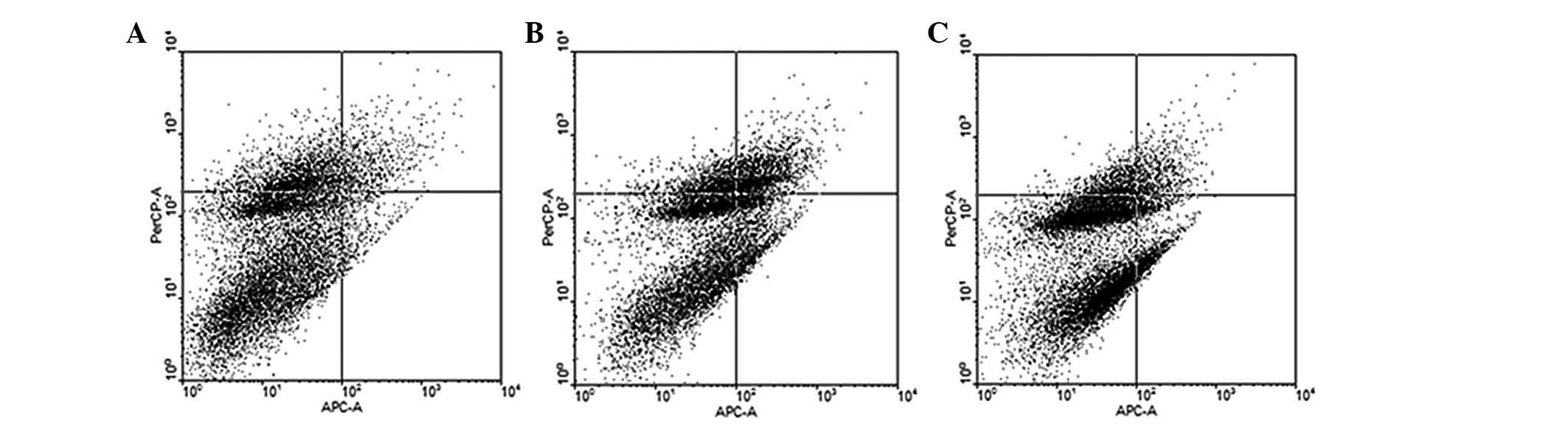
The results of the flow cytometric analysis are
shown in Fig. 6. The apoptotic
rate was 9.53±0.63% in the NC group, 30.22±0.56% in the
pGenesil-1-miR-145 group and 12.54±1.89% in the pGenesil-1-miR-NC
group. The apoptotic rate was significantly higher in the
pGenesil-1-miR-145 group compared with that in the NC group.
Discussion
miRNAs are small, endogenous molecular regulators of
gene expression that have critical roles in the body (26,27).
In early studies on the biological characteristics of miRNAs, the
pH1-RNApuro plasmid was used as a recombinant construct for
upregulating miRNA expression (22). The pGenesil-1 plasmid was selected,
which carries a U6 promoter, a kanamycin resistance gene and an
EGFP gene, which is used for selection following transfection. The
structure of pGenesil-1-miR-145 was determined via DNA sequencing
and restriction enzyme digestion. RT-qPCR detection indicated that
miR-145 is upregulated in the pGenesil- 1 -miR-145-transfected
group, which suggested that pGenesil--miR-145 should be used for
further investigation.
Spermatogenesis is a complex process, which involves
the development of spermatogonial stem cells into highly
differentiated spermatozoon. This process includes two stages:
Active proliferation of spermatogonia and meiosis of spermatocytes.
Certain studies have demonstrated the accelerated germ cell
apoptosis and decreased mitotic activity of spermatogonia in
infertile men during the spermatocyte stage (28,29).
A recent study revealed that normal active spermatogonia in testes
are arrested at the pachytene stage (30). This spermatogenic failure may be
due to meiotic failure, but the mechanism of genetic defects in
spermatogenesis remains to be elucidated (31). A microarray assay was used to
demonstrate the altered expression of different miRNAs, including
miR-145 in the testes of patients with NOA. The microarray assay
revealed that aberrant miRNA expression is associated with male
infertility. As a fertility biomarker, comparison of the DNA array
of abnormal human testis samples with normal human testis samples
exhibited SOX9 gene overexpression (17). The transcription factor SOX9, which
is required for Sertoli cell maturation and normal spermatogenesis,
regulates steroidogenic factor-1 promoter activity in Sertoli cells
(32). Studies on chondrogenic
differentiation demonstrate that miR-145 is a negative regulator
through directly targeting SOX9 during the early stage of
chondrogenic differentiation (33). Altered hsa-miR-145 expression in
testicular cells is involved in the regulation of target genes
associated with male infertility. In the present study, recombinant
plasmids were constructed, which express pGenesil-1-miR-145 to
transfect normal Hs 1.tes cells. The results of the RT-qPCR and
western blot analyses demonstrated that miR-145 suppressed SOX9
mRNA and protein expression in transiently transfected Hs 1.tes
cells compared with that in pGenesil-1-miR-NC-mediated cells and
untreated Hs 1.tes cells (P<0.05). The present results coincide
with those of previous studies, which indicated that miR-145
downregulates SOX9 protein expression. SOX9 mRNA expression was
also decreased in the present study. miR-145 inhibition during SOX9
transcription was greater than that during translation. This
difference may be due to miR-145 regulating the expression of
numerous genes, which subsequently inTERAct with SOX9.
Previous studies have revealed that miR-145 is
downregulated in numerous types of human cancer and its
transfection reduces cell proliferation in tumor cell lines
(34–39). Earlier studies revealed that
miR-145 directly targets the proto-oncogene c-Myc and insulin
receptor substrate-1, which are associated with cell proliferation
(11,40). In bladder cancer, miR-145 directly
controls FSCN1, which functions mainly in cortical cell
protrusions, which mediate inTERActions between cells and the
extracellular matrix, cell-to-cell inTERActions and cell migration.
FSCN1 also forms cytoplasmic microfilamentous bundles, which
contribute to cell architecture and intracellular movements
(12). In the present study,
pGenesil-1-miR-145 was transfected into NT-2 cells and it was
demonstrated that miR-145 overexpression significantly reduces the
growth of NT-2 cells. Flow cytometric analysis revealed that the
apoptotic rate increased in the pGenesil-1-miR-145 group. RT-qPCR
revealed that the mRNA levels of c-Myc and FSCN1 decreased in the
pGenesil-1-miR-145 transfection group.
miR-145s function by directly targeting the
pluripotency factors OCT4, SOX2 and KLF4 to inhibit the
pluripotency of stem cells and control embryonic stem cell
differentiation. miR-145 and OCT4 form a double-negative feedback
loop and the promoter of miR-145 is inhibited by OCT4 (10,41).
In the present study, RT-qPCR revealed that miR-145 inhibited the
mRNA expression of OCT4 and SOX2, but not that of KLF4. Earlier
studies revealed that the genes, which preserve stem cell
properties, including OCT4, SOX2, KLF4 and Nanog, have oncogenic
characteristics and are involved in tumor development. These
results demonstrated that miR-145 directly regulates the biological
properties of tumor cells, as well as regulating cancer stem
cells.
In conclusion, recombinant pGenesil-1-miR-145
significantly inhibited the mRNA and protein expression of SOX9 in
human testicular cells. These data have important implications in
studies on spermatogenesis. Differentially expressed molecules may
be used as biomarkers to provide insights into the mechanisms
underlying male infertility. The results of the present study may
assist in developing a gene therapy for azoospermia. miR-145
inhibits the expression of oncogenes, such as OCT4, SOX2, c-Myc and
FSCN1, as well as the proliferation of testicular embryonal
carcinoma cells. Thus, upregulating miR-145 expression is a
potential alternative treatment for testicular germ cell tumors.
Further investigation into the inTERActions of miR-145 with other
target genes are required to understand the occurrence and
development of spermatogenesis and the suppression of malignant
germ cell tumors.
Acknowledgments
The authors are grateful to Dr Liu Ying and Dr Sun
Huaqin in the Joint Laboratory for Reproductive Medicine of Sichuan
University - The Chinese University of Hong Kong. The present study
was supported by funds from the Science and Technology Department
of Sichuan Province, China.
References
|
1
|
Zhu YJ, Liu SY, Wang H, Wei P and Ding XP:
The prevalence of azoospermia factor microdeletion on the Y
chromosome of Chinese infertile men detected by multi-analyte
suspension array technology. Asian J Androl. 10:873–881. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Evenson DP, Larson KJ and Jost LK: Sperm
chromatin structure assay: its clinical use for detecting sperm DNA
fragmentation in male infertility and comparisons with other
techniques. J Androl. 23:25–43. 2002.PubMed/NCBI
|
|
3
|
Agarwal A and Said TM: Role of sperm
chromatin abnormalities and DNA damage in male infertility. Hum
Reprod Update. 9:331–345. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stark A, Bushati N, Jan CH, Kheradpour P,
Hodges E, Brennecke J and Kellis M: A single Hox locus in
Drosophila produces functional microRNAs from opposite DNA strands.
Genes Dev. 22:8–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Papaioannou MD and Nef S: microRNAs in the
testis: Building up male fertility. J Androl. 31:26–33. 2010.
View Article : Google Scholar
|
|
7
|
Hayashi K, Chava de Sousa Lopes SM, Kaneda
M, Tang F, Hanjkova P, Lao K, Surani MA, et al: MicroRNA biogenesis
is required for mouse primordial germ cell development and
spermatogenesis. PLoS One. 3:e17382008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lian J, Tian H, Liu L, Zhang XS, Li WQ,
Deng YM and Sun F: Downregulation of microRNA-383 is associated
with male infertility and promotes testicular embryonal carcinoma
cell proliferation by targeting IRF1. Cell Death Dis. 1:e942010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop N, Nagel R, Agami R, et al: A genetic screen implicates
miRNA-372 and miRNA-373 as oncogenes in testicular germ cell
tumors. Cell. 124:1169–1181. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu N, Papagiannakopoulos T, Pan G, Thomson
JA and Kosik KS: MicroRNA-145 regulates OCT4, SOX2 and KLF4 and
represses pluripotency in hum an embryonic stem cell. Cell.
137:647–658. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sachdeva M, Zhu S, Wu F, Walia V, Kumar S,
Elble R, Watabe K and Mo YY: p53 represses c-Myc through induction
of the tumor suppressor miR-145. Proc Natl Acad Sci USA.
106:3207–3212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chiyomaru T, Enokida H, Tatarano S,
Kawahara K, Uchiada Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N
and Nakagawa M: miR-145 and miR-133a function as tumour suppressors
and directly regulate FSCN1 expression in bladder cancer. Br J
Cancer. 102:883–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lian J, Zhang X, Tian H, Liang N, Wang Y,
Liang CZ, Li X and Sun F: Altered microRNA expression in patients
with non-obstructive azoospermia. Reprod Biol Endocrinol. 7:132009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Michael MZ, O’Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific MicroRNAs in colorectal neoplasia1. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
16
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rockett JC, Patrizio P, Schmid JE, Hecht
NB and Dix DJ: Gene expression patterns associated with infertility
in humans and rodent models. Mutat Res. 549:225–240. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee VM and Andrews PW: Differentiation of
NTERA-2 clonal human embryonal carcinoma cells into neurons
involves the induction of all three neurofilament proteins. J
Neurosci. 6:514–521. 1986.PubMed/NCBI
|
|
19
|
Kumi-Diaka J, Hassanhi M, Brown J,
Merchant K, Garcia C and Jimenez W: CytoregR inhibits growth and
proliferation of human adenocarcinoma cells via induction of
apoptosis. J Carcinog. 5:12006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Griffths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:140–144. 2006.
View Article : Google Scholar
|
|
21
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
Mitsudomi T and Takahashi T: Reduced expression of the let-7
microRNAs in human lung cancers in association with shortened
postoperative survival. Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cho WC, Chow AS and Au JS: Restoration of
tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung
adenocarcinoma patients with epidermal growth factor receptor
mutation. Eur J Cancer. 45:2197–2206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jung YH, Gupta MK, Shin JY, Uhm SJ and Lee
HT: MicroRNA signature in testes-derived male germ-line stem cells.
Mol Hum Reprod. 16:804–810. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu
Q, Deitch EA, Huo YQ, Delphin ES and Zhang C: MicroRNA-145, a novel
smooth muscle cell phenotypic marker and modulator, controls
vascular neointimal lesion formation. Circ Res. 105:158–166. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vamsi K: Gangaraju and Haifan Lin:
MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Bio.
10:116–125. 2009. View
Article : Google Scholar
|
|
27
|
Lakshmipathy U and Hart RP: Concise
review: MicroRNA expression in multipotent mesenchymal stromal
cells. Stem Cells. 26:356–363. 2008. View Article : Google Scholar
|
|
28
|
Lin WW, Lamb DJ, Lipshultz LI and Kim ED:
Demonstration of testicular apoptosis in human male infertility
states using a DNA laddering technique. Int Urol Nephrol.
31:361–370. 1999. View Article : Google Scholar
|
|
29
|
Steger K, Aleithe I, Behre H and Bergmann
M: The proliferation of spermatogonia in normal and pathological
human seminiferous epithelium: an immunohistochemical study using
monoclonal antibodies against Ki-67 protein and proliferating cell
nuclear antigen. Mol Hum Reprod. 4:227–233. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bar-Shira Maymon B, Yogev L, Yavetz H,
Lifschitz-Mercer B, Schreiber L, Kleiman SE, Botchan A, Hauser R
and Paz G: Spermatogonial proliferation patterns in men with
azoospermia of different etiologies. Fertil Steril. 80:1175–1180.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gonsalves J, Sun F, Schlegel PN, Turek PJ,
Hopps CV, Greene C, Martin RH and Pera RA: Defective recombination
in infertile men. Hum Mol Genet. 13:2875–2883. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schumacher V, Gueler B, Looijenga LH,
Becker JU, Amann K, Engers R, Dotsch J, Stoop H, Schulz W and
Royer-Pokora B: Characteristics of testicular dysgenesis syndrome
and decreased expression of SRY and SOX9 in frasier syndrome
regulation of the orphan nuclear receptor steroidogenic factor 1 by
sox proteins. Mol Reprod Dev. 75:1484–1494. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang B, Guo H, Zhang Y, Chen L, Ying D and
Dong S: MicroRNA-145 regulates chondrogenic differentiation of
mesenchymal stem cells by targeting SOX9. PLoS One. 6:e216792011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bandrés E, Cubedo E, Agirre X, Malumbres
R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M and
Garcia-Foncillas J: Identification by real-time PCR of 13 mature
microRNAs differentially expressed in colorectal cancer and
non-tumoral tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sempere LF, Christensen M, Silahtaroglu A,
Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S and Cole CN:
Altered microRNA expression confined to specific epithelial cell
subpopulations in breast cancer. Cancer Res. 67:11612–11620. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar
|
|
37
|
Szafranska AE, Davison TS, John J, Cannon
T, Sipos B, Maghnouj A, Labourier E and Hahn SA: MicroRNA
expression alTERAtions are linked to tumorigenesis and
non-neoplastic processes in pancreatic ductal adenocarcinoma.
Oncogene. 26:4442–4452. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ozen M, Creighton CJ, Ozdemir M and
Ittmann M: Widespread deregulation of microRNA expression in human
prostate cancer. Oncogene. 27:1788–1793. 2008. View Article : Google Scholar
|
|
39
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shi B, Sepp-Lorenzino L, Prisco M, Linsley
P, deAngelis T and Baserga R: MicroRNA-145 targets the insulin
receptor substrate-1 and inhibits the growth of colon cancer cells.
J Biol Chem. 282:32582–32590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tay Y, Zhang J, Thomson AM, Lim B and
Rigoutsos I: MicroRNAs to Nanog, Oct4 and Sox2 coding regions
modulate embryonic stem cell differentiation. Nature.
455:1124–1128. 2008. View Article : Google Scholar : PubMed/NCBI
|