Introduction
Osteosarcoma (OS) is the most common type of bone
cancer and the incidence of OS is higher in adolescents (1). There are no obvious clinical signs at
early stages of the disease only pain, swelling and joint
dysfunction (1). OS is a disease
with high local aggressiveness and a tendency to metastasize to the
lungs. Patients were amputated in early therapies but still 80% of
them inevitably succumbed to lung metastasis (2). Thus, identification of molecular
biomarkers that may provide precise diagnosis and treatment for OS
are required.
Cytokines, a family of protein or micro-molecule
polypeptides, are important in the regulation of cellular adhesion
molecules and exhibit antitumor activity (3,4).
Cytokines can regulate cell growth and differentiation, and enhance
immune responses to stimuli (5).
In tumor pathogenesis, cytokines inhibit chemical carcinogenesis,
lymphomas and breast cancer invasion (6–8). Li
et al (9) showed that
enhanced expression of bone morphogenetic protein in OS cells
exerted inhibitory effects on their growth and migration. Elevated
expression of chemokine (C-X-C motif) ligand 4 (CXCL4), CXCL6 and
CXCL12 in the OS serum from infants was screened using an antibody
microarray (5). This study aimed
to identify other cytokines that may be biomarkers for OS diagnosis
and therapy.
Protein expression profiling of serum obtained from
patients and healthy individuals has been analyzed by enzyme-linked
immunosorbent assay (ELISA), microRNA proofing and matrix-assisted
laser desorption/ionization mass spectrometry (10–12).
High-throughput screening has been demonstrated to be rapid and
effective for protein profiling, which has been applied in a simple
manner (13). Antibody microarray,
a high-throughput method, has been widely used for protein
expression profiling, such as profiling of cytokines and chemokines
(14–16). Thus far, numerous potential
biomarkers for cancer diagnosis have been identified in different
cancer types by antibody microarray (14,17,18).
In the present study, antibody microarray was used
to identify the cytokines that were significantly expressed in OS
and an ELISA method was used to confirm microarray results.
Finally, clustering analysis of the expressed cytokines was
performed for evaluation of classification accuracy.
Materials and methods
This study was approved by the ethics committee of
The Second Affiliated Hospital of Xuzhou Medical College and
Affiliated Huai’an Hospital of Xuzhou Medical College (Huaian,
China) and all participants provided written informed consent.
Serum samples
Blood samples were obtained from 20 patients
(between 2009 and 2013) with OS, including 10 male samples and 10
female samples at average age of 21.8 years. According to Dahlin’s
criteria (19), 12 cancer tissues
were classified as osteoblastoma and the other samples were
classified as chondroblastoma. Ten patients were detected with
metastasis and the rest of the patients were negative for
metastasis. In addition, 20 control serum samples were obtained
from healthy individuals and blood donors including 10 male samples
and 10 female samples at an average age of 23.9 years. Table I shows the characteristics of
patients in the OS group.
 | Table IClinical characteristics of the OS
group and control group. |
Table I
Clinical characteristics of the OS
group and control group.
| Clinical
parameter | Number
|
|---|
| OS group | Control group |
|---|
| Age (years) |
| 8–20 | 13 | 7 |
| 21–25 | 11 | 9 |
| Gender |
| Male | 10 | 10 |
| Female | 10 | 10 |
| Dahlin’s
criteria |
| Osteoblastoma
type | 12 | – |
| Chondroblastoma
type | 8 | – |
| Metastasis |
| No | 10 | – |
| Yes | 10 | – |
| Serum samples | 20 | 20 |
Antibody microarray
A high-throughput antibody microarray AAH CYT-G2000,
including AAH-CYT-G6, AAH-CYT-G7 and AAH-CYT-G8 was purchased from
RayBiotech Inc. (Atlanta, GA, USA). One hundred and seventy-four
antibodies have been immobilized on the slides in advance by the
manufacturer. Cytokines in each serum sample were detected twice.
Additionally, the negative control and the positive control for the
cytokines were also detected twice in each serum. The slides were
blocked in 3% (w/v) fat-free milk powder (blocking solution)
followed by washing in phosphate-buffered saline (PBS). Then, 100
μl serum samples (diluted four times with blocking solution)
were incubated at 4°C overnight. Subsequently, slides were rinsed
using PBST (containing PBS and 0.05% Tween-20, pH 7.4) and PBS
successively, four times for 5 min. Then biotin-labeled antibodies
were incubated for 2 h and washed with PBST for 30 min. After
drying, slides were incubated with fluorescent dye-conjugated
streptavidin (RayBiotech Inc., Norcross, GA, USA) for 1.5 h and
were scanned by an Axon GenePix 4000B scanner at 532 nm (Axon,
Union City, CA, USA).
ELISA
ELISA was performed using commercially available
kits from R&D Systems (Minneapolis, MN, USA) according to the
manufacturer’s instructions. Absorbance was detected at 450 nm by a
Bio-Rad model 680 microplate reader (Bio-Rad, Hercules, CA, USA).
Duplicated experiments were performed for each sample.
Statistical analysis
Axon GenePix Pro 6.0 software (Axon, Union City, CA,
USA) was used to analyze the scanning data and signals were
normalized by positive signal values. Data were analyzed by one-way
analysis of variance using SPSS 17.0 software (SPSS, Inc., Chicago,
IL, USA). P<0.05 and fold change >2 were considered to
indicate a statistically significant difference. The ratio of
signal values from the sample channel to reference channel was
calculated for each antibody spot.
In order to evaluate whether cytokine expression
screening by antibody microarray could distinguish OS samples from
control samples, unsupervised clustering analysis (20) of upregulated cytokines was
performed using Cluster 3.0 (21),
which was based on uncentered correlation and average linkage
algorithms (22).
Results
Screening of significantly upregulated
cytokines using antibody microarray
Twenty serum samples from patients with OS and 20
from healthy individuals were analyzed using a microarray
containing 174 antibodies. The sensitivity of the antibody
microarray is pg/ml and there were no cross reactions between
cytokines. The background noise was low and signals from positive
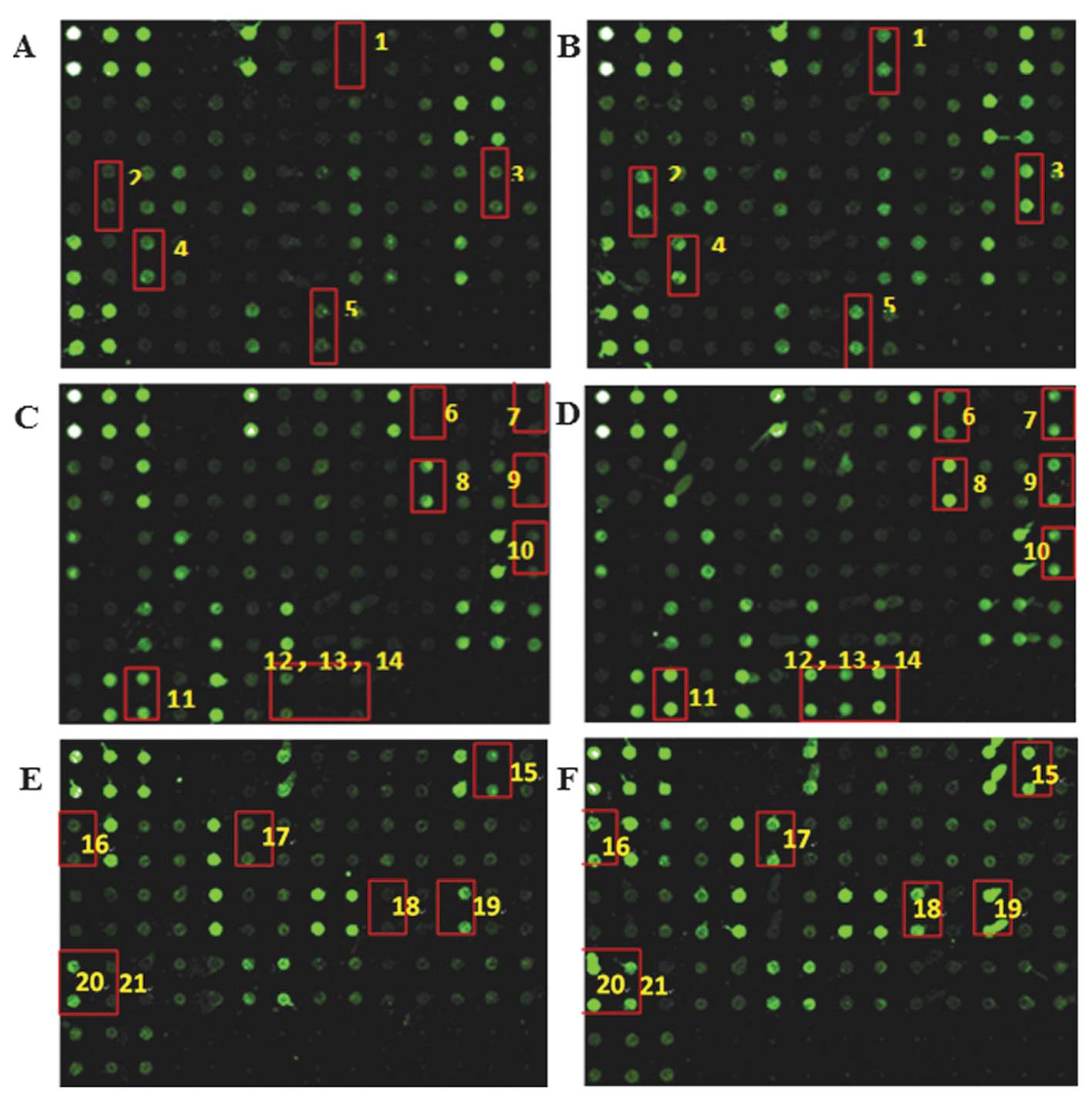
and negative control arrays were normal. Fig. 1 shows the expression levels of
cytokines in OS samples compared with that in with control samples.
The cytokines shown in Fig. 1 are
listed in Table II. Expression
levels of 21 cytokines were upregulated in OS samples when compared
with control samples (Table II).
The ratio of signal values from OS to control serum ranged from
2.15 to 12.14 fold. These results indicate clear differences in the
cytokine expression profile between the OS and control groups.
 | Table IIExpressed cytokines identified in OS
using antibody microarray. |
Table II
Expressed cytokines identified in OS
using antibody microarray.
| No. | Cytokine | Signal value
| P-value | Fold change |
|---|
| Control group | OS group |
|---|
| 1 | BMP-4 | 89 | 234 | 0.034 |
2.63 |
| 2 | IL-10 | 324 | 698 | 0.041 |
2.15 |
| 3 | IL-6 | 446 | 5421 | 0.011 | 12.14 |
| 4 | MCP-1 | 1324 | 4673 | 0.023 |
3.53 |
| 5 | TGF-β | 564 | 1321 | 0.009 |
2.34 |
| 6 | bFGF | 169 | 765 | 0.004 |
4.52 |
| 7 | CCL-28 | 125 | 897 | 0.02 |
7.18 |
| 8 | GRO | 2749 | 8754 | 0.006 |
3.18 |
| 9 | HGF | 165 | 806 | 0.007 |
4.88 |
| 10 | IL-8 | 234 | 786 | 0.021 |
3.36 |
| 11 | TIMP-2 | 3215 | 12497 | 0.034 |
3.89 |
| 12 | uPAR | 987 | 3215 | 0.008 |
3.26 |
| 13 | VEGF-A | 123 | 798 | 0.045 |
6.49 |
| 14 | VEGF-D | 132 | 1342 | 0.003 | 10.17 |
| 15 | CXCL-16 | 1400 | 5414 | 0.018 |
3.87 |
| 16 | Endoglin | 204 | 1189 | 0.003 |
5.82 |
| 17 | IGF-II | 214 | 1023 | 0.041 |
4.78 |
| 18 | MMP-1 | 234 | 1213 | 0.038 |
5.18 |
| 19 | MMP-9 | 2315 | 15478 | 0.029 |
6.69 |
| 20 | PDGF AA | 2984 | 32158 | 0.0004 | 10.78 |
| 21 | PDGF-AB | 101 | 1078 | 0.048 | 10.71 |
ELISA validation of antibody microarray
performance
Validation is important for interpreting and using
antibody microarrays. The following nine cytokines shown in
Table II were selected for ELISA
analysis: interleukin 6 (IL-6), monocyte chemoattractant protein-1
(MCP-1), transforming growth factor-β (TGF-β), growth-related
oncogene (GRO), hepatocyte growth factor (HGF), CXCL-16, Endoglin,
matrix metalloproteinase-9 (MMP-9) and platelet-derived growth
factor (PDGF)-AA. Table III
shows the cytokine expression profile analysis by antibody
microarray and ELISA. ELISA results showed that expression levels
of cytokines were significantly upregulated in OS samples compared
with control samples (P<0.05), which was consistent with the
microarray results. However, the ELISA concentration ratios between
the OS group and control group were not exactly the same as that in
the microarray results. ELISA results showed that the
concentrations of CXCL-16, TGF-β and Endoglin were 4.99-, 3.56- and
3.29-fold greater than that in the control group, respectively.
While antibody microarray results demonstrated that IL-6 and
PDGF-AA had higher signal values compared with the control serum
samples, 12.14- and 10.17-fold increases, respectively.
 | Table IIIEnzyme-linked immunosorbent assay
validation of nine cytokines. |
Table III
Enzyme-linked immunosorbent assay
validation of nine cytokines.
| Cytokine | Signal value
| ELISA concentration
(pg/ml)
|
|---|
| Control group | Osteosarcoma (OS)
group | Ratio (OS to
control) | Control group | OS group | Ratio (OS to
control) |
|---|
| IL-6 | 446±82.23 | 5421±236.98 | 12.14 | 10.8±2.23 | 21.65±9.34a | 2.00 |
| MCP-1 | 1324±257.51 | 4673±273.56 |
3.53 | 380.65±65.66 |
985.65±132.55b | 2.59 |
| TGF-β | 564±128.46 | 1321±142,87 |
2.34 | 12.12±3.09 | 43.14±10.11a | 3.56 |
| GRO | 2749±874.44 | 8754±486.66 |
3.18 | 98.24±16.23 |
221.23±45.98a | 2.25 |
| HGF | 165±22.77 | 806±45.66 |
4.88 | 897.12±132.12 | 2134±209.88b | 2.38 |
| CXCL-16 | 1400±285.63 | 5414±853.65 |
3.87 | 2.45±1.02 | 12.23±4.56b | 4.99 |
| Endoglin | 204±32.86 | 1189±174.80 |
5.82 | 3.88±2.32 | 12.78±3.89a | 3.29 |
| MMP-9 | 2315±443.74 | 15478±5974.92 |
6.49 | 456.12±89.24 |
879.31±129.05a | 1.93 |
| PDGF-AA | 2984±346.44 | 32158±1679.54 | 10.17 | 4321.93±736.88 |
7865.45±628.00a | 1.82 |
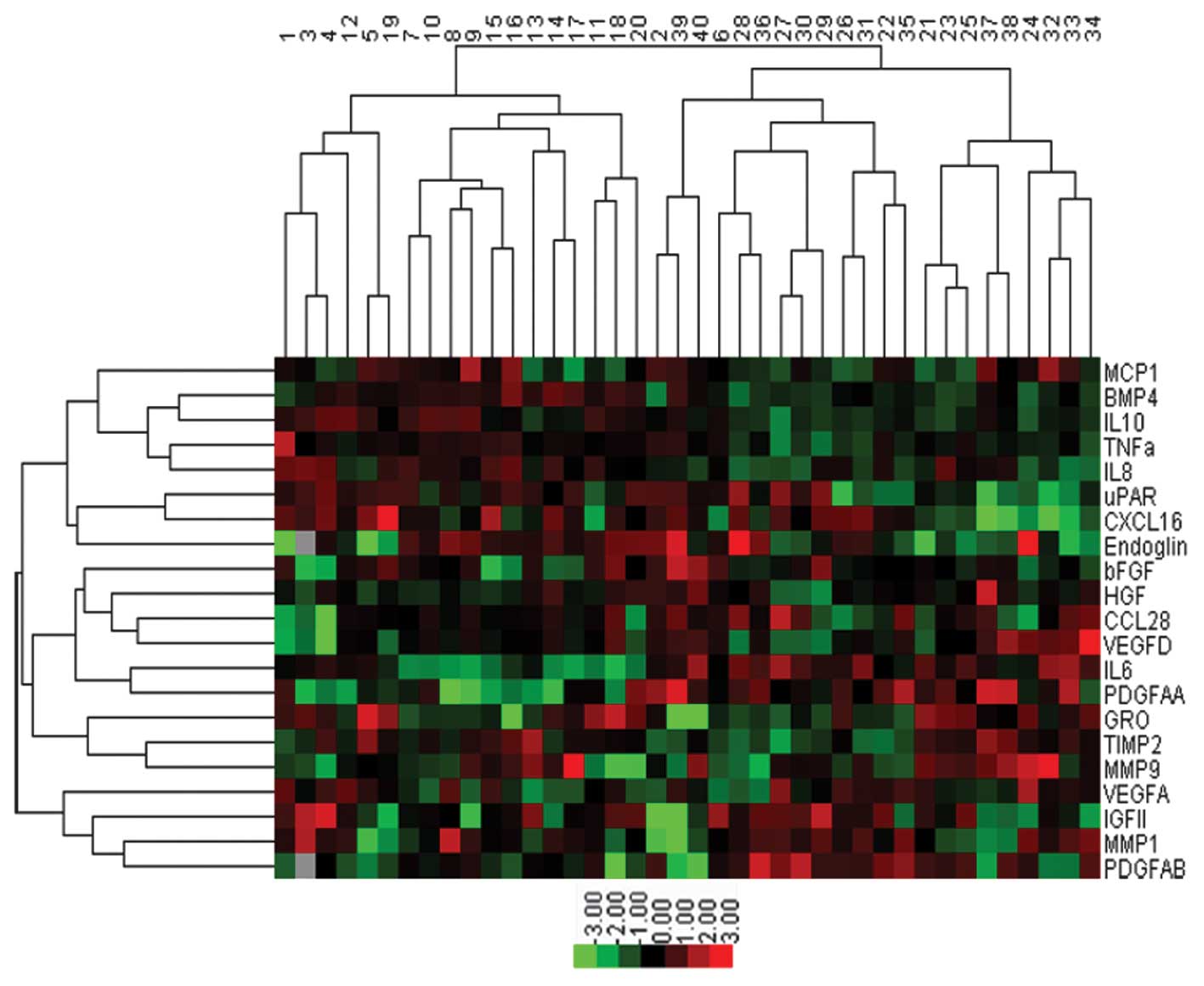
Sample classification
Cluster analysis was performed on the 21
significantly upregulated cytokines (P<0.05) screened by
antibody microarray (Fig. 2). The
control group included 20 samples (nos. 1–20) and OS group included
20 samples (nos. 21–40). In total, 18 serum samples in the control
group could be distinguished from OS group and all serum samples in
the OS group could be distinguished from the control group. The
accuracy of classification was 95% ((18+20)/40). The results showed
that cytokine screening was able to distinguish OS patients from
healthy individuals.
Discussion
In the present study, 21 cytokines were identified
that were significantly upregulated in OS serum samples, compared
with control serum samples. ELISA was then used to validate
antibody microarray results. Thus, the antibody microarray method
was observed to be reliable and significantly expressed cytokines
may be used as biomarkers for OA diagnosis and therapy. In
pediatric OS patients, antibody microarray identified elevated
expression of CXC chemokines and the results were validated by
ELISA (5). Similarly, antibody
microarray has been used to screen biomarkers in other types of
cancer, such as prostate (23),
lung (24) and pancreatic cancer
(25). The ratio of antibody
microarray signals and ELISA concentrations from OS to control
serum were not identical, however, the pattern of cytokine
expression was the same. Microarrays can be affected by numerous
factors, such as loss of antibody function, low antigen
concentration and inconsistent antigen labeling (23).
This study identified 21 cytokines with upregulated
expression in OS serum samples and expression of nine cytokines was
validated by ELISA. Among these cytokines, MMP-9 has been reported
to be expressed in childhood osseous OS (26). IL-6, HGF, Endoglin, TGF-β, PDGF-AA
and MCP-1 have been reported to be associated with the pathogenesis
of OS (27–32). CXCL16, a small protein that is a
member of the CXC chemokine family (33), has been reported to be expressed in
tumor cells and may be a prognostic marker in patients with
colorectal cancer (34) and renal
cell cancer (35). In the present
study, CXCL16 was differentially expressed in OS samples compared
with the control, and this was validated by ELISA. Conversely,
CXCL16 did not show any differential abundance in pediatric OS
patients compared with noncancerous disease controls in a study by
Li et al (5). These
opposite results may be due to the differences in serum samples and
antibody microar-rays. However, this requires confirmation in
future studies.
To the best of our knowledge, this is the first
study to show that GRO is upregulated in patients with OS. GRO, a
member of the CXC chemokine subfamily, associated with
tumorigenesis and metastasis (36), acted as growth factor in an
autocrine manner in a human pancreatic cancer cell line (37) and mediated the invasion of breast
cancer cells (38). GRO was also
observed to contribute to the proliferation of esophageal cancer
(36). Our results showed that
expression levels of GRO were higher in OS samples than in control
samples, which suggested that GRO may be a biomarker for OS
diagnosis and therapy. However, further investigation is required
for confirmation.
In conclusion, 21 upregulated cytokines were
identified in OS samples using antibody microarray and ELISA to
validate the results. Clustering analysis showed that expressed
cytokines could distinguish patients with OS from healthy
individuals. Thus, the expression of cytokines, screened by
antibody microarray, may be used as biomarkers for OS diagnosis and
therapy. However, the present data are limited to a relatively
small group of patient samples and large experiments are required
to investigate the association between cytokine expression and
OS.
References
|
1
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21:vii320–vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wafa H and Grimer RJ: Surgical options and
outcomes in bone sarcoma. Expert Rev Anticancer Ther. 6:239–248.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zetter B: Adhesion molecules in tumor
metastasis. J Exp Med. 4:219–229. 1993.
|
|
4
|
Wolf SF, Lee K, Swiniarski H, O’toole M
and Sturmhoefel K: Cytokines and cancer immunotherapy. Immunol
Invest. 29:143–146. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Y, Flores R, Yu A, et al: Elevated
expression of CXC chemokines in pediatric osteosarcoma patients.
Cancer. 117:207–217. 2011. View Article : Google Scholar
|
|
6
|
Smyth MJ, Thia KY, Street SE, et al:
Differential tumor surveillance by natural killer (NK) and NKT
cells. J Exp Med. 191:661–668. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davidson WF, Giese T and Fredrickson TN:
Spontaneous development of plasmacytoid tumors in mice with
defective Fas-Fas ligand interactions. J Exp Med. 187:1825–1838.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin EY, Nguyen AV, Russell RG and Pollard
JW: Colony-stimulating factor 1 promotes progression of mammary
tumors to malignancy. J Exp Med. 193:727–740. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li B, Yang Y, Jiang S, Ni B, Chen K and
Jiang L: Adenovirus-mediated overexpression of BMP-9 inhibits human
osteosarcoma cell growth and migration through downregulation of
the PI3K/AKT pathway. Int J Oncol. 41:1809–1819. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khan N, Cromer CJ, Campa M and Patz EF:
Clinical utility of serum amyloid A and macrophage migration
inhibitory factor as serum biomarkers for the detection of nonsmall
cell lung carcinoma. Cancer. 101:379–384. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Noo ME, Tollenaar RA, Özalp A, et al:
Reliability of human serum protein profiles generated with C8
magnetic beads assisted MALDI-TOF mass spectrometry. Anal Chem.
77:7232–7241. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oh Y-H, Hong M-Y, Jin Z, et al: Chip-based
analysis of SUMO (small ubiquitin-like modifier) conjugation to a
target protein. Biosens Bioelectron. 22:1260–1267. 2007. View Article : Google Scholar
|
|
14
|
Zhou H, Bouwman K, Schotanus M, et al:
Two-color, rolling-circle amplification on antibody microarrays for
sensitive, multiplexed serum-protein measurements. Genome Biol.
5:R282004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang R-P, Huang R, Fan Y and Lin Y:
Simultaneous detection of multiple cytokines from conditioned media
and patient’s sera by an antibody-based protein array system. Anal
Biochem. 294:55–62. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang CC, Huang RP, Sommer M, et al:
Array-based multiplexed screening and quantitation of human
cytokines and chemokines. J Proteome Res. 1:337–343. 2002.
View Article : Google Scholar
|
|
17
|
Pitha-Rowe I, Petty WJ, Feng Q, et al:
Microarray analyses uncover UBE1 L as a candidate target gene for
lung cancer chemoprevention. Cancer Res. 64:8109–8115. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tannapfel A, Anhalt K, Häusermann P, et
al: Identification of novel proteins associated with hepatocellular
carcinomas using protein microarrays. J Pathol. 201:238–249. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dahlin DC and Coventry MB: Osteogenic
sarcoma. A study of six hundred cases. J Bone Joint Surg Am.
49:101–110. 1967.PubMed/NCBI
|
|
20
|
Johnson SC: Hierarchical clustering
schemes. Psychometrika. 32:241–254. 1967. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
de Hoon MJ, Imoto S, Nolan J and Miyano S:
Open source clustering software. Bioinformatics. 20:1453–1454.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
D’haeseleer P: How does gene expression
clustering work? Nat Biotechnol. 23:1499–1501. 2005. View Article : Google Scholar
|
|
23
|
Miller JC, Zhou H, Kwekel J, et al:
Antibody microarray profiling of human prostate cancer sera:
antibody screening and identification of potential biomarkers.
Proteomics. 3:56–63. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao WM, Kuick R, Orchekowski RP, et al:
Distinctive serum protein profiles involving abundant proteins in
lung cancer patients based upon antibody microarray analysis. BMC
cancer. 5:1102005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Orchekowski R, Hamelinck D, Li L, et al:
Antibody microarray profiling reveals individual and combined serum
proteins associated with pancreatic cancer. Cancer Res.
65:11193–11202. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Himelstein BP, Asada N, Carlton MR and
Collins MH: Matrix metalloproteinase. 9 (MMP. 9) expression in
childhood osseous osteosarcoma. Med Pediatr Oncol. 31:471–474.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bian ZY, Fan QM, Li G, Xu WT and Tang TT:
Human mesenchymal stem cells promote growth of osteosarcoma:
Involvement of interleukin. 6 in the interaction between human
mesenchymal stem cells and Saos 2. Cancer Sci. 101:2554–2560. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Coltella N, Manara MC, Cerisano V, et al:
Role of the MET/HGF receptor in proliferation and invasive behavior
of osteosarcoma. FASEB J. 17:1162–1164. 2003.PubMed/NCBI
|
|
29
|
Postiglione L, Di Domenico G, Caraglia M,
et al: Differential expression and cytoplasm/membrane distribution
of endoglin (CD105) in human tumour cell lines: implications in the
modulation of cell proliferation. Int J Oncol. 26:1193–1201.
2005.PubMed/NCBI
|
|
30
|
Kloen P, Gebhardt MC, Perez-Atayde A,
Rosenberg AE, Springfield DS, Gold LI and Mankin HJ: Expression of
transforming growth factorbeta (TGF-beta) isoforms in
osteosarcomas: TGF-beta3 is related to disease progression. Cancer.
80:2230–2239. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sulzbacher I, Birner P, Trieb K, Träxler
M, Lang S and Chott A: Expression of platelet-derived growth
factor-AA is associated with tumor progression in osteosarcoma.
Modern Pathol. 16:66–71. 2003. View Article : Google Scholar
|
|
32
|
Lin SK, Kok SH, Yeh FT, et al: MEK/ERK and
signal transducer and activator of transcription signaling pathways
modulate oncostatin M-stimulated CCL2 expression in human
osteoblasts through a common transcription factor. Arthritis Rheum.
50:785–793. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lehrke M, Millington SC, Lefterova M, et
al: CXCL16 is a marker of inflammation, atherosclerosis and acute
coronary syndromes in humans. J Am Coll Cardiol. 49:442–449. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hojo S, Koizumi K, Tsuneyama K, et al:
High-level expression of chemokine CXCL16 by tumor cells correlates
with a good prognosis and increased tumor-infiltrating lymphocytes
in colorectal cancer. Cancer Res. 67:4725–4731. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gutwein P, Schramme A, Sinke N, et al:
Tumoural CXCL16 expression is a novel prognostic marker of longer
survival times in renal cell cancer patients. Eur J Cancer.
45:478–489. 2009. View Article : Google Scholar
|
|
36
|
Wang B, Hendricks DT, Wamunyokoli F and
Parker MI: A growth-related oncogene/CXC chemokine receptor 2
autocrine loop contributes to cellular proliferation in esophageal
cancer. Cancer Res. 66:3071–3077. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Takamori H, Oades ZG, Hoch OC, Burger M
and Schraufstatter IU: Autocrine growth effect of il-8 and gro
[alpha] on a human pancreatic cancer cell line, capan-1. Pancreas.
21:52–56. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li J and Sidell N: Growth related oncogene
produced in human breast cancer cells and regulated by Syk protein
tyrosine kinase. Int J Cancer. 117:14–20. 2005. View Article : Google Scholar : PubMed/NCBI
|