Introduction
Pancreatic cancer (PC) is an aggressive malignancy
with one of the highest mortality rates amongst cancers worldwide.
It is the sixth leading cause of mortality from malignant disease
in China and the fourth leading cause of cancer-associated
mortality in the USA (1–3). Rapid tumor progression, late
diagnosis, early and aggressive metastasis and high resistance to
conventional chemotherapy lead to exceptionally poor prognosis with
an overall five-year survival rate of <5% (4). Therefore, novel markers for early
diagnosis and novel therapeutic targets for PC require to be
identified. Although the etiology of PC is also attributed to
numerous environmental factors, the accumulation of genetic and
epigenetic changes remains the fundamental mechanism of
tumorigenesis (5–7).
MicroRNA (miRNA/miR) is a type of short non-coding
RNA that suppresses the expression of protein-coding genes by
partial complementary binding, particularly to the 3′-untranslated
regions (3′UTRs) of mRNAs. miRNA expression alterations are
involved in the initiation, progression, and metastasis of human
cancer and it is believed that miRNAs function as tumor suppressors
and oncogenes in cancer development (8,9).
Accumulating studies have shown that disturbed expression of miRNAs
is involved in the process of pathogenesis and drug resistance
(10,11).
PIM3 was initially identified as a novel gene that
is induced by membrane depolarization or forskolin in the rat
pheochro-mocytoma cell line PC12 and was designated as a kinase
induced by depolarization (KID)-1 (12). However, KID-1 was renamed PIM3
because it showed high sequence similarity with the proto-oncogenic
provirus integrating site Moloney murine leukemia virus (PIM)
family of proteins (13).
Recently, PIM3 was found to be aberrantly expressed in pancreatic
ductal adenocarcinoma (PDAC) cells and to phosphorylate the
pro-apoptotic protein B-cell lymphoma 2-associated death promoter
(14). In addition, PIM3 was shown
to be regulated by transcription factors such as ETS-1 and serve as
a positive regulator of signal transducer and activator of
transcription 3 signaling in PC cells (15,16).
The present study aimed to investigate the
post-transcriptional regulation of the expression of PIM3 in PC,
focusing on miRNAs directly targeting PIM3.
Materials and methods
Cell culture
PANC-1, MIAPaca-2 and HEK293T cells (China
Infrastructure of Cell Line Resources, Beijing, China) were
cultured in Dulbecco's modified Eagle's medium (DMEM) containing
10% fetal bovine serum (Hyclone, Logan, UT, USA), 100 IU/ml
penicillin and 10 mg/ml streptomycin (Hyclone). All cells were
maintained at 37°C under an atmosphere of 5% CO2.
Tissue samples
PC and matched adjacent normal tissues from 38
patients were obtained post-operatively from March to September in
2012 from the Department of Heptapobiliary Surgery, The First
Affiliated Hospital to the General Hospital of the PLA (Beijing,
China). The patients provided signed, informed consent for their
tissues to be used for scientific research. Ethical approval for
the study was obtained from the First Affiliated Hospital to the
General Hospital of the PLA. Diagnoses were based on pathological
and/or cytological evidence. Histological features of the specimens
were evaluated by two senior pathologists according to
classification criteria from the World Health Organization
(17). Tissues were obtained from
patients prior to chemotherapy or radiation therapy. Specimens were
immediately frozen and stored at −80°C prior to western blot and
reverse transcription quantitative polymerase chain reaction
(RT-qPCR) analyses.
Western blot analysis
Protein extracts were boiled in
SDS/β-mercaptoethanol sample buffer (Sigma-Aldrich, St. Louis, MO,
USA), and 30 µg of each sample was loaded onto a lane of a
12% polyacrylamide gel (Sigma-Aldrich). The proteins were separated
by electrophoresis, and the proteins in the gels were blotted onto
polyvinylidene difluoride membranes (GE Healthcare, Little
Chalfont, UK) by electrophoretic transfer. The membrane was
incubated with rabbit anti-PIM3 monoclonal antibody (1:1,000;
ab75776; Abcam, Cambridge, MA, USA), mouse anti-β-actin monoclonal
antibody (1:1,000; sc-58673; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) or rabbit anti-AGO2 monoclonal antiobdy (1:1,000;
ab186733; Abcam) for 1 h at 37°C. The specific protein-antibody
complex was detected by using horseradish peroxidase-conjugated
goat anti-rabbit (sc-2004) or rabbit anti-mouse (sc-358920)
immunoglobulin G (1:5,000; Santa Cruz Biotechnology, Inc.).
Detection by enhanced chemiluminescence (ECL) reaction was carried
using an ECL kit (Pierce Biotechnology, Rockford, IL, USA) and
X-ray films (Carestream Health, Inc., Xiamen, China). The β-actin
signal was used as a loading control. The band intensity was
analyzed by using Quantity One software, version 4.6.2 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
RT-qPCR analysis
RT-qPCR analysis was used to determine the relative
expression levels of 13 selected miRNAs. Total RNA was extracted
from tissues, using TRIzol (Invitrogen Life Technologies, Carlsbad,
CA, USA) according to the manufacturer's instructions. The
expression levels of candidate miRNAs were detected by TaqMan miRNA
RT-Real Time PCR (Applied Biosystems Life Technologies, Foster
City, CA, USA). Single-stranded cDNA was synthesized by using the
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems Life
Technologies, Waltham, MA, USA) and then amplified by using TaqMan
Universal PCR Master Mix (Applied Biosystems Life Technologies)
together with miRNA-specific TaqMan Minor Groove Binder probes
(Applied Biosystems Life Technologies). U6 small nuclear RNA was
used for normalization. The experiments were processed using an ABI
7300 PCR Thermal Cycler (Applied Biosystems Life Technologies). The
protocol for qPCR was a classic two step PCR: 95°C for 10 min; 95°C
for 15 sec followed by 60°C for 1 min for 35 cycles. Each sample in
each group was measured in triplicate and the experiment was
repeated at least three times. The relative expression was
calculated using the ΔΔCt method. The products were separated by 2%
agarose (Sigma-Aldrich) to confirm the specificity of the PCR
reaction.
3′UTR luciferase reporter assays
To generate the 3′UTR luciferase reporter, the
full-length 3′UTR from PIM3 was cloned into the downstream region
of the firefly luciferase gene in the pGL3-control vector (Promega,
Madison, WI, USA). The primer sequences for PIM 3′UTR cloning were
as follows: PIM3, forward CTCGAGGGAGCTGCACCTGACTGGGA and reverse
TCTAGATATGTACAAAAACATTTTAATTGAAATACC. The primers were synthesized
by BGI-GBI Biotech Co., Ltd. (Beijing, China). miRNA mimics and
inhibitor were synthesized by GenePharma Co., Ltd (Shanghai,
China). The sequence for the double strand miR-506 mimic was
5′-UAAGGCACCCUUCUGAGUAGA-3′, for the single strand miR-506
inhibitor was 5′-TCTACTCAGAAGGGTGCCTTA-3′, for the miR-506 control
was 5′-UUCUCCGAACGUGUCACGUTT-3′ and for the miR-506 inhibitor
control was 5′-CAGUACUUUUGUGUAGUACAA-3′. The pRL-TK vector
(Promega) containing Renilla luciferase was co-transfected for data
normalization. For luciferase reporter assays, HEK293T cells were
seeded in 48-well plates. Luciferase reporter vectors were
co-transfected with one of the miRNA mimics by using Lipofectamine
2000 (Invitrogen Life Technologies). Two days post-tranfection,
cells were harvested and assayed using the Dual-Luciferase Assay
system (Promega). Each treatment was performed in triplicate in
three independent experiments. The results were expressed as
relative luciferase activity (firefly luciferase light
units/Renilla luciferase light units). To identify the binding site
of miR-506, a plasmid with three nucleotide mutations in the
predicted miR-506 binding site was used.
AGO2 knockdown
Pancreatic cancer cell lines PANC-1 and MIAPaca-2
were transfected with AGO2 siRNA and the corresponding control
using Lipofectamine 2000 (Invitrogen Life Technologies). A total of
48 h subsequent to transfection, the cells were lysed by RIPA Lysis
and Extraction Buffer (Pierce Biotechnology) and the expression of
AGO2 and IPM3 was detected by western blot analysis. β-actin was
used as loading control.
Cell proliferation assay
PANC-1 and MIAPaca-2 cells were seeded in 96-well
plates at a low density (5×103) in DMEM culture and
allowed to attach overnight. The cells were then transfected with
miR-506 mimic or inhibitor, with a scrambled-sequence single strand
or double strand short hairpin RNA as the control. MTT (20
µl; 5 mg/ml; Sigma-Aldrich) was added into each well 48 h
after transfection, and the cells were incubated for a further 4 h.
Following addition of dimethyl sulfoxide (Sigma-Aldrich), the
absorbance was measured at 570 nm using a 96-well plate reader.
Statistical analysis
Data were analyzed by using SPSS Statistical Package
version 17 (SPSS, Inc., Chicago, IL, USA). Results were analyzed
using Student's t-test. P<0.05 was considered to indicate a
statistically significant difference between values. The results of
the luciferase and MTT assays were displayed as the mean ± standard
deviation. The results of miRNA expression in the clinical samples
were exhibited using box-and-whisker plots, where the whiskers
represented the range of the data.
Results
PIM3 expression in PC cells is regulated
by miRNAs
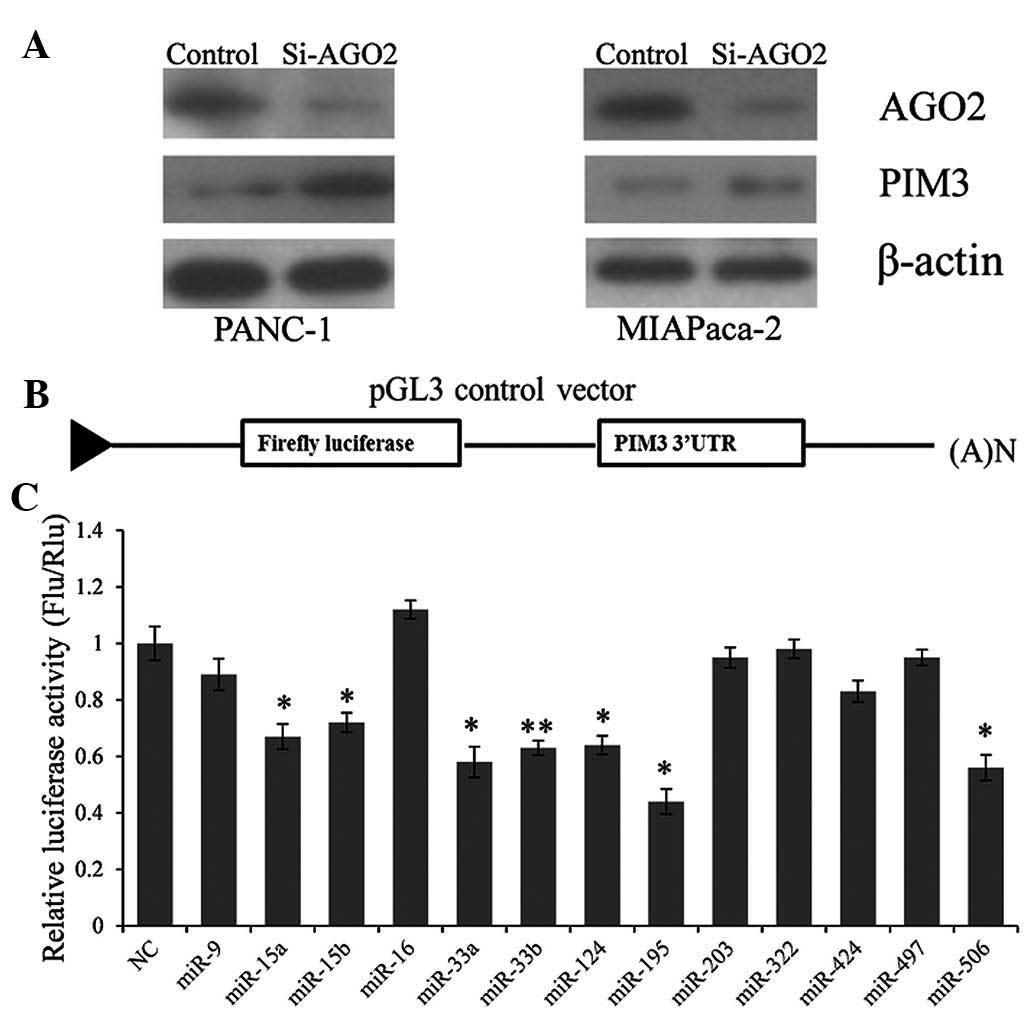
To explore whether the expression of PIM3 is
regulated by miRNAs, argonaute RNA-induced silencing complex (RISC)
catalytic component 2 (AGO2), the key component of the RISC complex
was knocked down in PANC-1 and MIAPaca-2 cells. It was observed
that the inactivation of the RISC complex caused a marked
upregulation of PIM3 expression (Fig.
1A). The result indicated that miRNAs participate in the
inverse control system of PIM3 expression.
Five out of thirteen selected miRNAs
target PIM3 directly
To identify which miRNAs repress PIM3 expression
directly, a reporter vector containing the full-length PIM3 3′UTR
downstream of the firefly luciferase coding region was constructed
(Fig. 1B). miRNAs that may target
the PIM3 3′UTR were predicted using the online bioinformatics tool
TargetScan (http://www.targetscan.org), and
thirteen miRNAs were selcted: miR-15a, miR-15b, miR-16, miR-33a,
miR-33b, miR-124, miR-195, miR-203, miR-322, miR-424, miR-497 and
miR-506. After screening by using the dual luciferase system, we
seven miRNAs (miR-15a, miR-15b, miR-33a, miR-33b, miR-124, miR-195
and miR-506) were identified to repress luciferase activity by
significantly targeting the PIM3 3′UTR (Fig. 1C).
Association between miRNAs and PIM3
expression
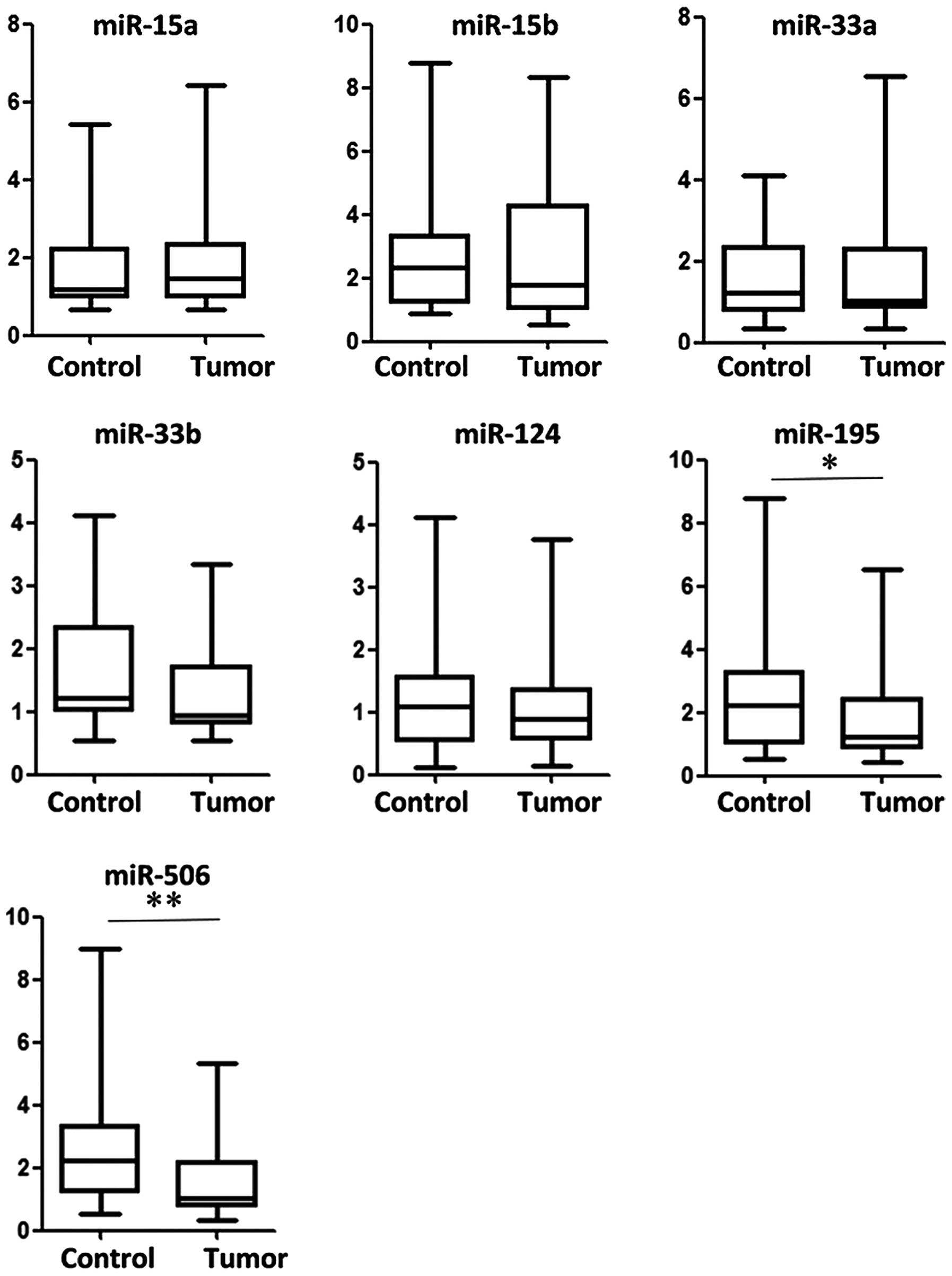
To explore the association between PIM3 and miRNAs
in PC samples, PIM3 expression was detected by western blot
analysis and the expression of seven selected miRNAs was detected
by RT-qPCR (Fig. 2). miR-195 and
miR-506 were significantly downregulated in tumor tissue samples
compared with normal adjacent tissues (P<0.05 and P<0.01,
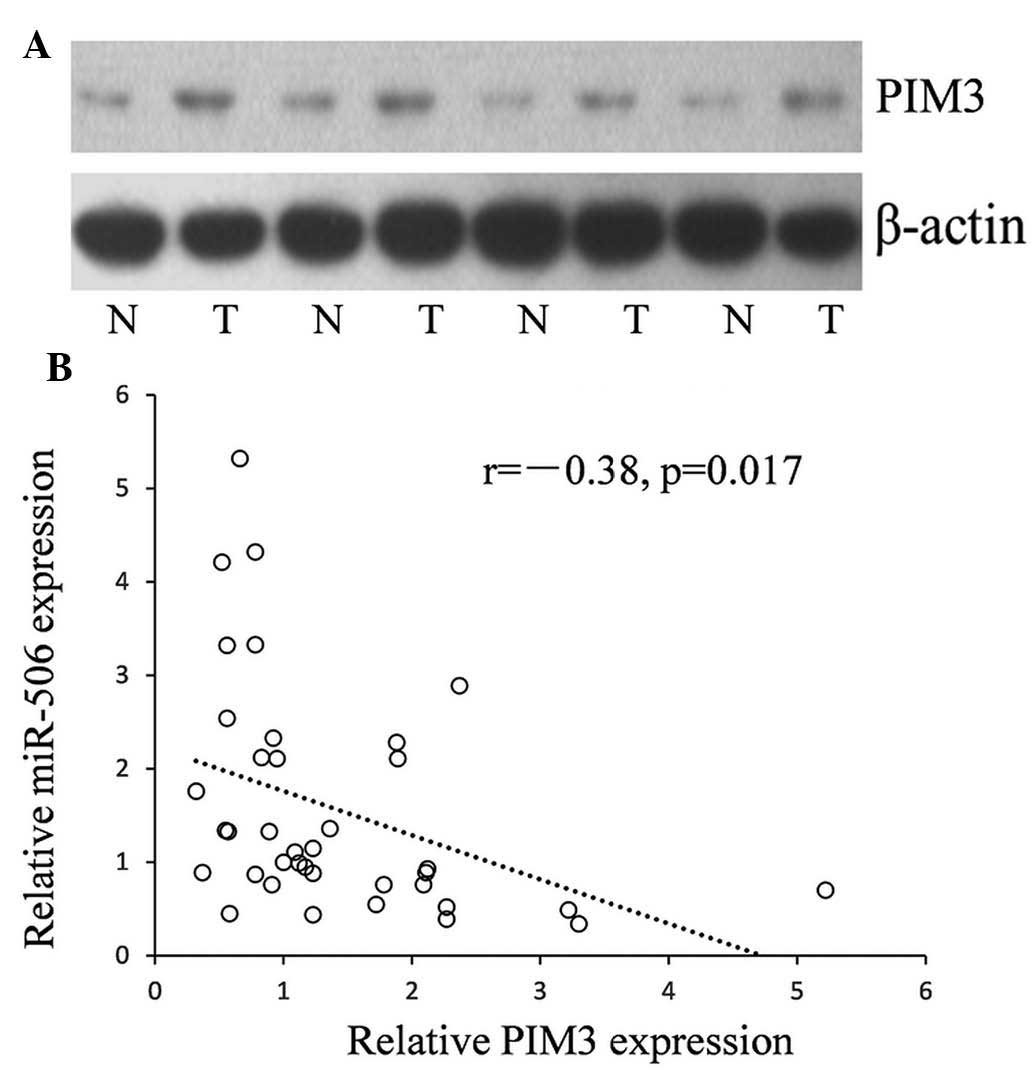
respectively). An example of PIM3 expression in PC and paired
normal control tissues is shown in Fig. 3A. The results indicated that ~74%
(29/38) of PC tissues displayed upregulated PIM3 expression and
~71% (27/38) or 63% (23/38) had downregulated miR-506 or miR-195
expression, respectively (Fig.
3B).
To reveal the correlation between downregulated
miRNAs (miR-195 and miR-506) and the PIM3 in PC tissues, PIM3
expression was assessed using western blot analysis. An inverse
correlation was identified between the expression levels of miR-506
and PIM3 in 38 clinical samples of PC. Low levels of miR-506 were
associated with high PIM3 expression (Pearson correlation, r=−0.38;
P=0.017; Fig. 3B).
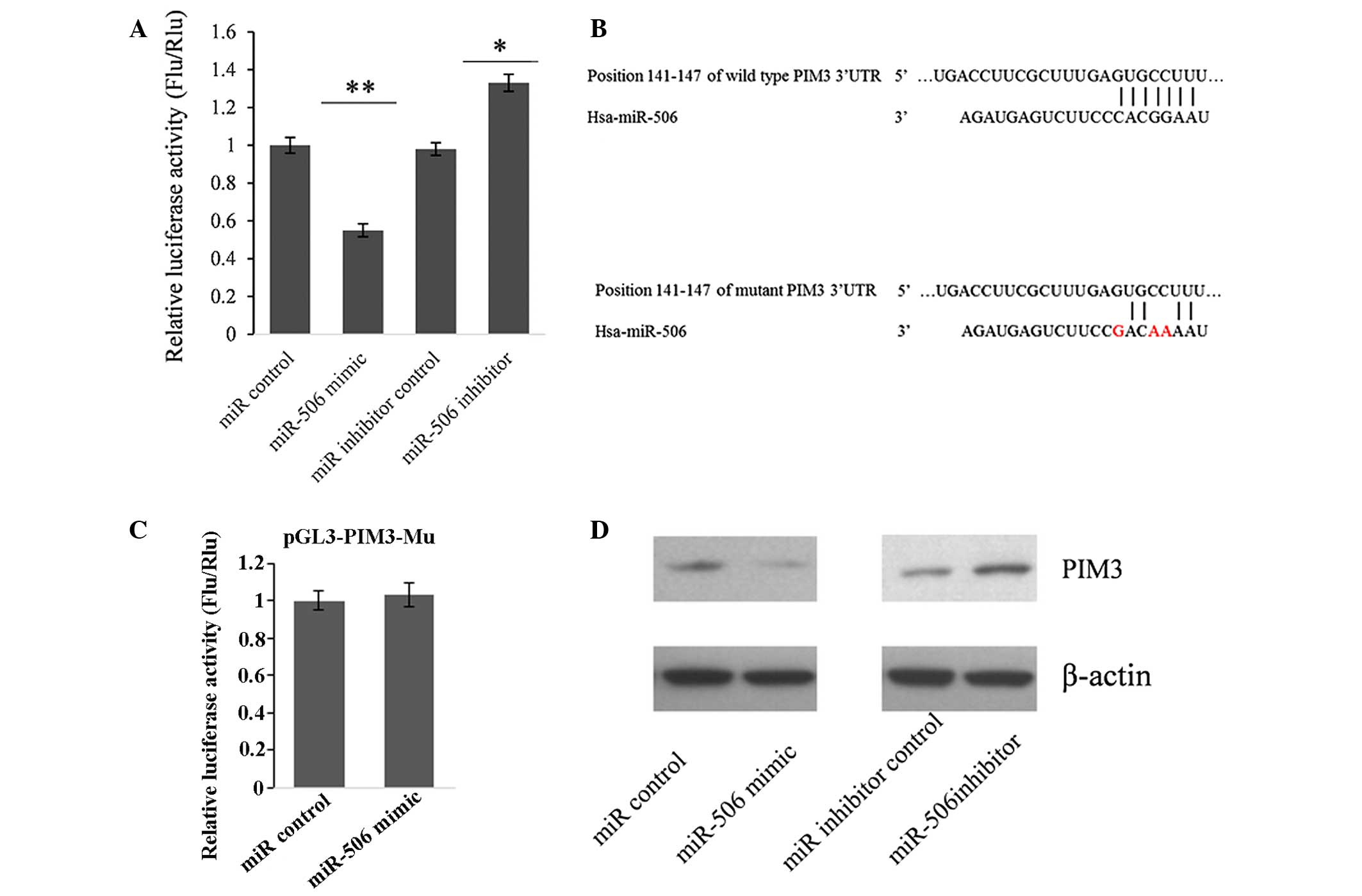
miR-506 represses PIM3 expression by
directly targeting its 3′UTR
To further confirm whether PIM3 is the target gene
of miR-506, the dual luciferase assay system was utilized again.
HEK293T cells were co-transfected with pGL3-PIM3 and miR-506 mimic
or inhibitor. As shown in Fig. 4A,
compared with the miRNA control, the miR-506 mimic significantly
suppressed the luciferase activity by 45.2% (P<0.01).
Furthermore, the luciferase activity was significantly upregulated
(by 35.7%) by the miR-506 inhibitor compared with that in the
miR-inhibitor control (P<0.05). These changes of firefly
luciferase translation indicated that miR-506 targets the 3′UTR of
PIM3.
A seed sequence mutation clone was also used to
further confirm the binding site for miR-506 (Fig. 4B). A vector containing a putative
miR-506 binding region in the 3′UTR of PIM3 with three mutant
nucleotides (designated as pGL3-PIM3-Mu) was constructed. The bar
graph in Fig. 4C shows that the
enzyme activity was not significantly reduced in cells transfected
with miR-control compared with that in cells transfected with
miR-506 mimic (P>0.05). This result indicated that miR-506 may
suppress PIM3 expression through binding to the seed sequence at
the 3′UTR of PIM3.
To further examine whether endogenous PIM3
expression is suppressed by miR-506, PANC-1 cells were transfected
with miR-506 mimic or inhibitor. PIM3 protein levels were detected
by western blot analysis 48 h post-transfection. Compared with the
corresponding control, the levels of PIM3 protein were
significantly suppressed by miR-506 mimic and upregulated by
miR-506 inhibitor in PANC-1 cells (Fig. 4D). These results indicated that
miR-506 repressed endogenous PIM3 expression in PC cells by
directly targeting the PIM3 3′UTR, and PIM3 is a target gene of
miR-506.
miR-506 suppresses PC cell
proliferation
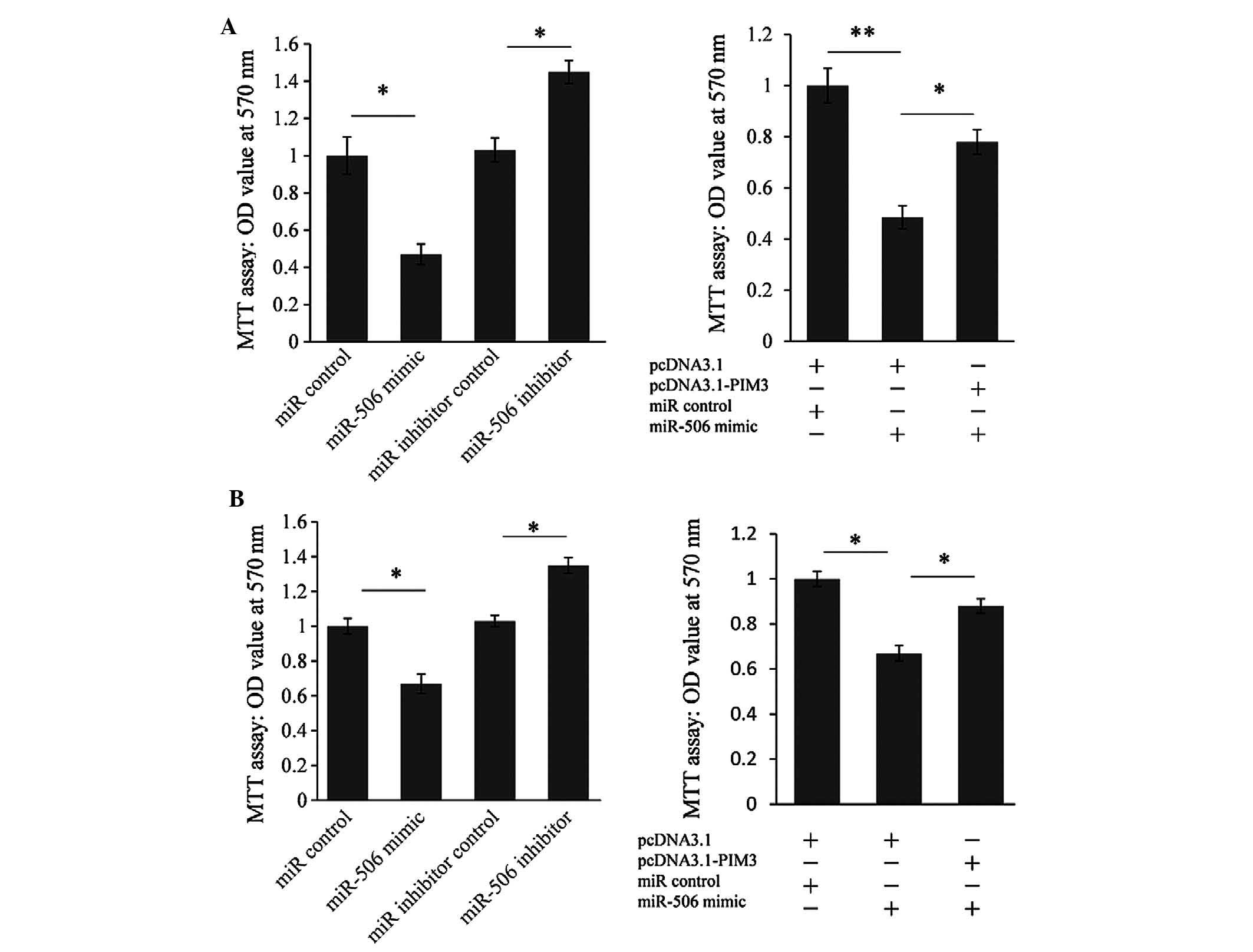
To further test whether miR-506 may execute
tumor-suppressive functions by targeting PIM3, the effect of
miR-506-mediated cell proliferation was assessed using an MTT assay
on PANC-1 (Fig. 5A) and MIAPaca-2
cells (Fig. 5B). The cell
proliferation ability was significantly reduced by the miR-506
mimic (by 52.8%) in PANC-1 cells and by 33.1% in MIAPaca-2 cells.
Furthermore, cell proliferation was significantly upregulated by
the miR-506 inhibitor (by 43.2%) in PANC-1 cells and by 32.3% in
MIAPaca-2 cells. To further confirm whether miR-506 represses cell
proliferation by targeting PIM3, a PIM3 expression vector was
co-transfected with miR-506 mimic into PANC-1 and MIAPaca-2 cells.
As shown in Fig. 5A and B (right),
the PIM3 expression vector partially reversed the repressed cell
proliferation caused by miR-506 overexpression, indicating that
miR-506 suppresses PC cell proliferation partially through
targeting PIM3.
Discussion
PIM3 is a member of the proto-oncogenic PIM family
that encodes serine/threonine kinases, and is aberrantly expressed
in human PC. Studies have indicated that overexpression of PIM3 is
associated with enhanced PC cell proliferation (18). In the present study, it was
confirmed that the expression of PIM3 is regulated by miRNAs
through an AGO2 knockout experiment. Subsequently, a dual
luciferase assay system was constructed and used for screening 13
selected miRNAs that may target the PIM3 3′UTR directly according
to a TargetScan analysis. The results indicated that miR-15a/b,
miR-16, miR-33a/b, miR-124, miR-195 and miR-506 repressed
luciferase activity by targeting the PIM3 3′UTR. However, only the
expression of miR-506 was negatively correlated with PIM3
expression in the PC tissues (r=−0.38, P=0.017). Furthermore, a
mechanistic study indicated that miR-506 acted as a tumor
suppressor by repressing PC cell proliferation and its
anti-proliferative function can be partially reversed by PIM3
overexpression.
Biological functions of miR-506, particularly in
cancer, have been studied; however, the roles of miR-506 in
carcinogenesis are yet to be elucidated (19,20).
In breast, cervical and ovarian cancer, miR-506 is confirmed to be
a tumor suppressor through targeting Ki-67, Gli3, CDK4 and CDK6
(20–22). Furthermore, downregulation of
miR-506 in cervical cancer was identified to be associated with the
cancer pathogenesis (20).
However, miR-506 overexpression was also reported in lung cancer
and melanoma; however, they appear to have opposite functions. Yin
et al (23) reported that
miRNA-506 was upregulated in lung cancer patients and its
overexpression selectively killed lung cancer cells through
inhibiting nuclear factor-κB p65 to evoke the generation of
reactive oxygen species and p53 activation. By contrast, Streicher
et al (24) reported that
the miRNA-506-514 cluster was overexpressed in almost all melanoma
samples that were assessed and had a positive role in initiating
melanocyte transformation and promoting melanoma growth. The
present study was the first, to the best of our knowledge, to
report that miR-506 was downregulated in PC tissues, which may be
applicable for clinical diagnosis. However, since one miRNA may
have tens or hundreds of target genes and its function may be
tissue-specific, further studies are required to fully unveil the
biological functions of miR-506.
References
|
1
|
Li D, Xie K, Wolff R and Abbruzzese JL:
Pancreatic cancer. Lancet. 363:1049–1057. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo X and Cui Z: Current diagnosis and
treatment of pancreatic cancer in China. Pancreas. 31:13–22. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang S, Bonaroti J, Unlu S, et al:
Sweating the small stuff: MicroRNAs and genetic changes define
pancreatic cancer. Pancreas. 42:740–759. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
van Kampen JG, Marijnissen-van Zanten MA,
Simmer F, van der Graaf WT, Ligtenberg MJ and Nagtegaal ID:
Epigenetic targeting in pancreatic cancer. Cancer Treat Rev.
40:656–664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wood LD and Hruban RH: Genomic landscapes
of pancreatic neoplasia. J Pathol Transl Med. 49:13–22. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu WK, Lee CW, Cho CH, et al: MicroRNA
dysregulation in gastric cancer: a new player enters the game.
Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs-the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ikenaga N, Ohuchida K, Mizumoto K, et al:
MicroRNA-203 expression as a new prognostic marker of pancreatic
adenocar-cinoma. Ann Surg Oncol. 17:3120–3128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Izumchenko E, Chang X, Michailidi C, et
al: The TGFβ-miR200-MIG6 pathway orchestrates the EMT-associated
kinase switch that induces resistance to EGFR inhibitors. Cancer
Res. 74:3995–4005. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feldman JD, Vician L, Crispino M, et al:
KID-1, a protein kinase induced by depolarization in brain. J Biol
Chem. 273:16535–16543. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Konietzko U, Kauselmann G, Scafidi J, et
al: Pim kinase expression is induced by LTP stimulation and
required for the consolidation of enduring LTP. EMBO J.
18:3359–3369. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li YY, Popivanova BK, Nagai Y, Ishikura H,
Fujii C and Mukaida N: Pim-3, a proto-oncogene with
serine/threonine kinase activity, is aberrantly expressed in human
pancreatic cancer and phosphorylates bad to block bad-mediated
apoptosis in human pancreatic cancer cell lines. Cancer Res.
66:6741–6747. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li YY, Wu Y, Tsuneyama K, Baba T and
Mukaida N: Essential contribution of Ets-1 to constitutive Pim-3
expression in human pancreatic cancer cells. Cancer Sci.
100:396–404. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang M, Kanwar N, Feng E, et al: PIM
kinase inhibitors downregulate STAT3 (Tyr705) phosphorylation. Mol
Cancer Ther. 9:2478–2487. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jass JR, Sobin LH and Watanabe H: The
World Health Organization's histologic classification of
gastrointestinal tumors. A commentary on the second edition.
Cancer. 66:2162–2167. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu B, Wang Z, Li HY, Zhang B, Ping B and
Li YY: Pim-3 promotes human pancreatic cancer growth by regulating
tumor vasculogenesis. Oncol Rep. 31:2625–2634. 2014.PubMed/NCBI
|
|
19
|
Yang FQ, Zhang HM, Chen SJ, Yan Y and
Zheng JH: MiR-506 is down-regulated in clear cell renal cell
carcinoma and inhibits cell growth and metastasis via targeting
FLOT1. PloS One. 10:e01202582015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wen SY, Lin Y, Yu YQ, et al: miR-506 acts
as a tumor suppressor by directly targeting the hedgehog pathway
transcription factor Gli3 in human cervical cancer. Oncogene.
34:717–725. 2015. View Article : Google Scholar
|
|
21
|
Liu G, Sun Y, Ji P, et al: MiR-506
suppresses proliferation and induces senescence by directly
targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol.
233:308–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arora H, Qureshi R and Park WY: miR-506
regulates epithelial mesenchymal transition in breast cancer cell
lines. PLoS One. 8:e642732013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yin M, Ren X, Zhang X, et al: Selective
killing of lung cancer cells by miRNA-506 molecule through
inhibiting NF-κB p65 to evoke reactive oxygen species generation
and p53 activation. Oncogene. 34:691–703. 2015. View Article : Google Scholar
|
|
24
|
Streicher KL, Zhu W, Lehmann KP, et al: A
novel oncogenic role for the miRNA-506–514 cluster in initiating
melanocyte transformation and promoting melanoma growth. Oncogene.
31:1558–1570. 2012. View Article : Google Scholar
|