Introduction
MicroRNAs (miRNAs) are a class of short non-coding
RNAs that regulate gene expression predominantly at the
post-transcriptional level. At present, almost 2,000 unique mature
human miRNAs have been identified, which serve critical roles in
tissue development and the pathogenesis of numerous diseases
(1–3). Greater than 250 miRNAs have been
reported to be expressed in the retina (4). It has been demonstrated that
miRNA-mediated gene regulation is a major mechanism underlying
retinal and optic nerve development, function and associated
diseases (5).
Radiation-induced retinopathy and radiation-induced
optic neuropathy (RION) are severe complications of radiotherapy
for intracranial, skull-base and sinus tumors, which may result in
irreversible visual loss. Approximately 85% of affected eyes have
final visual acuity of 6/60 or worse (6). However, the detailed molecular
mechanisms of radiation-induced retinopathy and RION remain
unclear. Previous studies have demonstrated that miRNA expression
is sensitive to ionizing radiation injury and alterations in miRNA
expression appear to be important in the mechanisms of
radiation-induced injury and associated disease (7,8).
Although radiation-induced retinopathy and RION have
been extensively investigated, the clinical therapeutic options
remain limited (9). According to
the characteristics of these diseases, early diagnosis is extremely
important for prevention and treatment of RION (10,11).
It has been reported that miRNAs can serve as improved biomarkers
for the early diagnosis of numerous diseases, including cancer
(12), autoimmune diseases
(13), cardiovascular diseases
(14), ectopic pregnancy (15) and systemic inflammatory response
syndrome (16). In contrast to
other biomarkers, miRNAs are protected from endogenous RNase
activity and are highly stable. For this reason, they have been
considered as ideal biomarkers for use in early diagnosis,
prediction of prognosis and therapeutic management (17).
The current study aimed to screen for altered
expression levels of miRNAs in RGC-5 cells exposed to ionizing
radiation, in order to identify the possible mechanisms of these
altered miRNAs in radiation injury, and to explore the possibility
of using specific miRNAs as potential biomarkers for
radiation-induced retinopathy and RION.
Materials and methods
RGC-5 cell culture
The RGC-5 cell line (American Type Culture
Collection, Manassas, VA, USA) was grown in modified RPMI 1640
medium (Gibco Life Technologies, Carlsbad, CA, USA), containing 10%
fetal bovine serum (Gibco Life Technologies) and 1%
penicillin-streptomycin (100 U/ml penicillin and 100 μg/ml
streptomycin; Sigma-Aldrich, St. Louis, MO, USA) at 37°C and 5%
CO2. Cells were usually passaged at a ratio of 1:20.
During cultivation, RGC-5 cells exhibited the same morphological
phenotype in different generations. For viability assays, cells
were plated onto positively charged 96-well plates. For microarray
and reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis, cells were harvested from 25 cm2
filter-capped cell culture flasks.
Radiation treatment and cell viability
assay
The Siemens Medical Linear Accelerator (6MV Oncor;
Siemens, Munich, Germany) was used in the current study to mimic
radiation injury in vitro. RGC-5 cells were irradiated with
250 cGy/min dose rate X-rays at final doses of 2, 4, 6 and 8 Gy.
Unexposed cells served as controls. Following radiation, cells were
cultured in a humidified incubator for 5 days and morphological
alterations were observed daily using a phase-contrast microscope
(Eclipse 90i; Nikon, Tokyo, Japan). Cell viability was evaluated by
an MTT assay according to the manufacturer's instructions. Briefly,
20 μl of 5 mg/ml MTT dye (Sigma-Aldrich) was added to each
well and cells were incubated at 37°C for 4 h. Subsequently, the
supernatant was removed and purple-colored precipitates of formazan
were dissolved by gently shaking for 10 min in 150 μl
dimethyl sulfoxide (Sangon Biotech Co., Ltd., Shanghai, China). The
microplate was read at 570 nm (Benchmark Microplate Reader; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The optical density (A
value) of each sample was measured and applied for cell viability
calculation.
RNA extraction and microarray assay
Total RNA of RGC-5 cells was extracted and purified
using the mirVana™ miRNA Isolation kit (Ambion Life Technologies,
Austin, TX, USA) according to the manufacturer's instructions, then
were checked for a RIN number to inspect RNA integration by an
Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara,
CA, USA). To analyze miRNA expression, total RNA samples (100 ng)
containing miRNAs were labeled with cyanine 3-pCp (Agilent
Technologies, Inc.) using the Agilent miRNAs Complete Labeling and
Hyb kit (Agilent Technologies, Inc.). Slides were then hybridized
for 20 h at 55°C using a hybridization system (G2535A; Agilent
Technologies, Inc.). Subsequent to hybridization, the slides were
washed in Agilent GE Wash Buffer 1 with Triton X-102, followed by
Agilent GE Wash Buffer 2 with Triton X-102. All slides were
immediately scanned using the Agilent Microarray Scanner and
Feature Extraction software, version 10.7 (Agilent Technologies,
Inc.) with the default settings. Raw data were normalized by the
Quantile algorithm with Gene Spring Software, version 11.0 (Agilent
Technologies, Inc.).
RT-qPCR
The expression levels of miRNAs were confirmed with
SYBR-based quantitative PCR (RT-qPCR). Briefly, a total of 1
μg RNA from each sample was transcribed into cDNA using the
PrimeScript RT reagent kit (GeneCopoeia, Inc., Guangzhou, China)
according to the manufacturer's instructions. The RT product (10
μl) was diluted with H2O up to 50 μl. cDNA
was then amplified by PCR with primers specific to the target
sequence (Table I). Amplification
conditions were as follows: 60 min incubation at 37°C, followed by
a 5 min termination reaction at 85°C. Dilutions of cDNA in the PCR
were adjusted for each gene with the aim of remaining within the
linear range of amplification. RT-qPCR was performed with SYBR
Green Realtime PCR Master mix (GeneCopoeia, Inc.) according to the
manufacturer's instructions. Cycling conditions were as follows:
Initial denaturation for 10 min at 95°C, 40 cycles of melting (95°C
for 20 sec), annealing (62.5°C for 20 sec) and extending (72°C for
20 sec). The relative alterations in expression for each miRNA were
calculated by the cycle threshold method (ΔΔCt method) as described
previously (17).
 | Table IPrimer sequences used for the miRNA
RT-qPCR analysis. |
Table I
Primer sequences used for the miRNA
RT-qPCR analysis.
| miRNAs | Primer sequence
(5′–3′) |
|---|
| U6 |
AAGGATGACACGCAAATTCG |
| rno-miR-192 |
CTGACCTATGAATTGACAGCC |
| rno-miR-210 |
CTGTGCGTGTGACAGCGGCTGA |
| rno-miR-212 |
TAACAGTCTCCAGTCACGGCCA |
| rno-miR-219-5p |
TGATTGTCCAAACGCAATTCT |
| rno-miR-22 |
AAGCTGCCAGTTGAAGAACTGT |
| rno-miR-338 |
TCCAGCATCAGTGATTTTGTTGA |
| rno-miR-34a |
TGGCAGTGTCTTAGCTGGTTGT |
|
rno-miR-34a* |
AATCAGCAAGTATACTGCCCTA |
| rno-miR-34c |
AGGCAGTGTAGTTAGCTGATTGC |
| rno-miR-503 |
TAGCAGCGGGAACAGTACTGCAG |
|
rno-miR-505* |
GGGAGCCAGGAAGTATTGATGTT |
| rno-miR-877 |
GTAGAGGAGATGGCGCAGGG |
Statistical analysis
Statistical analysis was performed with SPSS
software, version 11.3 (SPSS, Inc., Chicago, IL, USA). For all
assays, experiments were performed a minimum of three times. Data
are presented as the mean ± standard deviation and were evaluated
by one-way analysis of variance and Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Effect of radiation on RGC-5 cell
morphology and viability
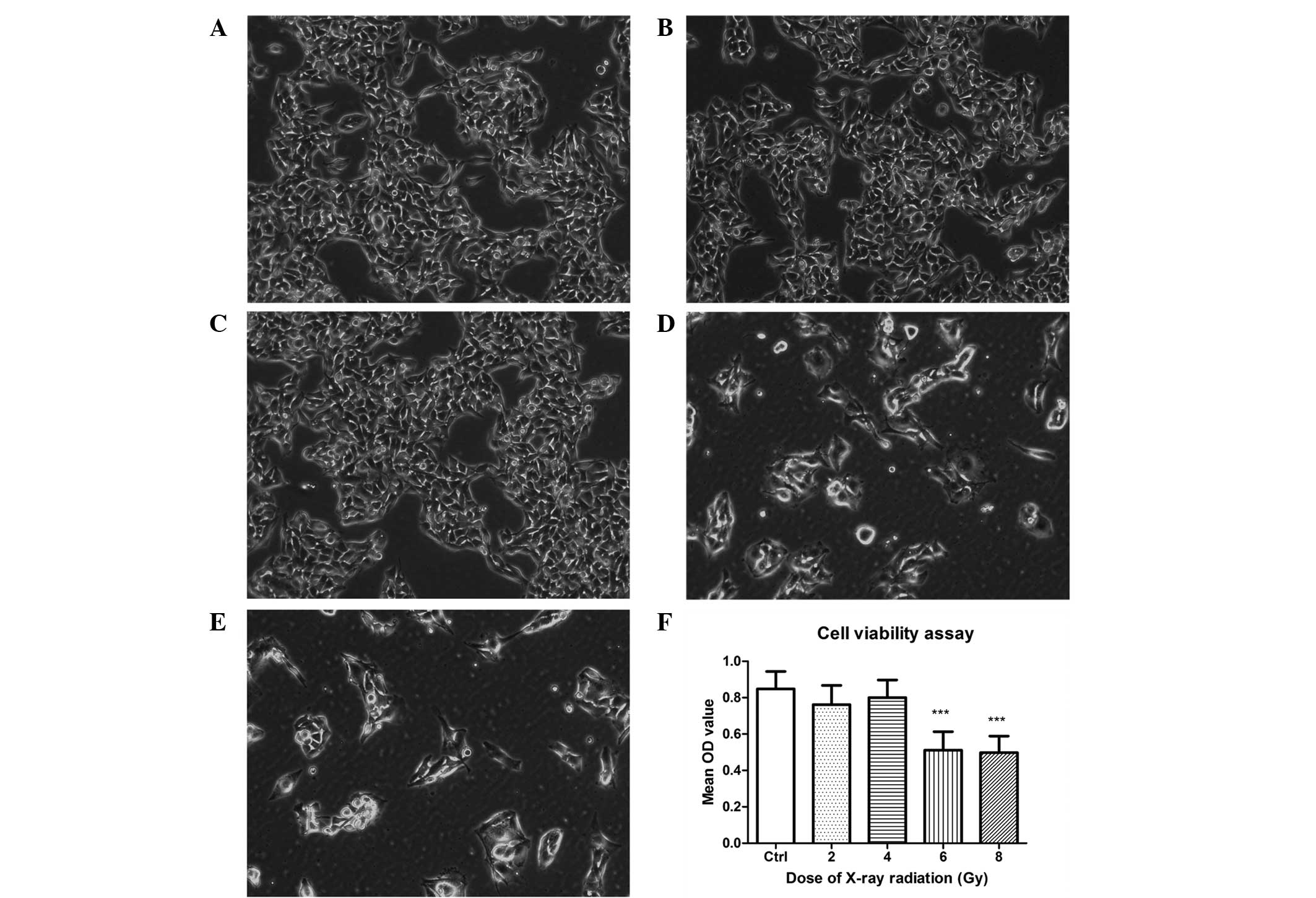
As demonstrated by phase contrast microscopy, the
majority of cells exhibited abnormal appearance in the 6 and 8 Gy
groups following 5 days exposure to X-ray radiation, with the
densities reduced and the cell shapes changing from a normal
appearance to rounded and swollen. Compared with the control group,
no significant morphological alterations were observed in the 2 and
4 Gy groups (Fig. 1A–E).
The MTT assay (Fig.
1F) indicated that the cell viability was significantly reduced
in the 6 and 8 Gy groups following 5 days of radiation treatment
(P<0.01). However, there was no significance difference between
the 6 and 8 Gy groups (P>0.05). No significant differences in
cell viability of the 2 and 4 Gy groups were identified compared
with the control group (P>0.05), which was consistent with the
morphological alterations described above.
Alterations in miRNA expression following
radiation injury
According to the results of the MTT assay, the 6 Gy
dose of radiation was applied in RGC-5 cells to evaluate miRNA
expression. In total, 677 miRNA probes were included in the miRNA
microarray. Subsequent to exposure to 6 Gy radiation for 5 days,
the expression levels of 37 miRNAs were statistically altered
compared with the control group (P<0.05), with 16 upregulated
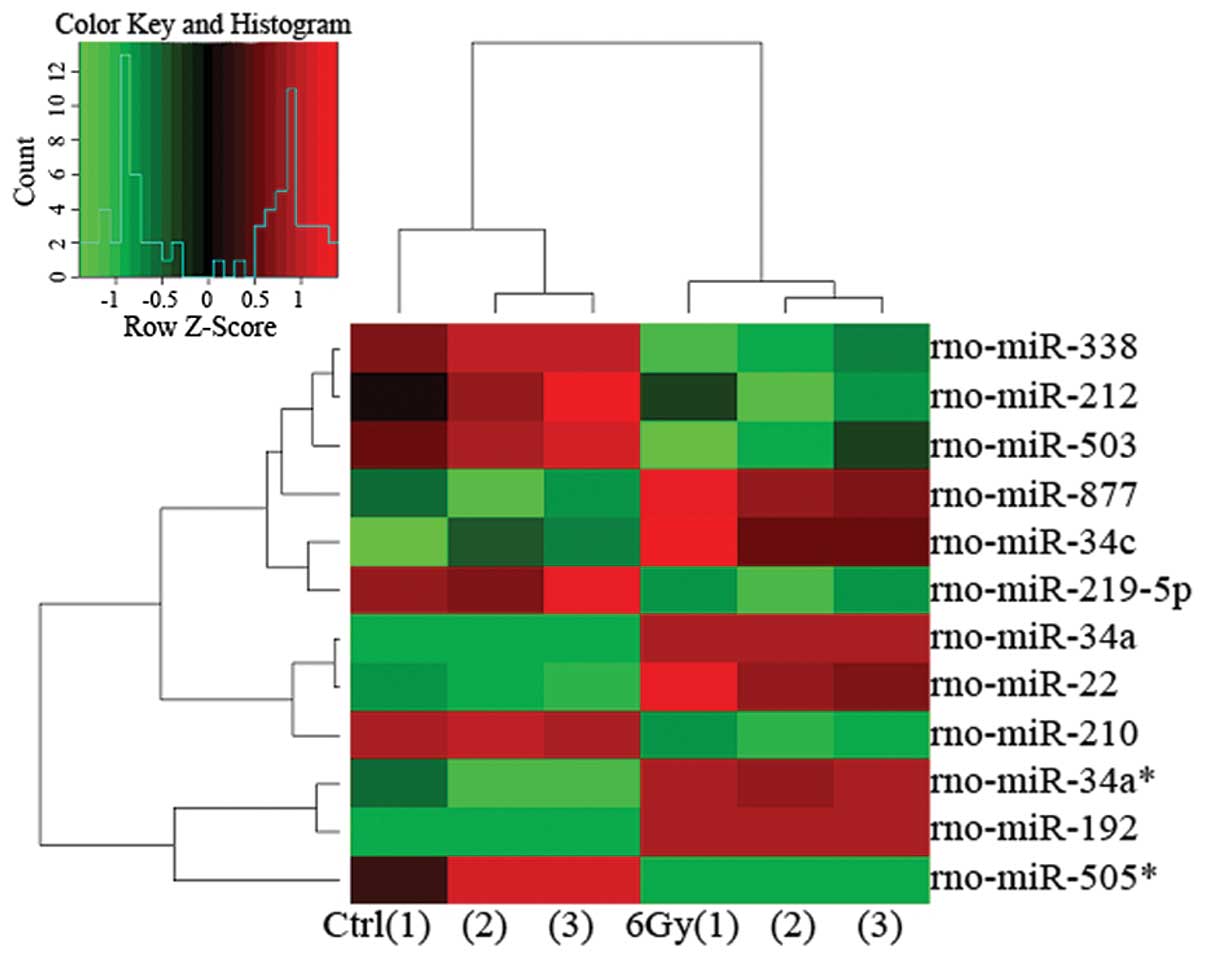
miRNAs and 21 downregulated miRNAs. Among these altered miRNAs, 12
were selected according to the fold change (>1.5 or <0.667)
and microarray repeatability (stable expression in triplicate
experiments) (Fig. 2; repeated 3
times). Of these 12 miRNAs, 6 were upregulated [miRNA-192
(miR-192), -34a*, -877, -34c, -34a and -22; P<0.05
and fold change >1.5] and 6 were downregulated (miR-212, -338,
-219-5p, -503, -210 and -505*; P<0.05 and fold change
<0.667) (Table II).
 | Table IIMicroarray and RT-qPCR results of
miRNA expression in RGC-5 cells subsequent to exposure to 6 Gy
radiation for 5 days (fold changes). |
Table II
Microarray and RT-qPCR results of
miRNA expression in RGC-5 cells subsequent to exposure to 6 Gy
radiation for 5 days (fold changes).
| miRNA name | Microarray | RT-qPCR |
|---|
| Upregulated |
| miR-192 | 72.75 | 1.76b |
|
miR-34a* | 46.13 | 2.10b |
| miR-877 | 6.97 | 1.68b |
| miR-34c | 2.71 | 1.70b |
| miR-34a | 1.84 | 2.26b |
| miR-22 | 1.82 | 1.96b |
| Downregulated |
| miR-212 | 0.66 | 0.38a |
| miR-338 | 0.64 | 0.48b |
| miR-219-5p | 0.50 | 0.41a |
| miR-503 | 0.38 | 0.65b |
| miR-210 | 0.28 | 0.58b |
|
miR-505* | 0.02 | 0.26b |
Independent RT-qPCR analysis to confirm
microarray results
In order to validate microarray data, RT-qPCR
analysis of miRNA expression was further performed for all of the
12 significantly altered miRNAs. The detailed RT-qPCR results
compared with the results of microarray analysis (described as fold
change) are presented in Table
II. The expression levels of the 12 miRNAs exhibited similar
alterations when measured by RT-qPCR (P<0.05), consistent with
the results of microarray analysis.
Discussion
miRNAs have been extensively investigated and have
been recognized to act as critical regulators of gene expression by
binding to their targets. miRNAs have revolutionized the
understanding of gene regulatory networks, providing novel tools to
manage the development and clinical therapy of numerous diseases.
It has been demonstrated that miRNAs are essential mediators and
serve key roles in retinal ganglion cell axon branching and optic
nerve formation (18,19). However, the alterations in miRNA
expression involved in radiation injury to RGCs and their detailed
functions remain to be fully elucidated. In the current study,
different doses of ionizing radiation were applied to induce
radiation injury in cultured RGC-5 cells. Alterations in the miRNA
expression levels were evaluated following radiation injury, which
may aid in understanding the molecular mechanisms of
radiation-induced retinopathy and RION.
The data of the present study revealed that exposure
to 6 Gy and 8 Gy doses of X-ray radiation resulted in radiation
injury in RGC-5 cells. The viability of RGC-5 cells was
significantly reduced, which was in accordance with previously
reported results (20,21). The microarray assay demonstrated
that there was a total of 37 miRNAs that were statistically altered
in the radiation-treated RGC-5 cells. It is known that when using
microarray to detect differentially expressed miRNA, multiple tests
are required to ensure that the results are stable and repeatable
(22). A minimum of 3 repeats of
each experiment were performed in the current study, following
which 12 significantly altered miRNAs were selected according to
the fold change and microarray repeatability, including 6
upregulated miRNAs (miR-192, -34a*, -877, -34c, -34a and
-22) and 6 downregulated miRNAs (miR-212, -338, -219-5p, -503, -210
and -505*). Microarray technology is a powerful tool
used to detect whether a set of miRNAs are differentially
expressed. However, microarray profiling of microRNA expression
raises a number of analytical challenges that must be addressed in
order to obtain reliable results, and RT-qPCR analysis has been
commonly used for further validation (23). In the current study, the data from
RT-qPCR analysis were consistent with that from microarray
analysis, indicating that the expression levels of these miRNAs
were regulated in response to the radiation-induced injury in RGC-5
cells.
Ocular damage is a common side effect of
radiotherapy, such as dry eye, epiphora, ectropion, scleral
necrosis, cataract, glaucoma, optic neuropathy and retinopathy.
Radiation-induced retinopathy and RION are the most serious
complications in terms of visual damage resulting from radiation
therapy for tumors adjacent to the retina and optic nerves
(24). The pathogenesis of
radiation-induced retinopathy and RION include vascular occlusion,
injury to the retinal ganglion cells and axons or a combination of
these effects (25). In a dog
model of radiation-induced retinal injury, retinal degeneration
with swelling and loss of the ganglion cells was observed following
radiation treatment for 1 year (26). In humans, retinopathy commonly
appears 6 months-3 years following radiation therapy, characterized
by irreversible ganglion and glial cell necrosis and vasculitis
(27). At present, the molecular
mechanisms of ionizing radiation-induced ganglion cell injury
remain to be fully elucidated. The data of the current study
indicated that there were 12 miRNAs involved in the process of
radiation-induced injury in RGC-5 cells. According to the reports,
the function of these miRNAs and their regulated target genes are
predominantly associated with the cell cycle and proliferation
(miR-192, -22, -212 and -219-5p) (28–31),
metabolism (miR-34a and -34c) (32) and apoptosis and anti-apoptosis
(miR-192, -34a, -34c, -338 and -210) (28,33,34).
For example, miR-192 is able to regulate p53 expression to induce
tubular epithelial cell growth cycle arrest and DNA damage
following kidney acute injury (28). miR-34a and miR-34c have been
demonstrated to be downstream effectors of p53-mediated human
fibroblast apoptosis and senescence (34). Further investigation of these
miRNAs may aid in the understanding of the pathogenesis and
development of radiation-induced retinopathy and RION.
Although numerous treatments have been suggested,
management of radiation-induced retinopathy and RION remains
controversial (9). At present,
clinical therapeutic options include the use of corticosteroids,
anticoagulants, antiaggregants and hyperbaric oxygen (HBO). A
retrospective literature review identified there was no favorable
effect of either corticosteroids or anticoagulation (35) and cases of radiation damage and
visual loss have been reported to develop despite anticoagulation
therapies (36). A previous study
reported that HBO was a beneficial therapy for patients with
radiation-induced retinopathy and RION (37). However, it has been suggested that
HBO therapy is only effective in certain cases when patients were
treated with HBO as early as possible following the onset of visual
loss (37). According to
characteristics of radiation-induced retinopathy and RION, the
onset of visual acuity loss may be as short as 3 months or as long
as 8 years following radiation exposure (38). Once radiotherapy has induced
retinal complications, visual damage has been reported to progress
quickly and be difficult to control (9). Early diagnosis is extremely important
for the prevention and treatment of radiation-induced retinopathy
(10,11). However, an effective method to
monitor the early stages of radiation-induced retinopathy and RION
remains to be developed.
A previous study demonstrated that miRNA expression
is significantly different between pathological tissues and normal
tissues (39). Understanding of
the potential use of miRNA expression levels as biomarkers for the
early detection of numerous diseases is on the increase (40). In the current study, 12 miRNAs,
which were all sensitive to ionizing radiation and were
differentially expressed between the experimental and control
groups, were screened using microarray analysis in RGC-5 cells
following radiation exposure. Due to the fact that it remains a
challenge to detect and treat radiation-induced retinopathy and
RION at an early stage, these 12 miRNAs are suggested as novel
potential biomarkers for diagnosis, prediction of prognosis and
therapeutic management.
Although the results of the current study are
promising, there are several limitations. Firstly, a clear drawback
of the present study is that it is an in vitro experiment.
The environment of cells in vivo is different to cells
cultured in vitro, however, it is easy to control the
experimental conditions using cultured cells and the results are
often more accurate. In addition, it has been identified that the
majority of cells release pathogenic miRNAs into the extracellular
environment of cells, where they are protected by RNase and remain
relatively stable (41).
Expression levels of miRNAs associated with diseases in biological
fluids have been suggested as promising biomarkers for diagnosis
and therapeutic monitoring (12–15).
Concerning the 12 miRNAs screened in the current study, it remains
unclear whether they are released by the retinal ganglion cells
into the extracellular environment following radiation exposure.
Thus, further studies are required to investigate the possibilities
of the 12 miRNAs as potential biomarkers for radiation-induced
retinopathy and RION.
In conclusion, the current study suggested that
cultured RGC-5 cells were sensitive to ionizing radiation injury.
The cell viability was significantly reduced following radiation
exposure, accompanied by alterations in miRNA expression. These
altered miRNAs may serve an important role in the mechanisms of
radiation-induced retinopathy and RION, and may be used as
potential biomarkers for the early detection of retinal and optic
nerve radiation injury.
Acknowledgments
The current study was financially supported by
research grants from the National Natural Science Foundation of
China (grant nos. 81372930 and 30700443) and the Zhejiang
Provincial Natural Science Foundation of China (grant nos.
LY12H12008 and LQ15H120001).
References
|
1
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Graves P and Zeng Y: Biogenesis of
mammalian microRNAs: A global view. Genomics Proteomics
Bioinformatics. 10:239–245. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang Y, Shen XJ, Zou Q and Zhao QL:
Biological functions of microRNAs. Bioorg Khim. 36:747–752.
2010.
|
|
4
|
Sundermeier TR and Palczewski K: The
physiological impact of microRNA gene regulation in the retina.
Cell Mol Life Sci. 69:2739–2750. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karali M, Peluso I, Marigo V and Banfi S:
Identification and characterization of microRNAs expressed in the
mouse eye. Invest Ophthalmol Vis Sci. 8:509–515. 2007. View Article : Google Scholar
|
|
6
|
Roden D, Bosley TM, Fowble B, Clark J,
Savino PJ, Sergott RC and Schatz NJ: Delayed radiation injury to
the retrobulbar optic nerves and chiasm. Clinical syndrome and
treatment with hyperbaric oxygen and corticosteroids.
Ophthalmology. 97:346–351. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chaudhry MA, Omaruddin RA, Kreger B, de
Toledo SM and Azzam EL: MicroRNA responses to chronic or acute
exposures to low dose ionizing radiation. Mol Biol Rep.
39:7549–7558. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Templin T, Paul S, Amundson SA, Young EF,
Barker CA, Wolden SL and Smilenov LB: Radiation-induced micro-RNA
expression changes in peripheral blood cells of radiotherapy
patients. Int J Radiat Oncol Biol Phys. 80:549–557. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee MS and Borruat FX: Should patients
with radiation-induced optic neuropathy receive any treatment? J
Neuroophthalmol. 31:83–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Levy RL and Miller NR: Hyperbaric oxygen
therapy for radiation-induced optic neuropathy. Ann Acad Med
Singapore. 35:151–157. 2006.PubMed/NCBI
|
|
11
|
Danesh-Meyer HV: Radiation-induced optic
neuropathy. J Clin Neurosci. 15:95–100. 2008. View Article : Google Scholar
|
|
12
|
Mo MH, Chen L, Fu Y, Wang W and Fu SW:
Cell-free circulating miRNA biomarkers in cancer. J Cancer.
3:432–448. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li JY, Yong TY, Michael MZ and Gleadle JM:
Review: The role of microRNAs in kidney disease. Nephrology
(Carlton). 15:599–608. 2010. View Article : Google Scholar
|
|
14
|
Hulsmans M and Holvoet P: MicroRNAs as
early biomarkers in obesity and related metabolic and
cardiovascular diseases. Curr Pharm Des. 19:5704–5717. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao Z, Zhao Q, Warrick J, Lockwood CM,
Woodworth A, Moley KH and Gronowski AM: Circulating microRNA
miR-323-3p as a biomarker of ectopic pregnancy. Clin Chem.
58:896–905. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schöler N, Langer C, Döhner H, Buske C and
Kuchenbauer F: Serum microRNAs as a novel class of biomarkers: A
comprehensive review of the literature. Exp Hematol. 38:1126–1130.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kroh EM, Parkin RK, Mitchell PS and Tewari
M: Analysis of circulating microRNA biomarkers in plasma and serum
using quantitative reverse transcription-PCR (qRT-PCR). Methods.
50:298–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pinter R and Hindges R: Perturbations of
microRNA function in mouse dicer mutants produce retinal defects
and lead to aberrant axon pathfinding at the optic chiasm. PLoS
One. 5:e100212010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marler KJ, Suetterlin P, Dopplapudi A,
Rubikaite A, Adnan J, Maiorano NA, Lowe AS, Thompson ID, Pathania
M, Bordey A, et al: BDNF promotes axon branching of retinal
ganglion cells via miRNA-132 and p250GAP. J Neurosci. 34:969–979.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lulli M, Witort E, Papucci L, Torre E,
Schiavone N, Dal Monte M and Capaccioli S: Coenzyme Q10 protects
retinal cells from apoptosis induced by radiation in vitro and in
vivo. J Radiat Res. 53:695–703. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Balaiya S, Malyapa R, Hsi W, Murthy RK and
Chalam KV: Evaluation of proton beam radiation sensitivity of
proliferating choroidal endothelial and retinal ganglion cells with
clonogenic assay. Cutan Ocul Toxicol. 31:14–19. 2012. View Article : Google Scholar
|
|
22
|
Wang B and Xi Y: Challenges for microRNA
microarray data analysis. Microarrays (Basel). pp. 22013
|
|
23
|
Sarkar D, Parkin R, Wyman S, Bendoraite A,
Sather C, Delrow J, Godwin AK, Drescher C, Huber W, Gentleman R, et
al: Quality assessment and data analysis for microRNA expression
arrays. Nucleic Acids Res. 37:e172009. View Article : Google Scholar :
|
|
24
|
Finger PT: Tumour location affects the
incidence of cataract and retinopathy after ophthalmic plaque
radiation therapy. Br J Ophthalmol. 84:1068–1070. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wolfensberger TJ, Zwingli M, Egger E,
Schnyder P and Zografos L: Subclinical experimental optic
neuropathy after accelerated proton beam irradiation.
Ophthalmologica. 216:420–425. 2002. View Article : Google Scholar
|
|
26
|
Pinard CL, Mutsaers AJ, Mayer MN and Woods
JP: Retrospective study and review of ocular radiation side effects
following external-beam Cobalt-60 radiation therapy in 37 dogs and
12 cats. Can Vet J. 53:1301–1307. 2012.
|
|
27
|
Gupta A, Dhawahir-Scala F, Smith A, Young
L and Charles S: Radiation retinopathy: Case report and review. BMC
Ophthalmol. 7:62007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hong JP, Li XM, Li MX and Zheng FL: VEGF
suppresses epithelial-mesenchymal transition by inhibiting the
expression of Smad3 and miR-192, a Smad3-dependent microRNA. Int J
Mol Med. 31:1436–1442. 2013.PubMed/NCBI
|
|
29
|
Li S, Hu R, Wang C, Guo F, Li X and Wang
S: miR-22 inhibits proliferation and invasion in estrogen receptor
α-positive endometrial endometrioid carcinomas cells. Mol Med Rep.
9:293–2399. 2014.
|
|
30
|
Tognini P and Pizzorusso T:
MicroRNA212/132 family: Molecular transducer of neuronal function
and plasticity. Int J Biochem Cell Biol. 44:6–10. 2012. View Article : Google Scholar
|
|
31
|
Huang N, Lin J, Ruan J, Su N, Qing R, Liu
F, He B, Lv C, Zheng D and Luo R: MiR-219-5p inhibits
hepatocellular carcinoma cell proliferation by targeting
glypican-3. FEBS Lett. 586:884–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu K, Huang J, Xie M, Yu Y, Zhu S, Kang
R, Cao L, Tang D and Duan X: MIR34A regulates autophagy and
apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy.
10:442–452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ragusa M, Majorana A, Banelli B,
Barbagallo D, Statello L, Casciano I, Guglielmino MR, Duro LR,
Scalia M, Magro G, et al: MIR152, MIR200B, and MIR338, human
positional and functional neuroblastoma candidates, are involved in
neuroblast differentiation and apoptosis. J Mol Med (Berl).
88:1041–1053. 2010. View Article : Google Scholar
|
|
34
|
Kumamoto K, Spillare EA, Fujita K,
Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S,
Yokota J and Harris CC: Nutlin-3a activates p53 to both
down-regulate inhibitor of growth 2 and up-regulate mir-34a,
mir-34b, and mir-34c expression and induce senescence. Cancer Res.
68:3193–3203. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guy J and Schatz NJ: Hyperbaric oxygen in
the treatment of radiation induced optic neuropathy. Ophthalmology.
93:1083–1088. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Danesh Meyer HV, Savino PJ and Sergott RC:
Visual loss despite anticoagulation in radiation-induced optic
neuropathy. Clin Experiment Ophthalmol. 32:333–335. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Levy RL and Miller NR: Hyperbaric oxygen
therapy for radiation induced optic neuropathy. Ann Acad Med
Singapore. 35:151–157. 2006.PubMed/NCBI
|
|
38
|
Demizu Y, Murakami M, Miyawaki D, Niwa Y,
Akaqi T, Sasaki R, Terashima K, Suqa D, Kamae I and Hishikawa Y:
Analysis of vision loss caused by radiation-induced optic
neuropathy after particle therapy for head-and-neck and skull-base
tumors adjacent to optic nerves. Int J Radiat Oncol Biol Phys.
75:1487–1492. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kress M, Hüttenhofer A, Landry M, Kuner R,
Favereaux A, Greenberg D, Bednarik J, Heppenstall P and Kronenberg
F: MicroRNAs in nociceptive circuits as predictors of future
clinical applications. Front Mol Neurosci. 6:332013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cheng L, Sharples RA, Scicluna BJ and Hill
AF: Exosomes provide a protective and enriched source of miRNA for
biomarker profiling compared to intracellular and cell-free blood.
J Extracell Vesicles. 26:32014.
|