Introduction
Cardiovascular disease is one of the major causes of
death worldwide. The pathologies of cardiovascular disease are
complex, although the most important are inflammation and oxidative
stress. Biomarkers of inflammation and oxidative stress may serve
to help identify patients at risk for cardiovascular disease, to
monitor the efficacy of treatments, and to develop novel
pharmacological tools. The use of serological biomarkers may
improve clinical decision making and a therapeutic strategy
setting. For primary cardiovascular events, markers with strong
predictive potential are mainly associated with lipids. For
secondary cardiovascular events, markers are associated more with
ischemia. However, there are fewer studies on microRNAs (miRNAs),
which as cardiovascular biomarkers. miRNAs are small, endogenous,
noncoding RNAs that regulate gene expression by targeting the
degradation or translational repression of mRNA. Previously, it has
been demonstrated that miRNAs circulating in the bloodstream are
useful biomarkers for cardiovascular disease (1–3).
Wang et al (4) reported that miR-208a is an excellent
diagnostic marker for acute myocardial infarction (AMI), which was
demonstrated by its sensitivity in detecting AMI in patients within
4 h of the onset of symptoms. Kuwabara et al (5) reported that circulating miR-133a
serves as a useful marker for cardiomyocyte death and can thus be
used for the detection of several cardiovascular diseases,
including AMI, unstable angina and takotsubo cardiomyopathy.
Few reports, however, have analyzed the differences
in miRNA expression profiles between patients with MI (with or
without heart failure) and individuals in a normal control group
using an miRNA array. Therefore, the aim of the present study was
to investigate specific miRNAs that were differentially expressed
in patients with MI (with or without heart failure) and individuals
in a normal control group. The results were used as a basis for
exploring novel circulating markers for MI and heart failure.
Materials and methods
Patients
A total of 25 consecutive patients, whose symptoms
were consistent with the established diagnostic criteria for AMI,
were selected and divided into two groups: AMI with no heart
failure [AMNHF; Killip class I and left ventricular ejection
fraction (LVEF) >50%] and the AMI with heart failure (AMHF;
Killip class ≥II and LVEF ≤40%) groups. A total of 10 patients who
had presented with AMI one year before the study was conducted were
recruited as the old MI with heart failure (OMHF) group (LVEF
>50%). For the control (N) group, 10 patients with normal
coronary angiography results and an LVEF >50% were selected. The
present study was approved by the ethics committee of Foshan Second
People’s Hospital (Foshan, China). Written informed consent was
obtained from the patients.
Plasma collection
Fasting blood samples were collected into EDTA
anticoagulant separator tubes, which were subsequently centrifuged
at 1,000 × g for 10 min at 4°C to separate the clots. The plasma
was then removed from the tubes and stored at −80°C until the time
of the assay.
RNA isolation and miRNA analysis
Total RNA was isolated using TRIzol®
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) and the
miRNeasy Mini kit (Qiagen, Hilden, Germany) in accordance with the
manufacturer’s instructions. All RNA species were efficiently
recovered using this method, including miRNA. The quality and
quantity of the isolated RNA was evaluated using a nanodrop
spectrophotometer (ND-1000; Nanodrop Technologies, Waltham, MA,
USA), and gel electrophoresis was utilized to assess the integrity
of the RNA.
Following RNA isolation from the samples, miRNA
labeling was conducted with the miRCURY™ Hy3™/Hy5™ Power Labeling
kit (Exiqon A/S, Vedbaek, Denmark) in accordance with the
manufacturer’s instructions. The 3′-end of each 1-μg sample
was subsequently labeled with a Hy3 fluorescent label, using T4 RNA
ligase. In brief, RNA in 2.0 μl water was combined with 1.0
μl calf intestinal alkaline phosphatase (CIP) buffer and
CIP. This mixture was then incubated for 30 min at 37°C, prior to
the reaction being terminated by incubation for 5 min at 95°C. A
total of 3.0 μl labeling buffer, 1.5 μl fluorescent
label (Hy3), 2.0 μl dimethylsulfoxide and 2.0 μl
labeling enzyme were subsequently added to the mixture. The
labeling reaction was incubated for 1 h at 16°C, and the
termination of the reaction was achieved by incubation for 15 min
at 65°C.
Following the termination of the labeling procedure,
the Hy3-labeled samples were hybridized on the miRCURY™ Locked
Nucleic Acid array (v. 16.0; Exiqon A/S) according to the
manufacturer’s instructions. The 25-μl mixture from the
Hy3-labeled samples was then combined with 25 μl
hybridization buffer for denaturation for 2 min at 95°C. Following
incubation on ice for 2 min, the mixture was hybridized to the
microarray for 16–20 h at 56°C in a 12-Bay Hybridization System
(Nimblegen Systems, Inc., Madison, WI, USA). This procedure was
designed to improve the uniformity of the hybridization and enhance
the signal by performing active mixing and maintaining a constant
incubation temperature. Once hybridization was complete, the slides
were washed multiple times using a wash buffer kit (Exiqon A/S),
prior to being dried by centrifugation for 5 min at 200 × g. The
Axon GenePix 4000B microarray scanner (Axon Instruments, Foster
City, CA, USA) was subsequently utilized to scan the slides.
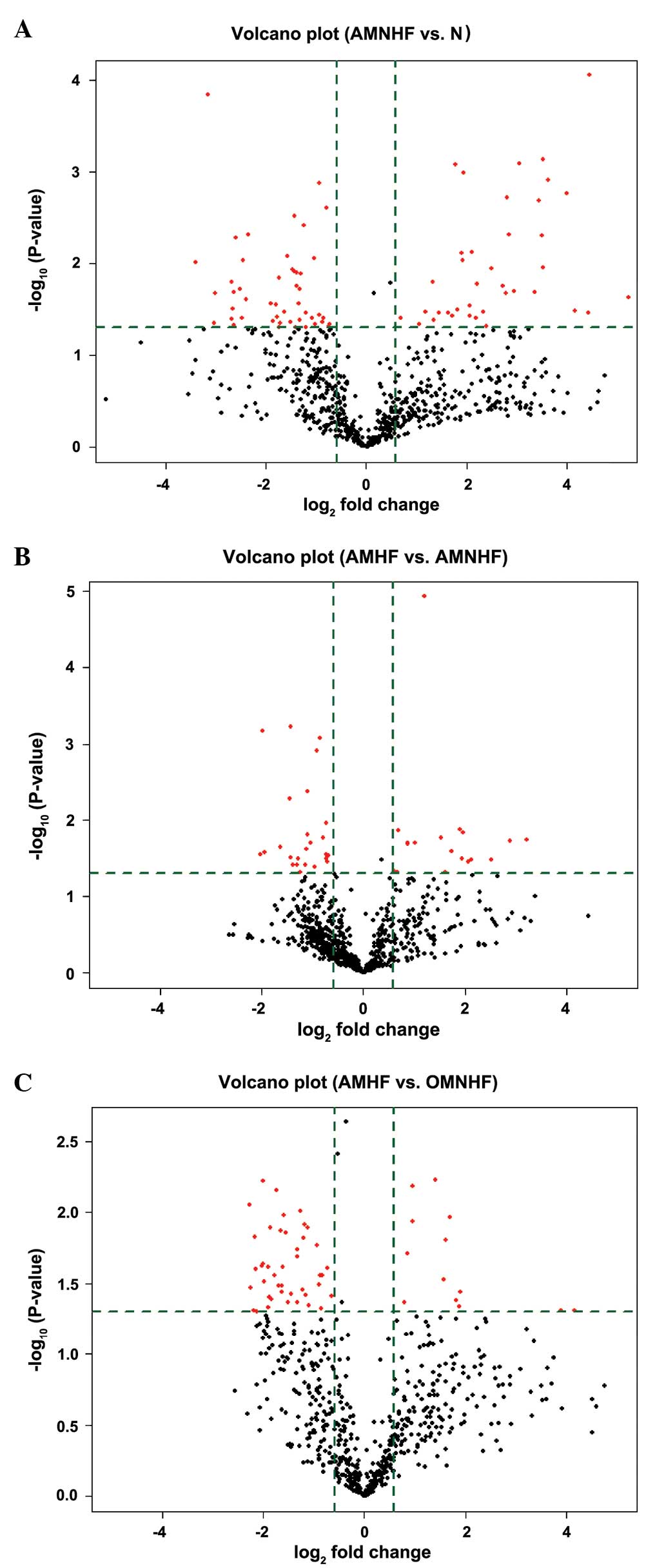
Volcano plots
Volcano plots enable the visualization of
differential expression between two different groups. Fold-change
and P-values are utilized to create the plots, thus facilitating
the visualization of the correlation between the fold-change (the
magnitude of the change) and the statistical significance (which
takes the magnitude of the change and variability into
consideration). These plots also allow subsets of genes to be
isolated based on different values.
Statistical analysis
The scanned images were imported into GenePix Pro
6.0 software (Axon Instruments) for grid alignment and data
extraction. The replicated miRNAs were averaged and miRNAs with
intensities of ≥50 in all samples were selected for the calculation
of the normalization factor. Expressed data were normalized using
median normalization. Following normalization, significant
differences in miRNA expression were identified using volcano plot
filtering, and hierarchical clustering was performed using
MultiExperiment Viewer 4.6 software (The Institute for Genomic
Research; http://www.tigr.org/software/tm4/mev.html). P<0.05
was considered to indicate a statistically significant difference
between values.
Results
Baseline characteristics
No significant differences were observed in age,
gender, body mass index or systolic blood pressure among the groups
(P>0.05). The majority of the participants in the patient groups
were taking angiotensin-converting enzyme inhibitors (ACEIs),
angiotensin receptor blockers (ARBs), statins or β-blockers. Only
one participant in the N group used statins and none of them were
taking ACEIs/ARBs or β-blockers (Table
I).
 | Table IBaseline characteristics of each
group. |
Table I
Baseline characteristics of each
group.
| Variable | AMNHF | AMHF | OMHF | Control | P-value |
|---|
| Age (years) | 59.2±6.0 | 60.5±6.6 | 58.7±7.3 | 58.4±6.5 | 0.852 |
| Gender, M/F
(n/n) | 6/4 | 8/7 | 6/6 | 5/5 | – |
| BMI
(kg/m2) | 24.00±2.38 | 24.58±2.55 | 24.10±3.43 | 23.86±1.64 | 0.904 |
| SBP (mmHg) | 125.8±5.1 | 120.8±5.6 | 122.5±4.2 | 121.9±4.9 | 0.123 |
| β-blocker, Y/N
(n/n) | 10/0 | 4/15 | 10/0 | 0/10 | – |
| ACEI, Y/N (n/n) | 9/10 | 15/15 | 8/2 | 0/10 | – |
| ARB, Y/N (n/n) | 1/10 | 0/15 | 2/8 | 0/10 | – |
| Statins, Y/N
(n/n) | 10/10 | 10/0 | 10/0 | 1/9 | – |
Volcano plots
In Fig. 1, the
vertical lines correspond to 1.5-fold positive and negative
differences, respectively, and the horizontal lines represent a
P-value of 0.05. Consequently, the red dots in the plot represent
the differentially expressed miRNAs with statistical
significance.
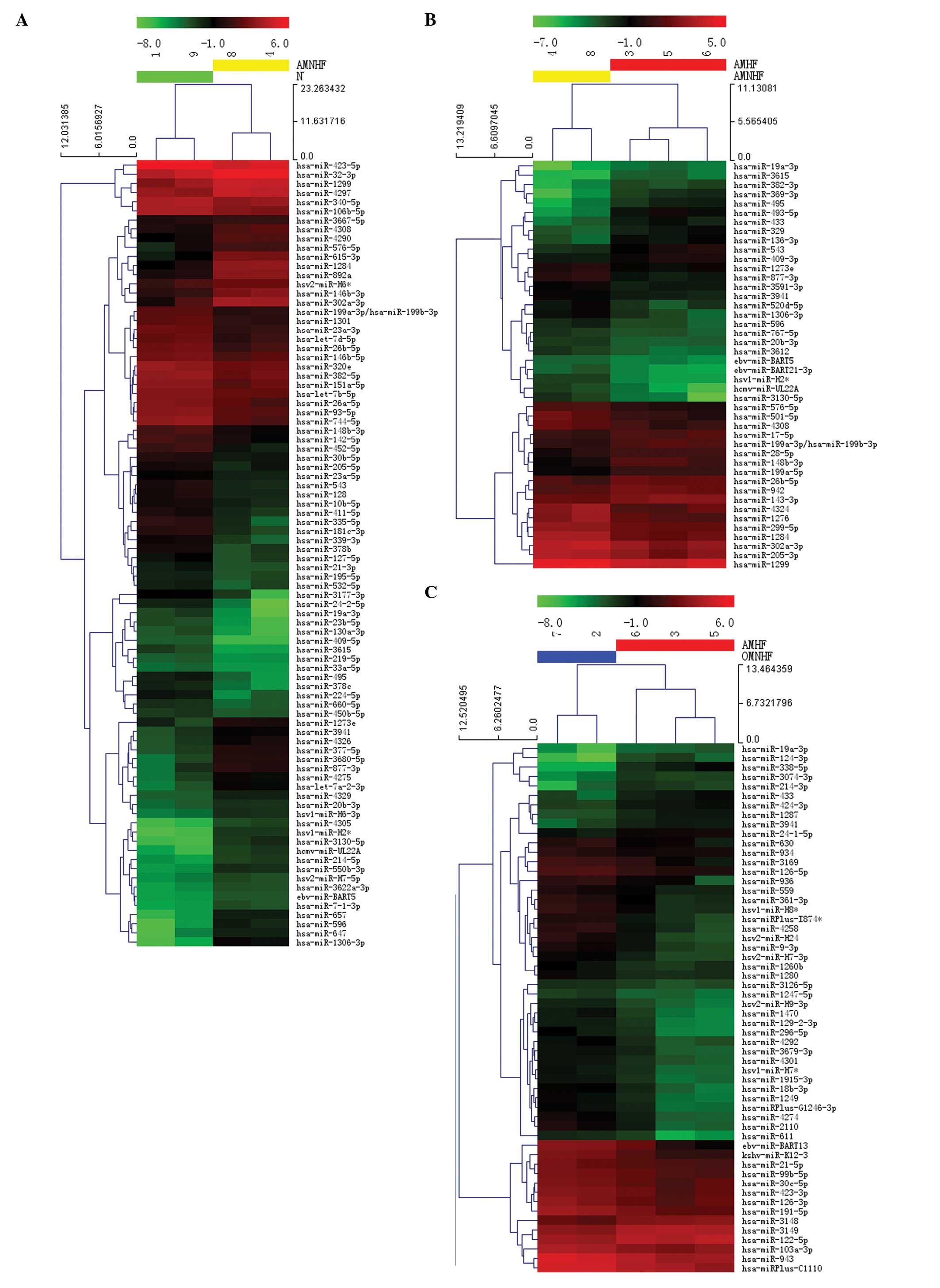
Heat map and hierarchical clustering
The heat map diagram shows the result of the two-way
hierarchical clustering of the miRNAs and samples. Each row
represents an miRNA and each column represents a sample. The miRNA
clustering tree is shown on the left and the sample clustering tree
appears at the top. The color scale shown at the top illustrates
the relative expression level of a miRNA in a specific slide: Red
represents a high relative expression level and green represents a
low relative expression level (Fig.
2).
Differential expression of miRNAs
In the AMHF group, the expression of 17 miRNAs was
upregulated and that of 21 miRNAs was downregulated by >1.5-fold
as compared with the expression in the AMNHF group. Compared with
the N group, the expression of miRNAs in the AMNHF group was
upregulated in 38 and downregulated in 48 cases by >1.5-fold.
Compared with the OMHF group, 13 miRNAs were upregulated and 43
were downregulated by >1.5-fold in the AMHF group (Fig. 1).
To confirm the results from the discovery phase, the
expression levels of candidate miRNAs were examined by quantitative
polymerase chain reaction (qPCR) using a 7900HT Fast Real-Time PCR
system (Life Technologies, Grand Island, NY, USA). Compared with
expression in the AMNHF group, the expression of hsa-miR-493-5p,
hsa-miR-369-3p, hsa-miR-495, hsa-miR-3615 and hsa-miR-433 was
upregulated, and that of hsa-miR-877-3p, hsa-miR-1306-3p,
hsv1-miR-H2, hsa-miR-3130-5p and hcmv-miR-UL22A was downregulated
in the AMHF group (P<0.05). In the AMNHF group, the expression
of hsa-miR-596, hsa-miR-657, hsa-miR-1306-3p, hsv1-miR-H2 and
hsa-miR-3130-5p was upregulated, and the expression of
hsa-miR-409-5p, hsa-miR-24-2-5p, hsa-miR-335-5p, hsa-miR-19a-3p and
hsa-miR-130a-3p was downregulated compared with that in the N group
(P<0.05). Compared with the expression in the OMHF group, the
expression of hsa-miR-338-5p, hsa-miR-124-3p, hsa-miR-214-3p,
hsa-miR-433 and hsa-miR-19a-3p was upregulated, and that of
hsv2-miR-H24, hsa-miR-1249, hsa-miR-4258, hsa-miR-1470 and
hsa-miR-2110 was downregulated in the AMHF group (P<0.05).
Discussion
To the best of our knowledge, this is the first
study that analyzes the differences in the miRNA expression profile
between patients with MI (with or without heart failure) and a
normal control group using a plasma miRNA array. Specifically, it
was found that, in the AMHF group, the expression of 17 miRNAs was
upregulated and the expression of 21 miRNAs was downregulated by
>1.5-fold as compared with that in the AMNHF group. Compared
with the expression in the N group, the expression of miRNAs in the
AMNHF group was upregulated in 38 and downregulated in 48 cases by
>1.5-fold. Compared with miRNAs in the OMHF group, 13 miRNAs
were upregulated and 43 were downregulated by >1.5-fold in the
AMHF group.
miRNAs are small, endogenous RNAs that have
important roles in living organisms and target mRNAs for
degradation or translational repression (6). Dysregulation and tissue-specific
patterns of intracellular miRNA expression have been reported for
various diseases, in particular for several types of cancer
(7). In addition, miRNAs appear to
circulate in the blood in a relatively stable form (8), suggesting that miRNAs may have
biological functions outside the cell; thus, they can potentially
serve as diagnostic or prognostic biomarkers for different types of
cancer, as well as therapeutic targets. With regard to
cardiovascular diseases, however, the potential of miRNAs as
diagnostic markers has only been proposed in the past few years
(1), and few prognostic features
or therapeutic potentials of circulating miRNAs have been reported.
Ai et al (9) found that the
miRNA-1 level was significantly higher in plasma from patients with
AMI compared with that in patients with no AMI, and the level
dropped to normal on discharge following the administration of
medication. Increased circulation of miRNA-1 was not associated
with age, gender, blood pressure, diabetes mellitus or the
established biomarkers for AMI. Wang et al (4) reported that elevated cardiac-specific
miR-208a in plasma may be a novel biomarker for the early detection
of myocardial injury in humans. miR-208a remained undetectable in
patients with no AMI, but was easily detected in 91% of patients
with AMI and in all of the patients with AMI within 4 h after the
onset of the symptoms. Receiver operating characteristic curve
analysis of the four miRNAs investigated revealed that miR-208a had
the highest sensitivity and specificity for diagnosing AMI. Tijsen
et al (10) reported
miR423-5p to be a novel circulating biomarker for heart failure.
Matsumoto et al (11) found
that the serum levels of miR-155 and miR-380 were approximately
four- and three-fold higher, respectively, in patients who died
within one year subsequent to discharge. Accordingly, a subset of
circulating miRNAs may be a predictor of cardiac mortality in
patients post-AMI.
In the present study, it was found that, compared
with expression in the AMNHF group, the expression of
hsa-miR-493-5p, hsa-miR-369-3p, hsa-miR-495, hsa-miR-3615 and
hsa-miR-433 was upregulated, whereas that of hsa-miR-877-3p,
hsa-miR-1306-3p, hsv1-miR-H2, hsa-miR-3130-5p and hcmv-miR-UL22A
was downregulated in the AMHF group; these miRNAs may be novel
diagnostic or prognostic biomarkers for heart failure following MI,
although they have not been reported previously and the underlying
mechanism has yet to be elucidated. The upregulated miRNAs,
including hsa-miR-59, hsa-miR-657, hsa-miR-1306-3p, hsv1-miR-H2 and
hsa-miR-3130-5p, as well as downregulated miRNAs, including
hsa-miR-409-5p, hsa-miR-24-2-5p, hsa-miR-335-5p, hsa-miR-19a-3p and
hsa-miR-130a-3p, may be novel diagnostic or prognostic biomarkers
for AMI. Compared with expression in the OMHF group, the expression
of hsa-miR-338-5p, hsa-miR-124-3p, hsa-miR-214-3p, hsa-miR-433 and
hsa-miR-19a-3p was upregulated, whereas that of hsv2-miR-H24,
hsa-miR-1249, hsa-miR-4258, hsa-miR-1470 and hsa-miR-2110 was
down-regulated in the AMHF group. Significant differences were
found in the miRNA expression profiles between patients with
different stages of heart failure following MI and the normal
control group. Thus, these specific miRNAs may be novel circulating
markers for MI and heart failure.
Several limitations of the present study should be
mentioned. Firstly, this was an initial screening analysis using a
small sample of patients with MI. Secondly, the regulation
mechanism and targets of the selected miRNAs were not elucidated.
Due to these limitations, further studies with larger sample sizes
are warranted to confirm the results of the present study.
Acknowledgments
The present study was supported by Guangdong
Provincial Nature Sciences and Technology Fund (no. S2012010008207)
and Foshan Municipal Medical Science and Technology Key Project
(no. 201108059).
References
|
1
|
D’Alessandra Y, Devanna P, Limana F, et
al: Circulating microRNAs are new and sensitive biomarkers of
myocardial infarction. Eur Heart J. 31:2765–2773. 2010. View Article : Google Scholar :
|
|
2
|
Stoner L, Lucero AA, Palmer BR, Jones LM,
Young JM and Faulker J: Inflammatory biomarkers for predicting
cardiovascular disease. Clin Biochem. 46:1353–1371. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Holten TC, Waanders LF, de Groot PG,
Vissers J, Hoefer IE, Pasterkamp G, Prins MW and Roest M:
Circulating biomarkers for predicting cardiovascular disease risk;
a systematic review and comprehensive overview of meta-analyses.
PLoS One. 22:e620802013. View Article : Google Scholar
|
|
4
|
Wang GK, Zhu JQ, Zhang JT, et al:
Circulating microRNA: a novel potential biomarker for early
diagnosis of acute myocardial infarction in humans. Eur Heart J.
31:659–666. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kuwabara Y, Ono K, Horie T, et al:
Increased microRNA-1 and microRNA-133a levels in serum of patients
with cardiovascular disease indicate myocardial damage. Circ
Cardiovasc Genet. 4:446–454. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ueda T, Volinia S, Okumura H, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: a microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar
|
|
8
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ai J, Zhang R, Li Y, et al: Circulating
microRNA-1 as a potential novel biomarker for acute myocardial
infarction. Biochem Biophys Res Commun. 391:73–77. 2010. View Article : Google Scholar
|
|
10
|
Tijsen AJ, Creemers EE, Moerland PD, et
al: MiR423-5p as a circulating biomarker for heart failure. Circ
Res. 106:1035–1039. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matsumoto S, Sakata Y, Nakatani D, et al:
A subset of circulating microRNAs are predictive for cardiac death
after discharge for acute myocardial infarction. Biochem Biophys
Res Commun. 427:280–284. 2012. View Article : Google Scholar : PubMed/NCBI
|