Introduction
Tuberculosis (TB) is an infectious disease commonly
caused by mycobacteria (1). TB is
considered to be an acute global health problem with ~9 million
novel TB cases and 1.4 million fatalities each year (2). TB commonly originates in the lungs,
but is able to spread to other parts of the body, leading to
extra-pulmonary diseases (3).
Among the patients infected with TB, ~10% progress to active TB
during their lifespan and the remaining individuals remain
asymptomatic (4). The immune
responses of TB patients are mainly regulated by T helper 1 cells,
which secrete interferon-γ (IFN-γ) (5). IFN-γ mediated immune responses
activate macrophages, which induce the secretion of other
cytokines, including interleukin (IL)-1, IL-12 and tumor necrosis
factor (TNF)-α (6). Previously,
genome-wide association studies have revealed that genetic
variation in genes involved in immune responses, including IL-1,
IL-12 and TNF-α, is associated with the risk of TB (7–9).
The IFN-γ-induced signaling pathway is activated by
interacting with its receptor composed of two subunits, IFN-γ
receptor (IFNGR) 1 and 2, which encode the ligand-biding chain
(α-chain) and the non-ligand binding chain, respectively. IFNGR is
involved in a positive feedback loop of IFN-γ expression (10). Genetic variation in
cytokine-associated genes, including IFNGR1 and
IFNGR2, have previously been found to be important in other
viral/host-mediated immune responses in TB (11–16).
Among the genetic variants in IFNGR1, the single nucleotide
polymorphism (SNP) rs2234711 has been revealed to be a major
marker of disease protection. In a recent Chinese study, patients
with rs2234711 had a significantly lower prevalence of TB
[odds ratio (OR)=0.82, P<0.001] (17). However, to date, an association
between the risk for TB and genetic variation in the IFNGR1
and IFNGR2 genes had not been demonstrated in a Korean
population. In the present study, the association of polymorphisms
in the IFNGR1 and IFNGR2 genes with the risk of TB in
the Korean population was investigated.
Patients and methods
Patients
A total of 673 patients with clinical manifestation
of pulmonary TB (mean age, 45.81 years; range, 16–92 years, 388
males and 285 females) were recruited from Soonchunhyang University
Bucheon Hospital (Bucheon, Republic of Korea). Polymerase chain
reaction was used to assess all sputum acid-fast bacillus
culture-positive samples to distinguish Mycobacterium
tuberculosis (MTB) from non-tuberculous mycobacteria (NTM). The
diagnosis of pulmonary TB was confirmed by the isolation of MTB
from the sputum or bronchoalveolar lavage fluid. Patients with an
NTM infection were excluded from the present study. Patients with
TB who had a family history of the disease were also excluded from
the study to eliminate the additional risk factors of added
exposure to TB. A total of 592 healthy controls (mean age, 50.22
years; range, 9–87 years, 277 males and 315 females) were
simultaneously recruited from a randomly sampled population who had
attended the clinic for routine health checkups in the same
regional area. Only patients above the age of 40 years were
included in the normal control group to exclude the possibility of
TB infection among young individuals who may subsequently develop
the condition. Individuals with other medical diseases/conditions,
including human immunodeficiency virus, hepatitis, diabetes,
alcoholism, autoimmune diseases and cancer were also excluded from
the present study.
The ethnicity of all patients and controls was
Korean. Written informed consent was obtained from all patients
prior to the start of the experiment. The experimental protocol was
approved by the Institutional Review Board of Soonchunhyang
University Bucheon Hospital (IRB no. schbc-biobank-2012-001).
SNP genotyping
Candidate SNPs of the IFNGR1 and
IFNGR2 genes were selected from Japanese and Han Chinese
data from the 1,000 Genomes database (http://browser.1000genomes.org/index.html) based on
the allele frequency and linkage disequilibrium (LD) status in the
Asian population. Additional SNPs which had been previously
investigated were also selected (14). A total of 11 SNPs of the
IFNGR1 gene and 11 SNPs of the IFNGR2 gene were
selected based on the following criteria: Minor allele frequency
(MAF; >5%) and LD (r2>0.98). A total of 22
polymorphisms were genotyped in 673 TB patients and 592 normal
controls using a TaqMan assay on the ABI prism 7900HT sequence
detection system (Applied Biosystems, Foster City, CA, USA)
(18). Quality control of the
genotyping was performed in 10% of the samples by duplicate
checking (rate of concordance in duplicates, >99.5%). Selected
SNPs and probe information on the polymorphisms is shown in
Table I.
 | Table IProbe information for IFNGR1 and
IFNGR2. |
Table I
Probe information for IFNGR1 and
IFNGR2.
| Gene | Loci | Assay on demand ID
or probe sequence |
|---|
| IFNGR1 |
rs28515059 | C__63095558_10 |
|
rs1327474 | C___2523634_10 |
|
rs2234711 | C__11693991_10 |
|
rs10457655 | C__30506149_10 |
|
rs9376269 | C__30272193_20 |
|
rs9376268 | C__30470198_10 |
|
rs9376267 | C__30182293_10 |
|
rs56251346 |
TGTTTACAAAGTGGGCACATCa |
| |
ATTGGAAACATTTCCCCATCb |
| | CATTACTTGCc |
| | CATTATTTGCd |
|
rs3799488 | C__25647358_10 |
| rs11914 | C___7578627_10 |
|
rs1887415 | C__11693851_30 |
| IFNGR2 |
rs4817565 |
GACATTGCCACAACATCCAGa |
| |
GAGCCTGGCCTCACTTTTTAb |
| | ACCTGTCCATc |
| | ACCTATCCATd |
|
rs73194070 |
ACTGTGAGGGAGCATTGACCa |
| |
CCGAAGGCAGACAGGTAAAGb |
| | ACCACCCCCCc |
| | ACCACACCCCd |
|
rs9808753 | C___2443413_1_ |
|
rs2834211 | C__16072862_10 |
|
rs2834213 | C___2443417_10 |
|
rs115346998 |
AGAAGGCTCCCTCATCATCAa |
| |
TCTTGCCTGTTGGATTCCTCb |
| | TGTCCATTACc |
| | TGTCCGTTACd |
|
rs8126735 |
TGAAGCATCTCCAGTGCCTAa |
| |
GAGCCAAACACAAAGGAAGCb |
| | TTATAATGGTc |
| | TTATGATGGTd |
|
rs8128483 |
GAAGAGGCACATGGAGGAAAa |
| |
CCTGGCAGACAACAGTTCACb |
| | TCATCGCTCCc |
| | TCATTGCTCCd |
|
rs143025663 |
GTTTCACACTCCACCAAGCAa |
| |
GCTGCAGTGAGCAGAGATTGb |
| | TTACAGATAGc |
| | TTACCGATAGd |
|
rs1059293 | C___2443435_10 |
|
rs17882754 |
TCATGGGAACTCAGCAAACAa |
| |
CTCAAGTGATCCACCCACCTb |
| | CAGGGCCTAGc |
| | CAGGACCTAGd |
Statistical analysis
The level of LD was obtained using Haploview version
4.2 software (Broad Institute, Cambridge, MA, USA; http://www.broadinstitute.org/mpg/haploview), with
examination of Lewontin’s D′ (|D′|) and the LD coefficient
r2 between all pairs of bi-allelic loci (19). Haplotypes were estimated using
PHASE version 2.1 software (Stephen Laboratory, University of
Chicago, Chicago, IL, USA) (20).
A comparison of genotype distributions between TB patients and
healthy controls was performed using a logistic regression model
adjusted for age (continuous value) and gender (male=0, female=1)
as co-variates using SAS software, version 9.3 (SAS Inc., Cary, NC,
USA). The effective number of independent marker loci was
calculated for multiple testing corrections using SNPSpD
(http://genepli.qimr.edu.au/general/daleN/SNPSpD/),
a program based on the spectral decomposition of matrices of
pair-wise LD between SNPs (21).
The total sum of independent marker loci in the gene was calculated
as 7.7553 for IFNGR1 and 9.3328 for IFNGR2, and this
value was applied to correct for multiple testing.
Results
Genotyping and haplotype analysis of
IFNGR1 and IFNGR2
In the present study, a total of 22 polymorphisms
(11 in IFNGR1 and 11 in IFNGR2) were selected, based
on their MAF, location and LD status, and genotyped in 673 TB cases
and 592 healthy controls. Detailed information regarding
polymorphisms, including allele, amino acid change, position, MAF,
heterozygosity and P-values for the Hardy-Weinberg equilibrium are
shown in Table II. LDs among SNPs
were obtained by calculating |D′| and r2 values.
Among the investigated polymorphisms, ten polymorphisms in
IFNGR1 and eight in IFGNR2 were used for LD block
construction of each gene. The genetic variants rs1887415,
rs115346998 and rs143025663 were excluded from LD
block construction due to its low frequency (MAF<5%). As a
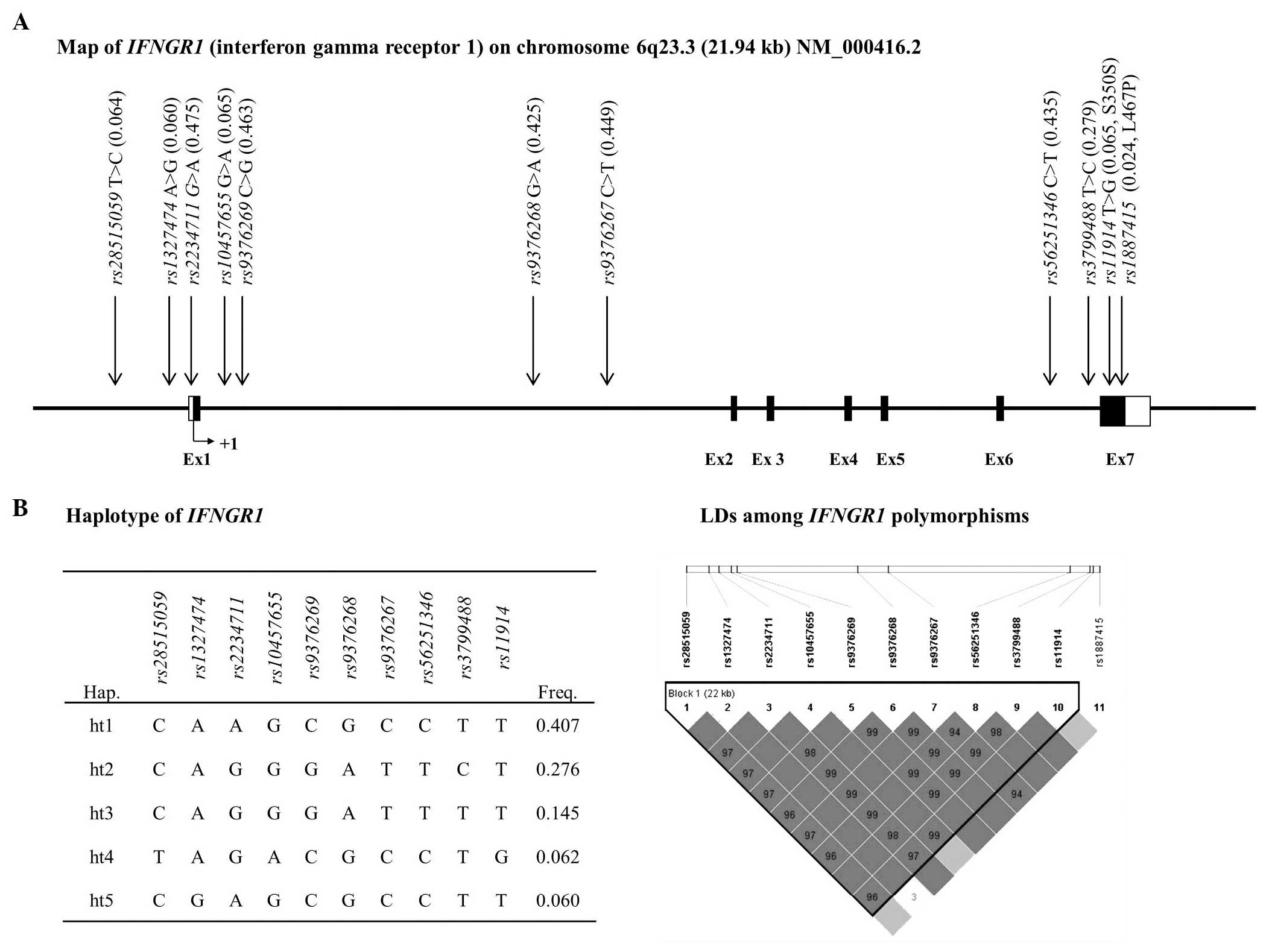
result, one LD block was constructed in IFNGR1 that
contained five major haplotypes (ht), which exhibited a MAF>5%
(Fig. 1). Among the IFNGR1
haplotypes, IFNGR1_ht4 and IFNGR1_ht5 exhibited
equivalence with rs28515059 and rs1327474,
respectively, and those haplotypes were excluded from the further
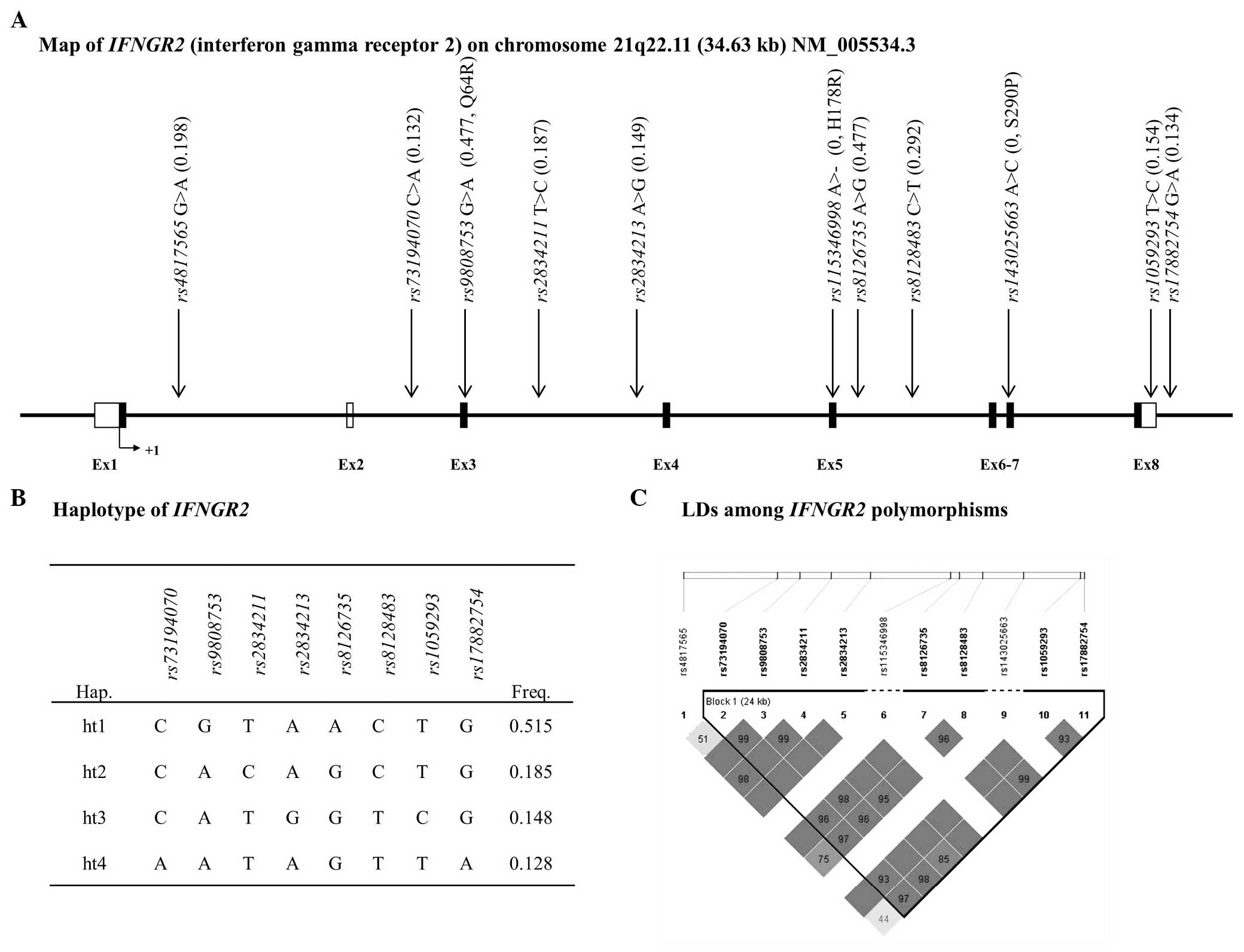
analysis. In the case of IFNGR2, one LD block was
constructed and it contained four major haplotypes, which exhibited
a MAF>5% (Fig. 2).
 | Table IIAllele information of IFNGR1
and −2 polymorphisms in Korean patients (n=1265). |
Table II
Allele information of IFNGR1
and −2 polymorphisms in Korean patients (n=1265).
| Gene | SNP | Allele | Position | AA Change | Genotype
| MAF | Heterozygosity | HWE
|
|---|
| C/C | C/R | R/R | TB | NC | Total |
|---|
| IFNGR1 |
rs28515059 | C>T | 5′ flanking | | 1,106 | 151 | 5 | 0.064 | 0.119 | 0.803 | 0.728 | 0.949 |
|
rs1327474 | A>G | 5′ flanking | | 1,117 | 145 | 3 | 0.060 | 0.112 | 0.374 | 0.859 | 0.451 |
|
rs2234711 | G>A | 5′ UTR | | 345 | 632 | 282 | 0.475 | 0.499 | 0.744 | 0.987 | 0.818 |
|
rs10457655 | G>A | Intron1 | | 1,101 | 154 | 5 | 0.065 | 0.122 | 0.841 | 0.669 | 0.876 |
|
rs9376269 | C>G | Intron1 | | 357 | 643 | 264 | 0.463 | 0.497 | 0.5 | 0.049 | 0.415 |
|
rs9376268 | G>A | Intron1 | | 411 | 633 | 221 | 0.425 | 0.489 | 0.939 | 0.157 | 0.396 |
|
rs9376267 | C>T | Intron1 | | 377 | 636 | 248 | 0.449 | 0.495 | 0.415 | 0.054 | 0.491 |
|
rs56251346 | C>T | Intron6 | | 395 | 640 | 230 | 0.435 | 0.491 | 0.904 | 0.136 | 0.296 |
|
rs3799488 | T>C | Intron6 | | 650 | 522 | 92 | 0.279 | 0.403 | 0.239 | 0.901 | 0.358 |
| rs11914 | T>G | Exon7 | S350S | 1,105 | 155 | 5 | 0.065 | 0.122 | 0.837 | 0.647 | 0.861 |
|
rs1887415 | T>C | Exon7 | L467P | 1,206 | 58 | 1 | 0.024 | 0.046 | 0.609 | 0.451 | 0.726 |
| IFNGR2 |
rs4817565 | G>A | Intron1 | | 813 | 402 | 50 | 0.198 | 0.318 | 0.488 | 0.424 | 0.972 |
|
rs73194070 | C>A | Intron2 | | 946 | 303 | 16 | 0.132 | 0.230 | 0.472 | 0.145 | 0.130 |
|
rs9808753 | G>A | Exon3 | Q64R | 341 | 642 | 282 | 0.477 | 0.499 | 0.645 | 0.682 | 0.540 |
|
rs2834211 | T>C | Intron3 | | 836 | 384 | 45 | 0.187 | 0.305 | 0.669 | 0.553 | 0.912 |
|
rs2834213 | A>G | Intron3 | | 917 | 315 | 30 | 0.149 | 0.253 | 0.754 | 0.726 | 0.634 |
|
rs115346998 | A> | Exon5 | H178R | 1,265 | – | – | – | – | – | – | – |
|
rs8126735 | A>G | Intron5 | | 339 | 645 | 281 | 0.477 | 0.499 | 0.402 | 0.798 | 0.436 |
|
rs8128483 | C>T | Intron5 | | 641 | 509 | 115 | 0.292 | 0.414 | 0.664 | 0.351 | 0.336 |
|
rs143025663 | A>C | Exon7 | Q290P | 1,264 | 1 | 0 | 0.000 | 0.001 | 0.985 | – | 0.989 |
|
rs1059293 | T>C | 3′ UTR | | 911 | 319 | 35 | 0.154 | 0.260 | 0.294 | 0.654 | 0.271 |
|
rs17882754 | G>A | 3′ flanking | | 942 | 1,248 | 17 | 0.134 | 0.233 | 0.607 | 0.124 | 0.158 |
Correlation analyses of SNPs in IFNGR1
and IFNGR2 with TB
The case-control analysis of the correlation between
IFNGR1 or IFNGR2 polymorphisms and the risk of TB was
conducted (Tables III and
IV). The correlation analysis
revealed that the two SNPs in IFNGR1, rs9376268 and
rs56251346, induced an increased risk for TB under a
co-dominant model (OR=1.18 and 1.19; P=0.05 and 0.04,
respectively). The two SNPs exhibited similar genetic effects with
a higher level of significance under a recessive model (OR=1.40;
P=0.03 for the two SNPs). Along with rs9376268 and
rs56251346, two SNPs in intron 1, rs9376269 and
rs9376267, also induced an increased risk for TB under a
recessive model (OR=1.40; P=0.02 for the two SNPs). However, the
level of significance was not retained following the correction for
multiple testing in all analysis models (P>0.05). Polymorphisms
in the coding region, rs11914 (S350S) and rs1887415
(L467P), were not associated with an increased risk for TB. In the
haplotype analysis, IFNGR1_ht2 exhibited a marginal
association with the risk for TB under a dominant model (P=0.04),
although its association was eradicated following the correction
for multiple testing. However, no genetic polymorphisms and
haplotypes in IFNGR2 exhibited significant correlations with
the risk of developing TB.
 | Table IIILogistic analysis of IFNGR1
polymorphisms. |
Table III
Logistic analysis of IFNGR1
polymorphisms.
| Loci | Allele | Position | AA change | MAF
| Codominant
| Dominant
| Recessive
|
|---|
| TB | NC | OR (95% CI) | P |
pcorr | OR (95% CI) | P |
pcorr | OR (95% CI) | P |
pcorr |
|---|
|
rs28515059 | C>T | 5′ flanking | | 0.062 | 0.065 | 0.94
(0.68–1.30) | 0.72 | 1 | 0.93
(0.66–1.31) | 0.67 | 1 | 1.25
(0.21–7.61) | 0.81 | 1 |
|
rs1327474 | A>G | 5′ flanking | | 0.058 | 0.062 | 0.93
(0.66–1.30) | 0.65 | 1 | 0.94
(0.66–1.33) | 0.71 | 1 | 0.47
(0.04–5.51) | 0.55 | 1 |
|
rs2234711 | G>A | 5′ UTR | | 0.464 | 0.487 | 0.90
(0.77–1.06) | 0.21 | 1 | 0.89
(0.69–1.15) | 0.37 | 1 | 0.85
(0.65–1.11) | 0.24 | 1 |
|
rs10457655 | G>A | Intron1 | | 0.063 | 0.067 | 0.93
(0.67–1.28) | 0.66 | 1 | 0.92
(0.65–1.28) | 0.61 | 1 | 1.26
(0.21–7.64) | 0.80 | 1 |
|
rs9376269 | C>G | Intron1 | | 0.476 | 0.448 | 1.13
(0.97–1.33) | 0.13 | 0.97 | 1.03
(0.80–1.32) | 0.80 | 1 | 1.40
(1.06–1.85) | 0.02 | 0.15 |
|
rs9376268 | G>A | Intron1 | | 0.442 | 0.405 | 1.18
(1.00–1.38) | 0.05 | 0.40 | 1.14
(0.90–1.45) | 0.27 | 1 | 1.40
(1.04–1.88) | 0.03 | 0.22 |
|
rs9376267 | C>T | Intron1 | | 0.461 | 0.435 | 1.13
(0.96–1.32) | 0.15 | 1 | 1.02
(0.80–1.30) | 0.88 | 1 | 1.40
(1.05–1.87) | 0.02 | 0.16 |
|
rs56251346 | C>T | Intron6 | | 0.452 | 0.415 | 1.19
(1.01–1.40) | 0.04 | 0.29 | 1.17
(0.92–1.48) | 0.22 | 1 | 1.40
(1.04–1.88) | 0.03 | 0.19 |
|
rs3799488 | T>C | Intron6 | | 0.295 | 0.262 | 1.19
(0.99–1.43) | 0.06 | 0.46 | 1.27
(1.02–1.60) | 0.04 | 0.28 | 1.12
(0.72–1.73) | 0.62 | 1 |
| rs11914 | T>G | Exon7 | S350S | 0.063 | 0.068 | 0.93
(0.67–1.28) | 0.64 | 1 | 0.91
(0.65–1.28) | 0.59 | 1 | 1.26
(0.21–7.68) | 0.80 | 1 |
|
rs1887415 | T>C | Exon7 | L467P | 0.019 | 0.029 | 0.67
(0.40–1.12) | 0.13 | 0.98 | 0.68
(0.40–1.15) | 0.15 | 1 | – | – | – |
| ht1 | | | | 0.399 | 0.416 | 0.92
(0.78–1.08) | 0.33 | 1 | 0.95
(0.75–1.21) | 0.69 | 1 | 0.82
(0.60–1.10) | 0.19 | 1 |
| ht2 | | | | 0.291 | 0.259 | 1.19
(0.99–1.43) | 0.06 | 0.40 | 1.26
(1.01–1.58) | 0.04 | 0.31 | 1.16
(0.74–1.80) | 0.52 | 1 |
| ht3 | | | | 0.146 | 0.144 | 1.03
(0.82–1.30) | 0.78 | 1 | 1.00
(0.77–1.28) | 0.97 | 1 | 1.65
(0.70–3.92) | 0.25 | 1 |
| ht4 | | | | 0.061 | 0.064 | 0.94
(0.68–1.30) | 0.71 | 1 | 0.93
(0.66–1.30) | 0.66 | 1 | 1.26
(0.21–7.68) | 0.80 | 1 |
| ht5 | | | | 0.058 | 0.062 | 0.93
(0.66–1.30) | 0.65 | 1 | 0.94
(0.66–1.33) | 0.71 | 1 | 0.47
(0.04–5.51) | 0.55 | 1 |
 | Table IVLogistic analysis of IFNGR2
polymorphisms. |
Table IV
Logistic analysis of IFNGR2
polymorphisms.
| Loci | Allele | Position | AA change | MAF
| Codominant
| Dominant
| Recessive
|
|---|
| TB | NC | OR (95% CI) | P |
pcorr | OR (95% CI) | P |
pcorr | OR (95% CI) | P |
pcorr |
|---|
|
rs4817565 | G>A | Intron1 | | 0.200 | 0.197 | 1.02
(0.84–1.24) | 0.84 | 1 | 1.06
(0.84–1.34) | 0.62 | 1 | 0.83
(0.47–1.47) | 0.52 | 1 |
|
rs73194070 | C>A | Intron2 | | 0.134 | 0.130 | 1.06
(0.83–1.35) | 0.63 | 1 | 1.05
(0.81–1.35) | 0.74 | 1 | 1.44
(0.51–4.03) | 0.49 | 1 |
|
rs9808753 | G>A | Exon3 | Q64R | 0.481 | 0.471 | 1.03
(0.88–1.21) | 0.72 | 1 | 1.04
(0.81–1.34) | 0.77 | 1 | 1.04
(0.79–1.36) | 0.78 | 1 |
|
rs2834211 | T>C | Intron3 | | 0.184 | 0.192 | 0.95
(0.78–1.17) | 0.64 | 1 | 0.97
(0.77–1.23) | 0.82 | 1 | 0.78
(0.43–1.44) | 0.43 | 1 |
|
rs2834213 | A>G | Intron3 | | 0.154 | 0.142 | 1.06
(0.85–1.32) | 0.61 | 1 | 1.07
(0.83–1.37) | 0.61 | 1 | 1.09
(0.52–2.30) | 0.82 | 1 |
|
rs115346998 | A>- | Exon5 | H178R | – | – | – | – | – | – | – | – | – | – | – |
|
rs8126735 | A>G | Intron5 | | 0.484 | 0.470 | 1.05
(0.89–1.23) | 0.57 | 1 | 1.10
(0.85–1.41) | 0.48 | 1 | 1.03
(0.79–1.35) | 0.84 | 1 |
|
rs8128483 | C>T | Intron5 | | 0.300 | 0.283 | 1.07
(0.90–1.27) | 0.45 | 1 | 1.11
(0.89–1.39) | 0.37 | 1 | 1.03
(0.70–1.53) | 0.87 | 1 |
|
rs143025663 | A>C | Exon7 | Q290P | 0.001 | – | – | – | – | – | – | – | – | – | – |
|
rs1059293 | T>C | 3′ UTR | | 0.160 | 0.146 | 1.07
(0.86–1.33) | 0.53 | 1 | 1.07
(0.83–1.38) | 0.59 | 1 | 1.21
(0.60–2.42) | 0.60 | 1 |
|
rs17882754 | G>A | 3′ flanking | | 0.137 | 0.132 | 1.06
(0.84–1.34) | 0.64 | 1 | 1.03
(0.80–1.34) | 0.80 | 1 | 1.66
(0.60–4.57) | 0.33 | 1 |
| ht1 | | | | 0.490 | 0.478 | 1.04
(0.89–1.22) | 0.65 | 1 | 1.07
(0.83–1.38) | 0.59 | 1 | 1.03
(0.79–1.34) | 0.84 | 1 |
| ht2 | | | | 0.181 | 0.189 | 0.96
(0.78–1.18) | 0.68 | 1 | 0.97
(0.76–1.22) | 0.77 | 1 | 0.86
(0.47–1.60) | 0.64 | 1 |
| ht3 | | | | 0.154 | 0.141 | 1.07
(0.86–1.33) | 0.56 | 1 | 1.08
(0.84–1.39) | 0.56 | 1 | 1.09
(0.52–2.29) | 0.82 | 1 |
| ht4 | | | | 0.131 | 0.126 | 1.07
(0.84–1.36) | 0.60 | 1 | 1.06
(0.82–1.38) | 0.66 | 1 | 1.33
(0.47–3.82) | 0.59 | 1 |
Discussion
In previous studies, genetic variations in the genes
involved in the IFN-γ signaling pathway have been associated with
the risk of developing several mycobacterial diseases, particularly
TB (13–15). Defects in the proper functioning of
IFN-γ meditated immune responses is a major cause of disease
susceptibility (22). IFN-γ
activates transcription of a large number of cytokines, including
those secreted by macrophages, including IL-12 and TNF-α, which
have roles in immune responses, thus the appropriate function of
the IFNGR appears to be important in host defense against
mycobacteria (23).
In the present study, a logistic analysis was
conducted to identify a possible significant association between
genetic variants in the IFNGR genes and TB in a Korean population.
Previous studies have revealed a correlation of the IFNGR1
polymorphisms rs2234711, rs1327474 and
rs11914, with TB (Table V)
(13,14,17,24,25).
Studies in African populations have revealed that the prevalence of
TB was lower in African populations with the minor alleles of
rs11914 (S350S) and rs2234711, suggesting a
protective effect (OR=0.66; P=0.022 and OR=0.75; P=0.041,
respectively) (13,25). The protective effect of
rs2234711 on TB prevalence has also been observed in a
Chinese population (OR=0.82, P<0.001) (17). In another Chinese study,
rs7749300, which revealed a marked LD with rs2234711
and rs1327474, were significantly associated with the risk
of TB (OR=3.96; P=0.0003, from haplotype analysis of three SNPs)
(14). However, rs7749300
was not investigated in the present study due to perfect LD with
rs2234711 in the 1,000 Genomes database. However, previously
demonstrated genetic effects were not replicated in the present
study, which may be due to differences in the genetic diversity
among the populations. In the case of IFNGR2, two
polymorphisms (rs2834213 and 1059293) exhibited a
protective effect against the risk of developing TB (OR=0.69–0.70;
P=0.0073–0.0088) (26); however,
these findings were not replicated in the present study.
 | Table VComparison of previous studies on
IFNGR1-TB association. |
Table V
Comparison of previous studies on
IFNGR1-TB association.
| Reference | Study
population | Study patients
(cases/control, n) | Studied allele
|
|---|
| rs1327474
P-value (OR) | rs2234711
P-value (OR) | rs11914
P-value (OR) | rs9376268
P-value (OR) | rs56251346
P-value (OR) |
|---|
| Awomoyi et
al (2004) (24) | Gambian | 320/320 | 0.34 (1.19) | 0.5 (1.01)a | 0.23 (1.41) | – | – |
| Cooke et al
(2006) (25) | African | 682/619 | – | 0.041
(0.75) | – | – | – |
| He et al
(2010) (14) | Chinese | 222/188 | NS | NS | – | – | – |
| de Wit et al
(2011) (13) | African | 505/318 | – | – | 0.022
(0.66) | – | – |
| Lu J et al
(2014) (17) | Chinese | 1434/1412 | – | <0.001
(0.82) | – | – | – |
| Present study
(2014)b | Korean | 673/592 | 0.65 (0.93) | 0.21 (0.90) | 0.64 (0.93) | 0.05
(1.18) | 0.04
(1.19) |
In order to investigate whether the present results
were due to ethnic differences or not, the genetic composition of
IFNGR genes were compared between ethnicities. Frequency analysis
and Fisher’s exact test were additionally conducted among the four
groups, which included a Korean population from the present study,
as well as African, Asian and Caucasian populations from the 1,000
Genomes database (Table VI). As a
result, the SNP rs11914 exhibited a significant difference
in allelic distribution between Korean and African individuals.
Genetic compositions of rs11914 in the Japanese and Chinese
populations also differed from that of Korean individuals. Along
with the rs11914 SNP, other investigated SNPs, including
rs10457655, rs9376269, rs9376268,
rs9376267, rs56251346 and rs3799488, have
demonstrated a wide degree of frequency variance depending on the
populations (P<0.05).
 | Table VIComparison of genetic distribution in
ethnic groups of polymorphisms in IFNGR1 and
IFNGR2. |
Table VI
Comparison of genetic distribution in
ethnic groups of polymorphisms in IFNGR1 and
IFNGR2.
| Gene | Loci | Allele | MAF
| Fisher’s exact test
|
|---|
| KOR | AS | AF | CA | KR vs. AS | KR vs. AF | KR vs. CA | AS vs. AF | AS vs. CA | AF vs. CA |
|---|
| IFNGR1 |
rs28515059 | C>T | 0.064 | 0.075 | 0.049 | 0.172 | 0.3703 | 0.0984 | <.0001 | 0.0277 | <.0001 | <.0001 |
|
rs1327474 | A>G | 0.060 | 0.054 | 0.045 | 0.38 | 0.6130 | 0.3154 | 0.4094 | 0.2682 | 0.3146 | 0.1509 |
|
rs2234711 | G>A | 0.475 | 0.497 | 0.49 | 0.443 | 0.9348 | 0.0458 | <.0001 | 0.2491 | <.0001 | <.0001 |
|
rs10457655 | G>A | 0.065 | 0.078 | 0.293 | 0.178 | <.0001 | <.0001 | <.0001 | <.0001 | <.0001 | <.0001 |
|
rs9376269 | C>G | 0.463 | 0.419 | 0.11 | 0.262 | 0.1787 | <.0001 | <.0001 | <.0001 | <.0001 | <.0001 |
|
rs9376268 | G>A | 0.425 | 0.39 | 0.059 | 0.265 | <.0001 | <.0001 | <.0001 | <.0001 | 0.0021 | <.0001 |
|
rs9376267 | C>T | 0.449 | 0.409 | 0.12 | 0.265 | 0.2480 | <.0001 | <.0001 | <.0001 | <.0001 | <.0001 |
|
rs56251346 | C>T | 0.435 | 0.401 | 0.059 | 0.265 | 0.1354 | <.0001 | <.0001 | <.0001 | <.0001 | <.0001 |
|
rs3799488 | T>C | 0.279 | 0.253 | 0.01 | 0.128 | <.0001 | <.0001 | <.0001 | <.0001 | <.0001 | <.0001 |
| rs11914 | T>G | 0.065 | 0.073 | 0.059 | 0.169 | <.0001 | <.0001 | <.0001 | 0.0874 | <.0001 | <.0001 |
|
rs1887415 | T>C | 0.024 | 0.016 | 0.02 | – | <.0001 | <.0001 | – | 0.7987 | – | – |
| IFNGR2 |
rs4817565 | G>A | 0.198 | 0.223 | 0.03 | 0.112 | 0.0077 | <.0001 | 0.0002 | <.0001 | 0.0006 | <.0001 |
|
rs73194070 | C>A | 0.132 | 0.134 | 0.083 | 0.213 | 0.2535 | 0.0019 | <.0001 | 0.0663 | 0.0217 | <.0001 |
|
rs9808753 | G>A | 0.477 | 0.465 | 0.222 | 0.112 | 0.1713 | <.0001 | <.0001 | <.0001 | <.0001 | 0.0003 |
|
rs2834211 | T>C | 0.187 | 0.204 | 0.018 | 0.112 | 0.0324 | <.0001 | 0.0044 | <.0001 | <.0001 | <.0001 |
|
rs2834213 | A>G | 0.149 | 0.177 | 0.033 | 0.243 | 0.3102 | <.0001 | <.0001 | <.0001 | 0.0901 | <.0001 |
|
rs115346998 | A>G | – | 0.005 | – | – | – | – | – | – | – | – |
|
rs8126735 | A>G | 0.477 | 0.46 | 0.242 | 0.087 | 0.2313 | <.0001 | <.0001 | <.0001 | <.0001 | <.0001 |
|
rs8128483 | C>T | 0.292 | 0.336 | 0.364 | 0.194 | 0.2078 | 0.0022 | 0.0001 | 0.4482 | <.0001 | <.0001 |
|
rs143025663 | A>C | 0.000 | 0.005 | – | – | 0.0449 | – | – | – | – | – |
|
rs1059293 | T>C | 0.154 | 0.199 | 0.171 | 0.454 | 0.0826 | 0.4749 | <.0001 | 0.4726 | <.0001 | <.0001 |
|
rs17882754 | G>A | 0.134 | 0.163 | 0.012 | 0.109 | – | – | – | – | – | – |
Previous studies have demonstrated that dysfunction
in the IFN-γ pathway caused by genetic variation may contribute to
a further impairment in cellular immune function in IFN-γ mediated
diseases, which may increase the susceptibility to disease. A
specific promoter polymorphism, rs1327474, and one coding
region polymorphism, rs11914 (S350S), were found to be
significantly associated with the risk of arthritis in a European
population (27). Other SNPs,
rs3799488 and rs10457655, exhibited associations with
the risk of rectal cancer prevalence and risk of atopic dermatitis,
respectively, in a Caucasian population (28,29).
Of note, functional analysis of IFNGR1
identified that the non-synonymous SNP rs1887415 (L467P)
does not functionally differ from the wild-type receptors (30). In addition, IFNGR1 L467P has
been reported to be associated with the high immunoprotein levels
against diseases (31,32). Previous studies of rs1887415 may be
a plausible explanation for the protective effect against TB
(OR=0.63) since IFNGR1 interacts with the IFN-γ immune
responses that induce secretion of other cytokines. The association
analyses demonstrated that genetic variants in the ligand-binding
chain of IFNGR (IFNGR1) affect the IFN-γ pathway, although
genetic variants in the signal-transducing chain of IFNGR (IFNGR2),
including three non-synonymous SNPs (Q64R, H178R, Q290P), do not
affect the IFN-γ pathway.
In conclusion, a correlation analysis between
polymorphisms in IFNGR genes and the risk of TB revealed that four
SNPs, rs9376269, rs9376268, rs9376267 and
rs56251346, were marginally associated with the development
of TB. The present study was the first to report, to the best of
our knowledge, the importance of IFNGR1 and IFNGR2 as
genetic factors in mycobacterial infectious disease, which may
prove useful for identifying the etiology of TB in a Korean
population.
Acknowledgments
The present study was supported by the Korean
Science and Engineering Foundation funded by the Korean government
(grant no. NRF-2011-0021659). The DNA samples were generously
provided by Soonchunhyang University, Bucheon Hospital Biobank and
a member of the National Biobank of Korea, supported by the
Ministry of Health, Welfare and Family Affairs, Republic of
Korea.
References
|
1
|
Vannberg FO, Chapman SJ, Khor CC, et al:
CD209 genetic polymorphism and tuberculosis disease. PLoS One.
3:e13882008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Organization WH: Global tuberculosis
report 2012: Geneva, Switzerland: World Moreno S, Jarrin I,
Iribarren JA, et al: Incidence and risk factors for tuberculosis in
HIV-Guyatt GH, Oxman AD, Vist GE, et al., GRADE: an emerging
consensus on rating quality of 0 Guyatt GH, Oxman AD, Kunz R, et
al, Incorporating considerations of resources use into 2012.
|
|
3
|
Lawn SD and Zumla AI: Tuberculosis.
Lancet. 378:57–72. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O’Garra A, Redford PS, McNab FW, Bloom CI,
Wilkinson RJ and Berry MP: The immune response in tuberculosis.
Annu Rev Immunol. 31:475–527. 2013. View Article : Google Scholar
|
|
5
|
Berry MP, Graham CM, McNab FW, et al: An
interferon-inducible neutrophil-driven blood transcriptional
signature in human tuberculosis. Nature. 466:973–977. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ferrara G, Losi M, Fabbri LM, Migliori GB,
Richeldi L and Casali L: Exploring the immune response against
Mycobacterium tuberculosis for a better diagnosis of the infection.
Arch Immunol Ther Exp (Warsz). 57:425–433. 2009. View Article : Google Scholar
|
|
7
|
Thye T, Vannberg FO, Wong SH, et al:
Genome-wide association analyses identifies a susceptibility locus
for tuberculosis on chromosome 18q11.2. Nat Genet. 42:739–741.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Z, Zhu H, Pu X, et al: Association
between tumor necrosis factor alpha-238 G/a polymorphism and
tuberculosis susceptibility: a meta analysis study. BMC Infect Dis.
12:3282012. View Article : Google Scholar
|
|
9
|
Morris GA, Edwards DR, Hill PC, et al:
Interleukin 12B (IL12B) genetic variation and pulmonary
tuberculosis: a study of cohorts from The Gambia, Guinea-Bissau,
United States and Argentina. PLoS One. 6:e166562011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schroder K, Hertzog PJ, Ravasi T and Hume
DA: Interferon-gamma: an overview of signals, mechanisms and
functions. J Leukoc Biol. 75:163–189. 2004. View Article : Google Scholar
|
|
11
|
Newport MJ, Huxley CM, Huston S, et al: A
mutation in the interferon-gamma-receptor gene and susceptibility
to mycobacterial infection. N Engl J Med. 335:1941–1949. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stein CM, Zalwango S, Chiunda AB, et al:
Linkage and association analysis of candidate genes for TB and
TNFalpha cytokine expression: evidence for association with IFNGR1,
IL-10 and TNF receptor 1 genes. Hum Genet. 121:663–673. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Wit E, van der Merwe L, van Helden PD
and Hoal EG: Gene-gene interaction between tuberculosis candidate
genes in a South African population. Mamm Genome. 22:100–110. 2011.
View Article : Google Scholar
|
|
14
|
He J, Wang J, Lei D and Ding S: Analysis
of functional SNP in ifng/ifngr1 in Chinese Han population with
tuberculosis. Scand J Immunol. 71:452–458. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Motsinger-Reif AA, Antas PR, Oki NO, Levy
S, Holland SM and Sterling TR: Polymorphisms in IL-1beta, vitamin D
receptor Fok1 and Toll-like receptor 2 are associated with
extrapulmonary tuberculosis. BMC Med Genet. 11:372010. View Article : Google Scholar
|
|
16
|
Fraser DA, Bulat-Kardum L, Knezevic J, et
al: Interferon-gamma receptor-1 gene polymorphism in tuberculosis
patients from Croatia. Scand J Immunol. 57:480–484. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu J, Pan H, Chen Y, et al: Genetic
polymorphisms of IFNG and IFNGR1 in association with the risk of
pulmonary tuberculosis. Gene. 543:140–144. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ: Allelic discrimination using
fluorogenic probes and the 5′ nuclease assay. Genet Anal.
14:143–149. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar
|
|
20
|
Stephens M, Smith NJ and Donnelly P: A new
statistical method for haplotype reconstruction from population
data. Am J Hum Genet. 68:978–989. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nyholt DR: A simple correction for
multiple testing for single-nucleotide polymorphisms in linkage
disequilibrium with each other. Am J Hum Genet. 74:765–769. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Naka I, Patarapotikul J, Hananantachai H,
Tokunaga K, Tsuchiya N and Ohashi J: IFNGR1 polymorphisms in Thai
malaria patients. Infect Genet Evol. 9:1406–1409. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Canedo P, Corso G, Pereira F, et al: The
interferon gamma receptor 1 (IFNGR1) -56C/T gene polymorphism is
associated with increased risk of early gastric carcinoma. Gut.
57:1504–1508. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Awomoyi AA, Nejentsev S, Richardson A, et
al: No association between interferon-gamma receptor-1 gene
polymorphism and pulmonary tuberculosis in a Gambian population
sample. Thorax. 59:291–294. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cooke GS, Campbell SJ, Sillah J, et al:
Polymorphism within the interferon-gamma/receptor complex is
associated with pulmonary tuberculosis. Am J Respir Crit Care Med.
174:339–343. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hijikata M, Shojima J, Matsushita I, et
al: Association of IFNGR2 gene polymorphisms with pulmonary
tuberculosis among the Vietnamese. Hum Genet. 131:675–682. 2012.
View Article : Google Scholar :
|
|
27
|
Nordang GB, Viken MK, Amundsen SS, et al:
Interferon regulatory factor 5 gene polymorphism confers risk to
several rheumatic diseases and correlates with expression of
alternative thymic transcripts. Rheumatology (Oxford). 51:619–626.
2012. View Article : Google Scholar
|
|
28
|
Slattery ML, Lundgreen A, Bondurant KL and
Wolff RK: Interferon-signaling pathway: associations with colon and
rectal cancer risk and subsequent survival. Carcinogenesis.
32:1660–1667. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Leung DY, Gao PS, Grigoryev DN, et al:
Human atopic dermatitis complicated by eczema herpeticum is
associated with abnormalities in IFN-gamma response. J Allergy Clin
Immunol. 127:965–973. e1–e5. 2011. View Article : Google Scholar
|
|
30
|
van de Wetering D, de Paus RA, van Dissel
JT and van de Vosse E: Functional analysis of naturally occurring
amino acid substitutions in human IFN-gammaR1. Mol Immunol.
47:1023–1030. 2010. View Article : Google Scholar
|
|
31
|
Aoki M, Matsui E, Kaneko H, et al: A novel
single-nucleotide substitution, Leu 467 Pro, in the
interferon-gamma receptor 1 gene associated with allergic diseases.
Int J Mol Med. 12:185–191. 2003.PubMed/NCBI
|
|
32
|
Thye T, Burchard GD, Nilius M,
Muller-Myhsok B and Horstmann RD: Genomewide linkage analysis
identifies polymorphism in the human interferon-gamma receptor
affecting Helicobacter pylori infection. Am J Hum Genet.
72:448–453. 2003. View
Article : Google Scholar : PubMed/NCBI
|