Introduction
Telomerase is a ribonucleoprotein complex, which
catalyzes the de novo addition of TTAGGG nucleotide repeat
sequences to prevent telomere shortening at the distal ends of
eukaryotic chromosomes. While telomerase may maintain chromosomal
integrity and stability during division of actively dividing cells,
it also enables cell proliferation, making it one of the primary
factors leading to carcinogenesis. Telomerase activation has been
detected in the vast majority of human carcinomas and in
vitro immortalized cells with no detectable expression in
normal stable human somatic cells (1,2). In
light of the characteristics above, human telomerase is one of the
most promising tumor markers and a potentially highly specific
molecular target for therapeutic interventions (3). Among several protein components of
human telomerase, human telomerase reverse transcriptase (hTERT),
as a catalytic subunit of the telomerase enzyme complex, has been
observed to be the key determinant of enzymatic activity in human
telomerase (4). By synthesizing
multiple tandem repeats of DNA (namely telomeric DNA), hTERT,
encoded by the TERT gene, compensates for the erosion of DNA
ends during replication and provides docking sites for telomeric
proteins that bind specifically to the ends of chromosomes
(5). Mutations in the TERT
gene regions may affect telomerase activity and it was observed in
several studies, which performed TERT single-nucleotide
polymorphism (SNP) analysis, that this gene had a role in
susceptibility to tumorigenesis in multiple types of cancer
(2,6,7).
Prostate cancer (PCa) is a significant health
problem for older males, with an estimated 233,000 novel cases and
29,480 cancer-associated fatalities expected in 2014 in the United
States alone (8). The widespread
use of prostate-specific antigen (PSA) screening, which may result
in a decrease in PCa mortality, has led to the overdetection,
overtreatment and increasing costs of this highly heterogeneous
disease with diverse clinical outcomes (9,10).
Side effects due to overtreatment and their negative impact on the
patient’s quality of life justify the importance of sparing
patients from unnecessary treatment and the requirement for
specific markers indicating disease prognosis (11). Although factors such as the Gleason
score and tumor stage are used to assess prognosis, there remains a
requirement for improved biomarkers to distinguish between PCa
cases that may likely recur, progress rapidly and be
life-threatening versus those that may not have a substantial
impact on mortality (12).
In two recent studies it was suggested that
quantification of TERT expression may be a valuable
non-invasive marker for discriminating between localized and
locally advanced PCa, as well as a useful tool for the early
prediction of biochemical recurrence of PCa (9,13).
Another study using immunohistochemistry demonstrated that the
immunoreactivity of hTERT may be used as a molecular marker for
high-grade prostate cancer (14).
However, the role of TERT genetic variations in PCa
progression remains to be elucidated. An aim of the present study
was to therefore clarify the association between TERT locus
polymorphisms and PCa aggressiveness in an in-patient Chinese
patient cohort. To the best of our knowledge, the present study was
the first to evaluate the effect of these variations on PCa
severity.
Materials and methods
Study population
Between February 2010 and April 2013, PCa patients
who were between 34 and 97 years old at the time of diagnosis were
recruited from the Departments of Urology at Xinhua Hospital
(School of Medicine, Shanghai Jiao Tong University) and Huashan
Hospital (Fudan University) in Shanghai, China. The study protocol
was approved by the Science and Technology Commission of Shanghai
Municipality and institutional review boards of Xinhua Hospital and
Huashan Hospital. All subjects received a detailed description of
the study protocol and provided informed consent. All eligible
subjects included in the present study were of Chinese Han
ancestry. The general eligibility criteria were: i) Newly diagnosed
PCa cases with histologically confirmed disease; ii) ability of the
patient to comprehend informed consent and iii) no previous
diagnosis of cancer. The exclusion criteria included patients with
chronic inflammatory conditions, infections within the past six
weeks and autoimmune diseases. A total of 1,210 individuals who met
the criteria were selected for genotyping. The age at diagnosis was
calculated from the date of the first positive biopsy and the serum
PSA levels (defined as the most recent PSA value within 1 year
prior to the diagnosis date) were obtained from a medical record
review.
Histopathological grading of biopsies and radical
prostatectomy specimens were performed according to the Gleason
scoring system (15). Clinical and
pathological stages were determined according to the 2010 American
Joint Committee on Cancer (AJCC) tumor, nodes and metastasis (TNM)
classification system. For Gleason scores and tumor stage
information, values from prostatectomy were used whenever
available; otherwise, biopsy values were used. The D’Amico risk
classification criteria were used to predict the prognosis of
patients with localized PCa (16),
and patients in the present study were grouped as low-, moderate-
or high-risk for clinical recurrence and rapid progression
following primary therapy for PCa. Patients diagnosed with N1
(involvement of regional lymph nodes) or M1 (distant metastasis)
PCa were included in the high-risk class. In addition, due to
comparably small numbers of low- and moderate-risk PCa cases, they
were combined into a single non-aggressive group. Thus, a total of
911 high-risk (aggressive) and 259 low/moderate-risk
(non-aggressive) PCa cases were included in the present study. The
remaining 40 patients could not be classified due to absent
phenotypic data.
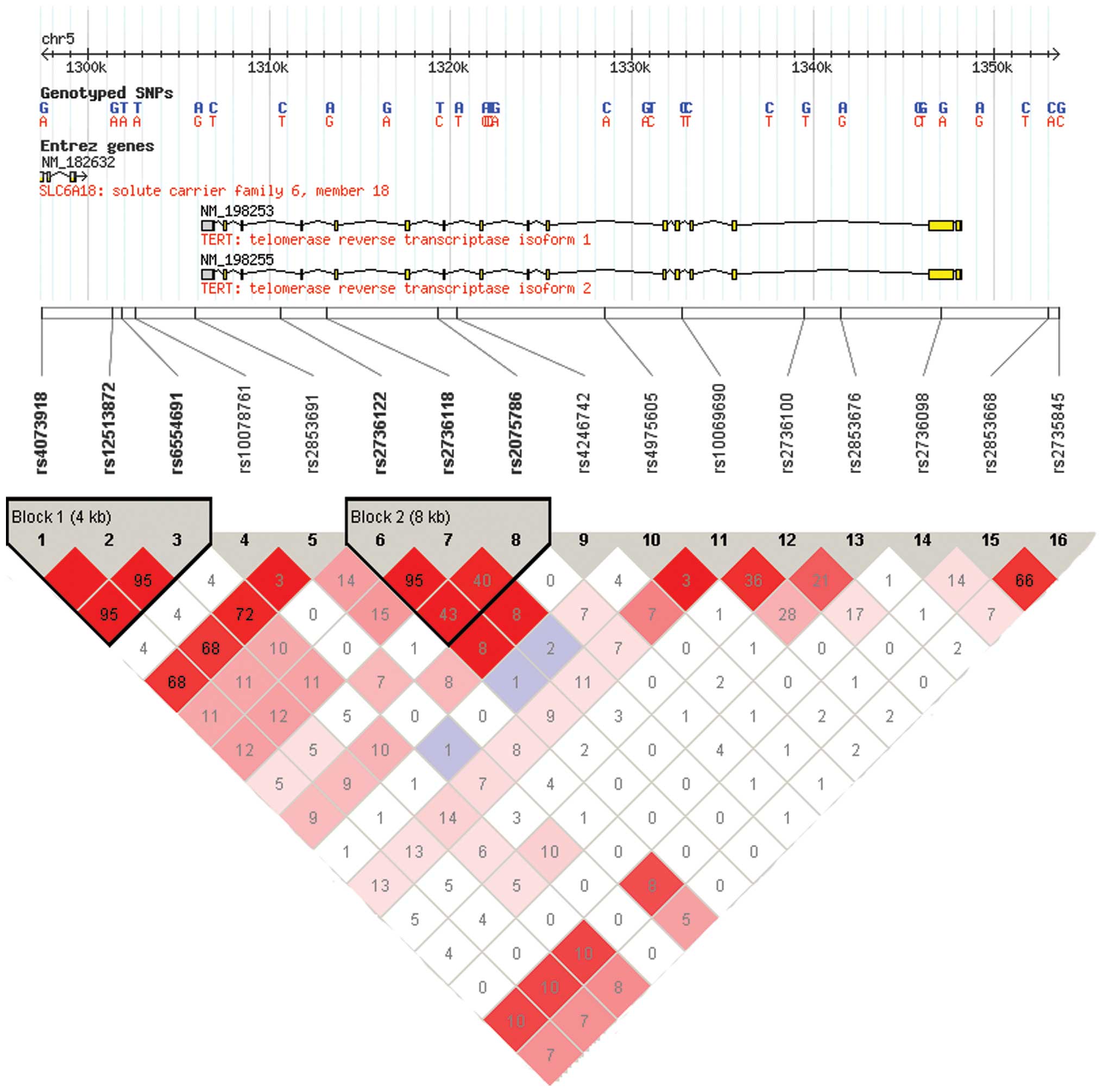
Selection of SNPs
A tagging approach was employed to perform a
comprehensive evaluation of genetic variants across the TERT
gene for their association with PCa aggressiveness. To include the
probable regulatory regions of the TERT gene, the upstream
of the initial gene region was extended for 10 kb and the
downstream for 10 kb and thus the peak signals were at 61.9 kb
(chr5:1, 296, 287–1, 358, 162, dbSNP b126). Subsequently, a greedy
algorithm was used, based on the r2 statistic to
identify tagging SNPs (tSNPs) using the Haploview program version
4.2 from the Broad Institute (http://www.broadinstitute.org/mpg/haploview) according
to the HapMap database (http://www.hapmap.org/, HapMap Data Rel 24/phaseII
Nov08, on NCBI B36 assembly, dbSNP b126; population: CHB+JPT) on
the basis of pairwise linkage disequilibrium (LD) r2
threshold of 0.8, Hardy-Weinberg Equilibrium (HWE)=0.05,
minor-allele frequency=0.01 and call rate=95%. As a result, two
SNPs (rs12513872 and rs6554691) in Block1 and one SNP (rs2736118)
in Block2 were excluded from the panel due to LD
(r2>0.8). In addition, one SNP (rs4246742) was
eliminated from the analysis due to difficulty in designing primers
for the genotyping assay. Finally, a total of 12 SNPs which met the
above criteria were analyzed for the present study (Fig. 1).
Genotyping
Blood samples were collected from all study subjects
and DNA was extracted using a whole blood genomic DNA extraction
kit (Qiagen, Chatsworth, CA, USA) and then diluted to the
concentration of 15–20 ng/l through the use of an ultraviolet
spectrophotometer (Nanodrop 8000; Thermo Fisher Scientific,
Waltham, MA, USA). Amplification of polymorphism flanking fragments
and single base extension were conducted by the polymerase chain
reaction (GeneAmp PCR Thermocycle Instrument 2720 and ABI PCR
Thermocycle Instrument 9700; Applied Biosystems, Inc., Carlsbad,
CA, USA). The 12 SNPs were genotyped for all subjects using a
MassARRAY iPLEX system (Sequenom, Inc., San Diego, CA, USA) at
Fudan University in Shanghai, China. A total of two duplicates and
two water samples were included in each 96-well plate as polymerase
chain reaction (PCR)-negative controls. All assays were performed
by technicians in a blinded manner. The average concordance rate
between samples was >99% among the duplicated quality control
samples and the genotyping missing rate was 2.5% for all
samples.
Statistical analysis
The genotype distribution for each tSNP was assessed
for the HWE using Pearson’s goodness-of-fit. To investigate the
association of genotypes with PCa aggressiveness (aggressive PCa
vs. non-aggressive PCa), the Gleason score (>7 vs. ≤7) and the
risk of developing early-onset PCa (≤60 vs. >60), the odds
ratios (ORs), 95% confidence intervals (CIs) as well as
corresponding P-values were calculated using unconditional logistic
regression with adjustment for age (as a continuous variable). Each
tSNP was analyzed using additive, dominant, recessive and
co-dominant models, respectively. All data were analyzed using
PLINK 1.07 software (17).
P-values were two-tailed and P<0.05 was considered to indicate a
statistically significant difference.
Results
Patient characteristics and clinical
features
The distribution of demographic characteristics and
clinical features of 1,210 PCa patients who were successfully
genotyped are presented in Table
I. The median age at diagnosis for these patients was 72 years
(range, 34–97 years). A total of 1,165 patients had available PSA
levels at diagnosis, with a median PSA of 6 ng/ml [Q1 (lower
quartile), (Q3 upper quartile): 5, 11 ng/ml]. Among 1,171 patients
with Gleason score information, 452 patients (38.6%) had Gleason
scores >7, 400 patients (34.2%) had scores of 7, and 319
patients (27.2%) had scores <7. Among patients who had AJCC
clinical stage information available, 648 patients (60.8%) had
organ-confined tumors (T1/T2) and 418 patients (39.2%) had
extraprostatic (T3/T4) disease. In addition, 291 patients (30.4%)
had lymphatic metastasis, while 330 patients (31.1%) had distant
metastasis. In total, sufficient information for the modified
criteria of the D’Amico risk classification was available for 1,170
patients, of whom 911 (77.9%) were high-risk and 259 (22.1%) were
low/moderate-risk.
 | Table IPatient characteristics and clinical
features. |
Table I
Patient characteristics and clinical
features.
| Patient
characteristic | Cases, n (%)
|
|---|
| High risk
(n=911) | Low/moderate risk
(n=259) | All cases
(n=1,210)a |
|---|
| Patients with
available age, n | 906 | 258 | 1,204 |
| Age at diagnosis
(years; mean ± SD) | 71.36±8.26 | 71.21±7.37 | 71.32±8.06 |
| ≤60, n (%) | 89 (9.8) | 19 (7.4) | 113 (9.39) |
| >60, n (%) | 817 (90.2) | 239 (92.6) | 1,091 (90.61) |
| Patients with PSA
levels at diagnosis, n (%) | 890 (76.4) | 256 (22.0) | 1,165 |
| 0–4 | 28 (2.4) | 22 (1.9) | 51 (4.38) |
| 4.01–10 | 61 (5.2) | 90 (7.7) | 156 (13.4) |
| 10.01–20 | 116 (10.0) | 144 (12.4) | 273 (23.4) |
| >20 | 685 (58.8) | 0 | 685 (58.8) |
| Patients with
Gleason score, n (%) | 883 (75.4) | 248 (21.2) | 1,171 |
| >7 | 452 (38.6) | 0 | 452 (38.6) |
| 7 | 287 (24.5) | 98 (8.4) | 400 (34.2) |
| <7 | 144 (12.3) | 150 (12.8) | 319 (27.2) |
| Pathological tumor
stage, n (%)b | | | |
| T stage | 808 (75.8) | 258 (24.2) | 1,066 |
| T1-T2 (%) | 390 (36.6) | 258 (24.2) | 648 (60.8) |
| T3-T4 (%) | 418 (39.2) | 0 | 418 (39.2) |
| N stage | 708 (73.9) | 250 (26.1) | 958 |
| N0 | 417 (43.5) | 250 (26.1) | 667 (69.6) |
| N1 | 291 (30.4) | 0 | 291 (30.4) |
| M stage | 808 (76.2) | 253 (23.8) | 1,061 |
| M0 | 478 (45.1) | 253 (23.8) | 731 (68.9) |
| M1 | 330 (31.1) | 0 | 330 (31.1) |
Association of TERT tSNPs with prostate
cancer aggressiveness
All tSNPs were within the HWE (all P>0.05) and
had a missing rate <0.05. Initially, the association of the
tSNPs across the TERT gene with PCa aggressiveness was
assessed (Table II). A total of
two SNPs (rs2736100 and rs10069690) were significantly associated
with aggressiveness, assuming an additive effect (P=0.037 and
0.030, respectively). Individuals that carried the homozygous C
allele of the TERT intron 2 SNP (rs2736100) had an OR of
0.81 (95% CI: 0.66–0.99), indicating reduced PCa aggressiveness in
comparison to those that carried the A allele. Males that carried
the homozygous T allele of the TERT intron 4 SNP
(rs10069690) had an OR of 0.76 (95% CI: 0.59–0.97), indicating
reduced PCa aggressiveness in comparison to those that carried the
C allele. Individuals that carried the TC and TT genotypes of
rs10069690 had a further reduced risk of developing an aggressive
form of PCa (OR=0.69, 95% CI: 0.52–0.93), compared with males that
carried the CC genotype, assuming a dominant model. Subsequently,
it was further evaluated whether these two SNPs conferred an
independent effect on PCa aggressiveness. A multivariate logistic
regression, which included SNPs and age in the model, revealed
non-significant associations for the two SNPs (P=0.203 and P=0.188
for rs2736100 and rs10069690, respectively), which indicated a
non-independent effect between these two SNPs (r2=0.36;
D′=0.91).
 | Table IIAssociation between TERT tSNPs
and prostate cancer aggressiveness (aggressive vs.
nonaggressive). |
Table II
Association between TERT tSNPs
and prostate cancer aggressiveness (aggressive vs.
nonaggressive).
| SNP | Genomic
positona | P-valuec OR (95%CI)
|
|---|
| Allelesb | Additive | Dominant | Recessive | Heterozygous | Homozygous |
|---|
| rs4073918 | 1297425 | C/T | 0.853 0.98
(0.79–1.21) | 0.446 0.90
(0.68–1.18) | 0.347 1.29
(0.76–2.19) | 0.267 0.85
(0.64–1.13) | 0.511 1.10
(0.83–1.44) |
| rs10078761 | 1302594 | T/A | 0.878 1.04
(0.62–1.82) | 0.878 1.04
(0.60–1.82) | – | 0.878 1.04
(0.62–1.82) | – |
| rs2853691 | 1305950 | T/C | 0.354 0.86
(0.62–1.19) | 0.320 0.82
(0.55–1.22) | 0.780 0.87
(0.34–2.24) | 0.334 0.82
(0.54–1.24) | 0.706 0.91
(0.57–1.47) |
| rs2736122 | 1310621 | G/A | 0.816 0.95
(0.63–1.44) | 0.829 0.95
(0.62–1.47) | 0.879 0.84
(0.09–8.12) | 0.848 0.96
(0.62–1.19) | 0.864 0.91
(0.39–2.82) |
| rs2075786 | 1319310 | A/G | 0.620 1.07
(0.82–1.39) | 0.810 1.03
(0.76–1.41) | 0.396 1.43
(0.63–3.26) | 0.971 0.99
(0.72–1.37) | 0.409 1.19
(0.79–1.80) |
| rs4975605 | 1328528 | C/A | 0.630 0.90
(0.60–1.37) | 0.633 0.90
(0.58–1.39) | 0.884 0.84
(0.09–8.18) | 0.646 0.90
(0.58–1.40) | 0.863 0.91
(0.29–2.82) |
| rs10069690 | 1332790 | C/T | 0.030 0.76
(0.59–0.97) | 0.014 0.69
(0.52–0.93) | 0.964 0.98
(0.42–2.30) | 0.011 0.68
(0.50–0.92) | 0.744 0.93
(0.61–1.43) |
| rs2736100 | 1339516 | A/C | 0.037 0.81
(0.66–0.99) | 0.118 0.79
(0.58–1.06) | 0.060 0.72
(0.50–1.01) | 0.293 0.84
(0.61–1.16) | 0.031 0.80
(0.66–0.98) |
| rs2853676 | 1341547 | C/T | 0.489 0.81
(0.45–1.46) | 0.390 0.77
(0.42–1.41) | – | 0.323 0.73
(0.40–1.35) | – |
| rs2736098 | 1347086 | C/T | 0.740 0.97
(0.79–1.18) | 0.531 0.91
(0.68–1.22) | 0.840 1.04
(0.71–1.54) | 0.672 0.92
(0.61–1.38) | 0.910 0.99
(0.80–1.22) |
| rs2853668 | 1353025 | G/T | 0.649 0.88
(0.50–1.54) | 0.942 1.03
(0.53–1.98) | 0.086 0.22
(0.04–1.24) | 0.560 1.24
(0.60–2.54) | 0.107 0.51
(0.23–1.16) |
| rs2735845 | 1353584 | C/G | 0.409 1.23
(0.75–2.00) | 0.399 1.30
(0.71–2.38) | 0.676 1.31
(0.37–4.68) | 0.459 1.28
(0.67–2.46) | 0.881 0.97
(0.65–1.45) |
Association of TERT tSNPs with Gleason
score
The association between TERT tSNPs and
Gleason score was estimated (Table
III). Compared with the A allele, the C allele of rs2736100
conferred a reduced risk of developing high-grade PCa with an OR of
0.83 (95% CI: 0.61–0.99; P=0.039). Rs10069690 was not significantly
associated with high-grade PCa. In addition, no rare homozygotes
for rs10078761, which is located 3′ of the TERT gene, were
observed in the present study cohort. The TT, AT and AA genotype
distributions for rs10078761 were 1,105, 79 and 0, respectively,
among the 1,184 samples that had genotyping data available. In
addition, this SNP was significantly associated with high-grade
tumors. Individuals that carried the A allele of rs10078761 had a
significantly decreased risk for developing high-grade PCa
(OR=0.48; 95% CI: 0.28–0.81; P=0.006).
 | Table IIIAssociation between TERT tSNPs
and gleason grade (gleason score >7 vs. ≤7). |
Table III
Association between TERT tSNPs
and gleason grade (gleason score >7 vs. ≤7).
| SNP | Location | MAF | P-valuea OR (95%CI)
|
|---|
| Additive | Dominant | Recessive | Heterozygous | Homozygous |
|---|
| rs4073918 | slc6a18 exon
11 | 0.28 | 0.069 0.84
(0.70–1.01) | 0.050 0.79
(0.62–1.00) | 0.489 0.86
(0.56–1.32) | 0.066 0.79
(0.62–1.02) | 0.265 0.88
(0.71–1.10) |
| rs10078761 | 3′ near gene | 0.03 | 0.006 0.48
(0.28–0.81) | 0.006 0.48
(0.28–0.81) | – | 0.006 0.48
(0.28–0.81) | – |
| rs2853691 | 3′ near gene | 0.16 | 0.592 0.92
(0.69–1.24) | 0.451 0.87
(0.62–1.24) | 0.787 1.12
(0.49–2.56) | 0.372 0.84
(0.58–1.22) | 0.852 1.04
(0.69–1.58) |
| rs2736122 | intron 13 | 0.06 | 0.781 1.05
(0.74–1.50) | 0.697 1.08
(0.74–1.56) | 0.599 0.54
(0.06–2.26) | 0.629 1.10
(0.75–1.60) | 0.591 0.73
(0.24–2.28) |
| rs2075786 | intron 10 | 0.16 | 0.108 0.83
(0.67–1.04) | 0.253 0.86
(0.66–1.12) | 0.063 0.50
(0.24–1.04) | 0.548 0.92
(0.70–1.21) | 0.051 0.70
(0.48–1.00) |
| rs4975605 | intron 6 | 0.06 | 0.836 1.04
(0.72–1.49) | 0.752 1.06
(0.73–1.55) | 0.605 0.55
(0.06–5.31) | 0.682 1.08
(0.74–1.59) | 0.594 0.73
(0.24–2.28) |
| rs10069690 | intron 4 | 0.16 | 0.083 0.82
(0.65–1.03) | 0.039 0.76
(0.58–0.99) | 0.832 1.08
(0.53–2.21) | 0.028 0.74
(0.56–0.97) | 0.967 0.99
(0.69–1.42) |
| rs2736100 | intron 2 | 0.42 | 0.039 0.83
(0.70–0.99) | 0.048 0.78
(0.61–0.99) | 0.186 0.81
(0.58–1.11) | 0.098 0.80
(0.62–1.04) | 0.053 0.84
(0.70–1.00) |
| rs2853676 | intron 2 | 0.07 | 0.592 0.92
(0.69–1.24) | 0.451 0.87
(0.62–1.24) | 0.787 1.12
(0.49–2.56) | 0.372 0.84
(0.58–1.22) | 0.852 1.04
(0.69–1.58) |
| rs2736098 | c.915 G>A | 0.40 | 0.387 0.80
(0.48–1.33) | 0.539 0.93
(0.72–1.18) | 0.305 1.19
(0.86–1.64) | 0.308 0.75
(0.44–1.30) | 0.797 1.20
(0.30–4.82) |
| rs2853668 | 5′ near gene | 0.11 | 0.560 0.86
(0.52–1.43) | 0.752 0.91
(0.51–1.62) | 0.326 0.34
(0.04–2.94) | 0.977 0.99
(0.55–1.79) | 0.324 0.58
(0.20–1.71) |
| rs2735845 | 5′ near gene | 0.18 | 0.493 1.14
(0.78–1.68) | 0.810 1.06
(0.65–1.74) | 0.212 1.85
(0.70–4.86) | 0.804 0.93
(0.55–1.60) | 0.255 1.22
(0.87–1.70) |
In addition, no significant association between TERT
tSNPs and the risk of developing an early-onset PCa (age ≤60 at
diagnosis) was observed in the present study (data not shown).
Discussion
The incidence of PCa in China has risen rapidly in
recent years. It is well established that males diagnosed with low
or moderate-risk PCa, based on the D’Amico classification criteria,
are less likely to experience progression to metastasis. Due to the
growing popularity of active surveillance and minimally invasive
therapies, it is extremely important to identify which cases are to
follow a more indolent course, by contrast to those that require
aggressive treatment to improve prognosis. Such knowledge would
enable clinicians to optimize the quality of life of patients who
are at a lower risk for disease aggressiveness and thus may be
spared unnecessary therapy and direct more radical therapies to
those with the greatest requirement. Genetic factors offer a
potentially promising avenue for further clarification of PCa
aggressiveness (18). Accordingly,
there is an urgent requirement for molecular biomarkers enabling
improved prediction of PCa behavior and identification of patients
with PCa who harbor potentially aggressive disease and those that
do not. Due to the important role of the TERT gene in PCa
progression, as previously reported (9,12,13),
it was hypothesized that TERT SNPs may be associated with
PCa aggressiveness. Therefore, the genetic variations across the
TERT gene were systematically evaluated for their impact on
PCa severity in a Chinese Han population of 1,210 cases in the
present study.
The TERT gene is located on the short (p) arm
of chromosome 5 at position 15.33 and consists of 16 exons and 15
introns spanning 35 kb of genomic DNA (19). TERT, as the reverse transcriptase
component of telomerase, was found to be rate-limiting for
telomerase activity and a tight regulator of telomerase activity at
the transcriptional and post-translational levels (4,20).
March-Villalba et al (9)
used quantitative RT-PCR to determine plasma hTERT mRNA levels in
patients with localized and locally advanced PCa, respectively. The
authors observed that patients with locally advanced disease had
significantly higher plasma hTERT mRNA expression than those with
localized disease. Sabaliauskaite et al (13) confirmed that TERT-positive PCa
cases had elevated levels of ETS-related gene (ERG), of
which the fusion with trans-membrane protease, serine 2 was not
only a significant event of prostate tissue malignization but also
associated with more aggressive disease and worse prognosis
(21), suggesting a possible
association between aberrant expression of ERG and
reactivation of TERT in prostate tumors. In addition,
Iczkowski et al (14) used
immunohistochemistry to detect the association between a polyclonal
antibody to TERT and the Gleason score of cancer, where it
was demonstrated that nuclear anti-TERT reactivity was restricted
to high-grade carcinoma (Gleason primary pattern ≥4). The above
studies all suggested a correlation between TERT and PCa
aggressiveness.
At present, introns are becoming increasingly
recognized as having significant roles in gene regulation,
including containing silencer or enhancer elements, alternative
splicing and exon shuffling (22).
SNP rs2736100, which is situated in intron 2 of TERT, lies
in a putative regulatory region according to the Evolutionary and
Sequence Pattern Extraction through Reduced Representation score
(23). In the present study, it
was identified that rs2736100 was associated with decreased PCa
aggressiveness and degree of differentiation (i.e. the major A
allele of rs2736100 was associated with a poorer degree of
differentiation of prostate cancer compared with the minor C
allele). A recent functional study demonstrated that the mutational
CC genotype of this SNP was associated with lower telomerase
activity and longer telomere length (TL) compared with the
wild-type, as elucidated using a TRAPeze telomerase detection kit
and quantitative RT-PCR-based assays, respectively (24). The lower telomerase activity
observed in the aforementioned study is consistent with the results
of the present study, indicating that the CC genotype is associated
with suppression of PCa progression. As for the TL, a previous
study demonstrated the inverse association between TL and cancer
incidence and mortality (25).
Telomere shortening may cause telomere dysfunction, ongoing
chromosomal instability and ultimately lead to an increased risk of
cancer development (26).
Although a population-based case-control study
failed to observe a statistically significant association between
leukocyte TL and PCa risk using quantitative PCR (27), two meta-analyses revealed that
shorter telomeres were significantly associated with an increased
overall risk of cancer compared with longer telomeres (28). This may explain the present result
in which the mutational CC genotype of SNP rs2736100, which is
associated with longer TL, may reduce the risk of developing a more
aggressive form of PCa. In addition, rs10069690, which is mapped to
intron 4 of TERT, has been observed to increase the risk of
developing ER-negative breast and ovarian cancer and also increase
the risk for those carrying breast cancer 1 mutations; however,
this occurs independently of altered TL. The present study
demonstrated that the minor T allele (minor allele in Caucasian and
Chinese populations) of rs10069690 was associated with longer TL,
as with the SNP rs2736100, which indicated that the T allele was
expected to inhibit the cancer development, while the evidence that
it increased the risk of cancer was to the contrary. Such notable
contradictions reflect the complexity of associations among genetic
variations, telomere structures and clinical phenotypes. Another
study demonstrated that the non-T allele of rs10069690 may increase
the risk of development and metastasis in primary hepatocellular
carcinoma in Chinese individuals (29). The different results of the two
studies may be reflective of the different roles genetic variations
have between tumorigenesis and tumor prognosis, and it may also
reflect the tumor-specific effect and genetic heterogeneity effect
among different ethnic populations with regards to cancer risk. In
addition, the present study observed specific residual LD even when
the tSNPs approach was used and this revealed the potential
disadvantage of this approach. Thus, due to the LD between these
two intron region tSNPs (rs2736100 and rs10069690), it was very
difficult to separate their own effect in the genetic association
study.
Furthermore, to the best of our knowledge, no
previous studies have reported the SNP rs10078761 which is located
3′ of the TERT gene and none of the mutational homozygotes
with the AA genotype of this SNP were detected in these subjects.
Possible explanations for this result may be that the individuals
carrying the AA genotype may be less likely to develop PCa; these
individuals may not be viable beyond the embryonic period, or this
may be attributed to the genotyping failure of the remaining 26
samples in the study population. This notable finding requires
further comprehensive investigation to examine this intergenic
variation in the future.
There are several limitations in the present study
that should be discussed. Firstly, although the established D’Amico
classification criteria has been generally accepted to predict the
prognosis of PCa patients, PCa aggressiveness using this criteria
may be affected by healthcare practices, including screening time
and the frequency of physical examination. For instance, frequently
examined individuals are more likely to be diagnosed with PCa at
lower PSA levels and earlier tumor stages and thus more are
classified as having a less aggressive disease. However, the course
of the disease in a number of these individuals may progress very
rapidly, which is only evident following subsequent clinical
observations. Thus, there is the probability of misclassification
of PCa aggressiveness in the present study. The ongoing collection
of clinicopathological variables, including biochemical recurrence,
clinical metastases and cancer-specific mortality for study
subjects appears necessary for more accurate classification in the
future (30). In addition, none of
the observed associations may survive when the most stringent
criteria to correct for multiple assessment (the Bonferroni
correction) is taken into account, in which the corrected α-value
would be 0.0042. Finally, this analysis of genotype profiling
should be coupled with complementary studies aimed at setting a
complete molecular signature of individuals, including epigenetic
modifications and gene expression profiles in RNA and protein
levels. Such a combination may be more precise in the comprehensive
classification of disease severity than the current systems.
In conclusion, the present results indicated that
genetic polymorphisms in the TERT gene are associated with
PCa aggressiveness in a Chinese Han population. This finding
provides evidence that TERT gene variations may be involved
in PCa development, progression and metastasis, and may be used as
a prognostic indicator. If further confirmed, these identified
genetic variations may assist to clarify which carcinomas are more
likely to progress rapidly and require more intensive treatment
versus those that may not have a severe impact on mortality.
Further validation in a larger set of PCa samples and subsequent
functional studies of TERT polymorphisms are required to
further evaluate the present findings.
Acknowledgments
The present study was funded by the Science and
Technology Commission of Shanghai Municipality (grant no.
11ZR1424100) and partly supported by the Key Project of the
National Natural Science Foundation of China (grant no. 81130047).
The authors would like to thank all the study subjects who were
involved in the present study for their time, effort and
cooperation. The authors would also like to thank all staff members
in the Department of Urology at Xinhua Hospital and Huashan
Hospital for their cooperation during data collection.
References
|
1
|
Shay J and Bacchetti S: A survey of
telomerase activity in human cancer. Eur J Cancer. 33:787–791.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mocellin S, Verdi D, Pooley KA, et al:
Telomerase reverse transcriptase locus polymorphisms and cancer
risk: a field synopsis and meta-analysis. J Natl Cancer Instit.
104:840–854. 2012. View Article : Google Scholar
|
|
3
|
Harley CB: Telomerase and cancer
therapeutics. Nat Rev Cancer. 8:167–179. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Horikawa I and Barrett JC: Transcriptional
regulation of the telomerase hTERT gene as a target for cellular
and viral oncogenic mechanisms. Carcinogenesis. 24:1167–1176. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nandakumar J and Cech TR: Finding the end:
recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol.
14:69–82. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baird DM: Variation at the TERT locus and
predisposition for cancer. Expert Rev Mol Med. 12:1–21. 2010.
View Article : Google Scholar
|
|
7
|
Rafnar T, Sulem P, Stacey SN, et al:
Sequence variants at the TERT-CLPTM1 L locus associate with many
cancer types. Nat Genet. 41:221–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
March-Villalba JA, Martínez-Jabaloyas JM,
Herrero MJ, Santamaría J, Aliño SF and Dasí F: Plasma hTERT mRNA
discriminates between clinically localized and locally advanced
disease and is a predictor of recurrence in prostate cancer
patients. Expert Opin Biol Ther. 12:69–77. 2012. View Article : Google Scholar
|
|
10
|
Heijnsdijk E, Der Kinderen A, Wever E,
Draisma G, Roobol M and De Koning H: Overdetection, overtreatment
and costs in prostate-specific antigen screening for prostate
cancer. Br J Cancer. 101:1833–1838. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng I, Plummer SJ, Neslund-Dudas C, et
al: Prostate cancer susceptibility variants confer increased risk
of disease progression. Cancer Epidemiol Biomarkers Prev.
19:2124–2132. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
FitzGerald LM, Kwon EM, Conomos MP, et al:
Genome-wide association study identifies a genetic variant
associated with risk for more aggressive prostate cancer. Cancer
Epidemiol Biomarkers Prev. 20:1196–1203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sabaliauskaite R, Jarmalaite S, Petroska
D, et al: Combined analysis of TMPRSS2-ERG and TERT for improved
prognosis of biochemical recurrence in prostate cancer. Gene
Chromosome Canc. 51:781–791. 2012. View Article : Google Scholar
|
|
14
|
Iczkowski KA, Pantazis CG, McGregor DH, Wu
Y and Tawfik OW: Telomerase reverse transcriptase subunit
immunoreactivity. Cancer. 95:2487–2493. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gleason DF and Mellinger GT: Prediction of
prognosis for prostatic adenocarcinoma by combined histological
grading and clinical staging. J Urol. 111:58–64. 1974.PubMed/NCBI
|
|
16
|
D’Amico AV, Whittington R, Malkowicz SB,
et al: Biochemical outcome after radical prostatectomy, external
beam radiation therapy, or interstitial radiation therapy for
clinically localized prostate cancer. JAMA. 280:969–974. 1998.
View Article : Google Scholar
|
|
17
|
Purcell S, Neale B, Todd-Brown K, et al:
PLINK: a tool set for whole-genome association and population-based
linkage analyses. Am J Hum Gen. 81:559–575. 2007. View Article : Google Scholar
|
|
18
|
Witte JS: Prostate cancer genomics:
towards a new understanding. Nat Reviews Genet. 10:77–82. 2009.
View Article : Google Scholar
|
|
19
|
Wick M, Zubov D and Hagen G: Genomic
organization and promoter characterization of the gene encoding the
human telomerase reverse transcriptase (hTERT). Gene. 232:97–106.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang S, Wu J, Hu L, et al: Common genetic
variants in TERT contribute to risk of cervical cancer in a Chinese
population. Mol Carcinog. 51:E118–E122. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar-Sinha C, Tomlins SA and Chinnaiyan
AM: Recurrent gene fusions in prostate cancer. Nat Rev Cancer.
8:497–511. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Landi MT, Chatterjee N, Yu K, et al: A
genome-wide association study of lung cancer identifies a region of
chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum
Gene. 85:679–691. 2009. View Article : Google Scholar
|
|
23
|
Taylor J, Tyekucheva S, King DC, Hardison
RC, Miller W and Chiaromonte F: ESPERR: learning strong and weak
signals in genomic sequence alignments to identify functional
elements. Genome Res. 16:1596–1604. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sheng X, Tong N, Tao G, et al: TERT
polymorphisms modify the risk of acute lymphoblastic leukemia in
Chinese children. Carcinogenesis. 34:228–235. 2013. View Article : Google Scholar
|
|
25
|
Willeit P, Willeit J, Mayr A, et al:
Telomere length and risk of incident cancer and cancer mortality.
JAMA. 304:69–75. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feldser DM, Hackett JA and Greider CW:
Telomere dysfunction and the initiation of genome instability. Nat
Rev Cancer. 3:623–627. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mirabello L, Huang WY, Wong JY, et al: The
association between leukocyte telomere length and cigarette
smoking, dietary and physical variables and risk of prostate
cancer. Aging cell. 8:405–413. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma H, Zhou Z, Wei S, et al: Shortened
telomere length is associated with increased risk of cancer: a
meta-analysis. PLoS One. 6:e204662011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dong J, Wang L, Tian Y, Guo Y and Liu H:
hTERT single nucleotide polymorphism is associated with increased
risks of hepatocellular carcinoma and tumor metastasis. Nan Fang Yi
Ke Da Xue Xue Bao. 31:49–52. 2011.In Chinese. PubMed/NCBI
|
|
30
|
Bensen JT, Xu Z, Smith GJ, Mohler JL,
Fontham ET and Taylor JA: Genetic polymorphism and prostate cancer
aggressiveness: A case-only study of 1,536 GWAS and candidate SNPs
in African-Americans and European-Americans. Prostate. 73:11–22.
2013. View Article : Google Scholar
|