Introduction
According to statistics, one in every 1,000 newborns
suffers from pre-lingual deafness and more than half of these cases
result from genetic factors (1).
Numerous cases of late-onset deafness are additionally due to
genetic defects and/or susceptibility to environmental factor
(2). The existing medical
interventions available for the restoration of hearing are
inadequate; thus, the key to the clinical control of deafness
remains predominantly dependent on prevention (3). The first step in the prevention of
deafness is to identify the causes of deafness for the existing
deaf populations (4). The domestic
large-scale molecular epidemiological survey of deafness revealed
that there were a number of mutation hot-spots in non-syndromic
deaf patients. The carrying rates of GJB2 and SLC26A4
gene mutations have been previously identified to be 21 and 14.5%,
respectively, and those of mitochondrial 12S RNA 1555 adenine
(A)>guanine (G) and 1494 cytosine (C)>thymine (T) mutations
are 3.8 and 0.6%, respectively (5–7). The
mutations of these three genes have been identified to be present
in humans of various ethnicity (5–7);
thus, the discovery of these mutation hot-spots provided a
theoretical basis for Chinese researchers to perform the screening,
diagnosis and blocking of deafness gene mutations.
Yongchuan district, which is located in the heart of
West Chongqing, and has a population of over one million. The
health of newborns can be controlled by pre-natal diagnosis and
genetic counseling. The early diagnosis of non-syndromic hearing
loss is particularly important, and preimplantation genetic
diagnosis technology for hereditary deafness, as well as the
refusal of ototoxic drugs, hearing aids, and cochlear implants may
improve the health of local populations (8). To achieve this, gaining a preliminary
understanding of the spectrum and frequency of deafness gene
mutation hot-spots in this region is the first step. To the best of
our knowledge, no studies have been conducted involving the
screening of local non-syndromic hearing loss-associated genes.
In the present study, the Nine Deafness Gene
Mutations Detection kit (Boao Biological Co., Ltd., Beijing, China)
was applied to perform a microarray-based genetic etiology
detection on adolescents with non-syndromic hearing loss in the
Yongchuan district (Chongqing, China) (CQ-YC ANSHL), aiming to
understand the spectrum and frequency of deafness gene mutation
hot-spots in this region, thus providing a theoretical basis for
the screening, diagnosis and blocking of deafness gene mutations
occurring in this area, and reducing the occurrence and development
of deafness in this region.
Materials and methods
Subjects
In the present study, the CQ-YC ANSHL subjects were
selected from the Special Education School (Yongchuan, Chongqing,
China) between October 2011 and October 2012. Among the 60
subjects, nos. 47 and 55 were siblings and the follow-up statistics
solely included the proband; therefore, only no. 47 was included in
the statistical analysis. The resulting total sample size was 59,
including 30 boys and 29 girls, aged 6–17 years, with a median age
of 13 years. The subjects were all Han descendents and residents of
Yongchuan district (Chongqing, China). Subsequent to obtaining
consent of the guardians, general details regarding the family,
family history of deafness, any medical complications of the mother
during pregnancy, the patients' medication history, as well as the
presence of any head trauma, diseases, deafness development,
associated symptoms, and the circumstances of diagnosis and
treatment were collected from the patients. In addition, a routine
otological examination, overall physical examination and a
pure-tone hearing threshold test were performed to exclude
syndromic deafness. The included subjects had non-syndromic hearing
loss and the deafness degrees were graded according to the results
of the pure-tone hearing threshold test, according to the Deaf
Classification Standard of the World Health Organization (9). According to whether a family history
of deafness was present or not, the 59 patients were divided into
two groups. The present study was performed in accordance with the
declaration of Helsinki and with approval from the Ethics Committee
of Chongqing Medical University (Yongchuan, China). Written
informed consent was obtained from the guardians of all
participants.
Specimen collection and processing
Subsequent to obtaining informed consent from the
guardians, 3–5 ml peripheral venous blood was drawn from each
subject and EDTA was added as an anticoagulant. All the specimens
were processed using the DP319 Blood Genomic DNA Extraction system
(Tiangen Biotech Co., Ltd., Beijing, China) on the same day. The
extracted genomic DNA was then quantified with the NanoDrop 2000
Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) to
ensure that the DNA concentration was >100 ng/µl and the
purity was confirmed by optical densities at 260/280 nm of 1.7–2.0.
The genomic DNA was then stored at −80°C.
Microarray analysis and polymerase chain
reaction (PCR) amplification
The reagents of the Nine Deafness Gene Mutations
Detection microarray kit (Boao Biological Co., Ltd., Beijing,
China) were mixed with the PCR-amplified primer mixtures and the
amplification reagent mixtures of the two groups in accordance with
the manufacturer's instructions. The resulting solution was added
into the pre-extracted genomic DNA to prepare the 20 µl
reaction system for the multiplex PCR. The PCR was conducted with
the primers provided in the kit with the following cycling
conditions: 37°C for 10 min, 95°C for 15 min and 96°C for 1 min,
followed by 94°C for 30 sec, 55°C for 30 sec and 72°C for 45 sec
for 32 cycles, then 70°C for 45 sec and 60°C for 10 min. The
cooling speed from 94°C for to 55°C was 0.4°C/sec, while the speed
of temperature increase from 55°C to 72°C was 0.2°C/sec. The PCR
procedure lasted 3 h 20 min, and an CFX96 Quantitative PCR
Instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was
used.
Hybridization
The PCR products were then placed in a hot bath
(95°C) for 5 min in order to denature the nucleotides, which were
then immediately placed into an ice-water mixture for 3 min.
Subsequently, 2.5 µl PCR products of each amplification
system were added into a tube containing 10 µl
hybridization-containing buffer, respectively. Following adequate
mixing, samples were centrifuged at 157 × g and added into the
chip-array area. The hybrid box was then closed and horizontally
placed into the 50°C-preheated BioMixer™ II chips hybridization
instrument (Boao Biological Co., Ltd.) for 1 h with a speed of 80 ×
g.
Chip washing and drying
The chips were then removed and washed within the
SlideWasher™ 8-chip washing instrument (Boao Biological Co., Ltd.).
Washing liquid I (42°C) was used to wash the chips once for 120 sec
with a cleaning force of 5, then the chips were washed twice with
washing liquid II (42°C) for 60 sec with a cleaning force of 5. The
chips were then put into a drying chamber for centrifugal drying at
241 × g for 2 min.
Scanning and interpretation of
results
The dried deafness gene chips were put into the
chip-reading window and scanned using the Jingxin
LuxScan™ 10 K/B microarray scanner (Boao Biological Co.,
Ltd.). The appropriate hereditary genetic testing chip
identification system (Jingxin Hereditary Genetic Testing Chip
Identification system, version 1.0.2.9; Boao Biological Co., Ltd.)
was used to read and automatically interpret the signals.
Statistical analysis
To compare the detection rate of deafness gene
mutation hot-spots between groups with or without a clear family
history the χ2 test was used. All statistical analyses
were conducted using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA).
Results
Disease history
The deafness degrees of the 59 CQ-YC ANSHL were
graded according to the pure-tone hearing threshold test, by which
49 were classified as being of the very severe deafness grade and
10 were classified with the severe deafness grade (the deafness
degrees of both ears varied and statistics were determined with
regard to the side with better hearing). The onset of deafness
ranged between the day of birth and 6 years of age. 77.97% (46/59)
of cases completely lost their language skills and 22.03% (13/59)
cases retained limited language skills. A total of 10 cases were
identified to have a family history of deafness, with 1–2 deaf
individuals within each family.
Genetic testing
The application of the Nine Deafness Gene Mutations
Detection microarray kit detected 22 patients with positive
deafness gene mutation hot-spots, and the positive rate was 37.29%.
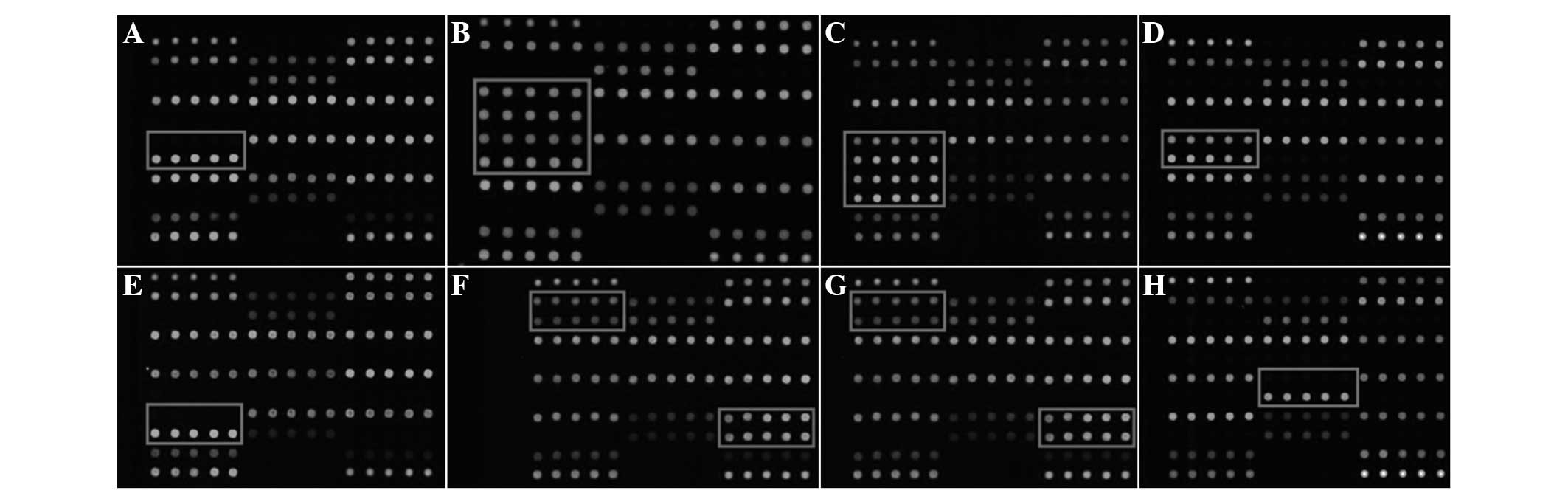
Among these, two cases of the GJB2 235 deletion (del) C
homozygous mutation (Fig. 1A); one
case of the GJB2 235 del C/176 del 16 complex heterozygous
mutation (Fig. 1B); one case of
the GJB2 235 del C/299 del AT complex heterozygous mutation
(Fig. 1C); eight cases of the
GJB2 235 del C heterozygous mutation (Fig. 1D); one case of the GJB2 299
del AT homozygous mutation (Fig.
1E); one case of the GJB2 35 del G/SLC26A4 IVS
7–2 A>G complex heterozygous mutation (Fig. 1F); two cases of the SLC26A4
IVS 7–2 A>G heterozygous mutation (Fig. 1G) and six cases of mitochondrial
12S rRNA 1555 A>G homogeneous mutation (Fig. 1H) were identified. The results of
the deafness mutation hot-spots screening, history of ototoxic drug
application and family history of deafness are presented in
Table I and the deafness chip
scanning results of certain selected subjects are presented in
Fig. 1. The comparison between the
groups with or without a clear family history gave a P-value
>0.05 (χ2=0.306, P=0.580); thus, no statistical
significance was observed.
 | Table IDeafness-associated factors in
adolescents with non-specific hearing loss in Yongchuan district
(Chongqing, China). |
Table I
Deafness-associated factors in
adolescents with non-specific hearing loss in Yongchuan district
(Chongqing, China).
| Results of deafness
mutation hotspot screening | Figure | Cases (n) | Subject nos. | History of ototoxic
drug application | Family history |
|---|
| Blank control | / | / | / | / | / |
| Normal control | / | / | / | / | / |
| Wild-type | / | 37 | 44, 45, 51, 30 | Gentamicin | No |
| | 53 | Streptomycin | No |
| | 3 | Uncertain | No |
| | 54 | Uncertain | Yes (mother) |
| | 55 | Uncertain | Yes (brother) |
| | 11 | No | Yes (brother) |
| | 32 | No | Yes (father and
mother) |
| | 47 | No | Yes (sister) |
| | 1, 2, 4, 6–9, 12,
16–20, 22, 25, 28, 29, 31, 34, 35, 37–41, 58 | No | No |
| GJB2 235 del C
homozygous mutation | Fig. 1A | 2 | 56 | Uncertain | No |
| | 10 | | Yes (brother) |
| GJB2 235 del
C/176 del 16 complex heterozygous mutation | Fig. 1B | 1 | 43 | Gentamicin | No |
| GJB2 235 del
C/299 del AT complex heterozygous mutation | Fig. 1C | 1 | 27 | No | No |
| GJB2 235 del C
heterozygous mutation | Fig. 1D | 8 | 24, 26, 50 | Gentamicin | No |
| | 14, 15, 33, 36,
60 | No | No |
| GJB2 299 del
AT homozygous mutation | Fig. 1E | 1 | 5 | No | No |
| GJB2 35 del
G/SLC26A4 IVS 7–2 A>G complex heterozygous mutation | Fig. 1F | 1 | 57 | Uncertain | No |
| SLC26A4 IVS
7–2 A>G heterozygous mutation | Fig. 1G | 2 | 13 | No | No |
| | 46 | No | Yes (mother) |
| Mitochondrial 12S
rRNA 1555 A>G homogeneous mutation | Fig. 1H | 6 | 49, 52 | Gentamicin | No |
| | 42 | Gentamicin | Yes (mother and
cousin) |
| | 48 | Gentamicin | Yes (brother) |
| | 23 | No | Yes (mother) |
| | 21 | No | No |
| Total | | 59 | | 13 (certain), 5
(uncertain), | 10 (family
history); |
| | | 41 (no
medication) | 49 (no family
history) |
Discussion
Genetic factors have been reported to influence the
development of deafness; thus, through identifying genetic factors
of deaf patients and high-risk groups and giving appropriate
guidance and interventional measures, the new-onset and aggravation
of deafness may be prevented to a certain extent (10). To date, numerous deafness genes
have been identified and the mutation spectrum is known to be
widespread. The identification of deafness gene mutation hot-spots
in domestic non-syndromic deaf patients has made it feasible to
investigate the associated causes.
GJB2 was the first disease-causing gene
identified for Chinese non-syndromic hearing loss (11) and in the present study, the
detection rate of the GJB2 mutation in CQ-YC ANSHL was
23.73% (14/59), accounting for 63.64% of all positive mutations
(14/22). A total of five cases of the GJB2 homozygous or
complex heterozygous mutation were identified to be the cause of
deafness, and the remaining nine cases of the GJB2
heterozygous mutation (including one case of GJB2 35 del
G/SLC26A4 IVS 7–2A>G complex heterozygous mutation) had
no family history of deafness. According to Mendelian inheritance
(12), these cases were identified
as the autosomal recessive heredity mode; however, the single
GJB2 allelic mutation could not be determined as the cause
of deafness, as the co-existence of additional possible factors
associated with deafness should be considered. GJB2 235 del
C was observed to be the most common deafness-associated mutation
in Yongchuan district in the cohort of the present study,
accounting for 73.68% (14/19) of all gene mutation loci detected,
followed by 299 del AT, accounting for 15.79% (3/19). The present
study detected one case of a35 del G mutation, and this patient's
family had lived in the local area for numerous years, with no
descent from any ethnic minority and no family history of deafness;
thus, it was suggested that a rare recessive genetic cause of
deafness was present in this patient within the Yongchuan
district.
A previous study observed that in one patient, the
mitochondrial 12S rRNA gene mutation was associated with maternal
inheritance, and the application of aminoglycosides resulted in
irreversible hearing loss (13).
In the present study, the detection rate of mitochondrial 12S rRNA
1555A>G mutation was 10.17% (6/59), while the 1494 C>T
mutation was not detected; among the six positive patients, three
patients had a family history of deafness and four cases had a
history of application of aminoglycoside drugs (gentamicin). This
result was higher than that reported by Dai et al (5) and Yuan et al (6), suggesting that the frequency of the
mutation of the mitochondrial 12S rRNA gene was increased in the
Yongchuan district. Screening towards deafness-associated mutations
in Yongchuan may aid in effectively identifying a large number of
individuals with this gene mutation, sensitivity to aminogly-coside
drugs and normal hearing abilities, who inherited the mutation from
maternal relatives. Thus, through education and application of
medication, the deafness of high-risk groups may be avoided, and in
addition, scientific genetic counseling, pre-natal diagnosis and
intervention may be performed over several generations, thus
blocking the passing on of this gene mutation in the family.
The SLC26A4 gene mutation has been previously
reported to result in non-syndromic hearing loss and syndromic
deafness (14,15). The present study detected two cases
of the SLC26A4 IVS 7–2A>G single heterozygous mutation
and one case of the GJB2 35 del G/SLC26A4 IVS 7–2
A>G complex heterozygous mutation in CQ-YC ANSHL. The detection
rate of the SLC26A4 IVS 7–2A>G mutation was 5.08% (3/59),
accounting for 13.64% (3/22) of all positive mutations, while the
2168 A>G mutation was not detected. A total of three cases had
no history of significant head trauma and high fever, only one case
of the SLC26A4 IVS 7–2A>G single heterozygous mutation
had a family history of deafness. To date, >150 cases of the
SLC26A4 gene mutation have been reported (16); however, the test results were
limited by the numbers and types of microarray deafness gene
mutation points and thus, not all known SLC26A4 gene
mutations could be detected. Further gene sequencing of the
patients with the SLC26A4 IVS 7–2A>G single heterozygous
mutation may be beneficial in elucidating the cause of their
deafness. In the present study, the detection rate of the
SLC26A4 mutation was lower than that reported in previous
studies (17,18), and the possible causes are as
follows: i) The SLC26A4 mutation is involved in the development of
large vestibular aqueduct syndrome (19), which commonly manifests as acquired
deafness. This deafness is progressive, and hearing aids and speech
training are capable of significantly improving hearing and speech
ability; thus, these patient may not have to enrolled in a special
education school and were thus not recruited for the present study
(20). ii) Differences may exist
in the deafness gene mutation spectrum in Yongchuan, as reported by
Dai et al (5) and Yuan
et al (6).
The present study analyzed the impact of the genetic
background on the detection rate of deafness gene mutation
hot-spots. The results demonstrated that there was no statistical
significance between the groups with or without a family history of
deafness (P>0.05; χ2=0.306). In addition, the genetic
background had no effect towards the detection rate of deafness
gene mutation hot-spots. This was considered to be due to the fact
that as deafness gene mutations detected by the microarray were
predominantly recessive or from maternal inheritance, the
participants were often carriers, but were not necessarily
deaf.
Previous studies have demonstrated that the
detection rate of single deafness gene mutation hot-spot screening
is not high, and for >50% of deaf patients, the genetic causes
of deafness are not clear; thus, the detection method and
integrated assessment tools require improvement (21,22).
In the present study, the mutation-positive rate was 37.29%
(22/59), while 62.71% (37/59) of patients carried the respective
wild-type genes. Subjects no. 47 and 55 were siblings, and the
deafness gene mutation hot-spot screening showed that they were the
wild-type; furthermore, there was no other deaf patient in their
family, and there fore, their mutation is likely to be of a
recessive hereditary type. However, mutations of genes other than
the nine common deafness-associated genes examined in the present
study may have been the underlying cause, and therefore, further
investigation of the causes of the deafness of patients no. 47 and
55 is required. The present study hypoth-esized that with the
advances in gene chip technology and in-depth research of the
genetic causes of deafness, additional deafness-associated
mutations may be covered, and individual screening against the
mutation spectrum may be undertaken in order to assess the
occurrence frequency in various regions and in patients of
different ethnicities. Therefore, using these techniques may aid in
the determination of the genetic causes of deafness.
The Yongchuan area is located in West Chongqing and
has a large population; genetic counseling with regard to
mutation-associated deafness is important to block the inheritance
of genetic deafness and improve the health of the local population.
In the present study, questionnaire-based statistics regarding the
CQ-YC ANSHL showed that in 11.86% (7/59) of cases, a history of
deafness was present in the family, the mother did not receive any
genetic counseling or risk assessment prior to the pregnancy. There
were numerous neglected children in the region whose abnormalities
were being ignored during their growth and development. Thus,
deafness was not identified early and due to the lack of formal
training in hearing aids and speech therapy, the patients'
language-communication skills and quality of life were severely
affected. Medical staff and parents are required to take early
symptoms of deafness in children and young people seriously, as
once they are diagnosed as deaf, a timely and appropriate treatment
should be performed. At the same time, the genetic cause of their
deafness should be determined to prevent the progression of
deafness in future generations. In addition, the present study
showed that the rate of non-syndromic hearing loss-associated gene
mutations among adolescents in the Yongchuan district was elevated
compared with that in other regions, suggesting that the government
and medical institutions should intensify genetic counseling and
pre-natal diagnosis of genetic diseases using existing tools,
including genetic diagnosis, in order to screen deaf patients and
high-risk groups for deafness gene mutations. The combination of
guidance on medication, pre-natal diagnosis and clinical
intervention may reduce deafness in the population.
Acknowledgments
The authors would like to thank Ms Dan Zhang, Ms
Jianmin Wang and Ms Juan Liao (Molecular Biology Laboratory and
Central Laboratory of Yongchuan Hospital, Chongqing Medical
University, Chongqing, China) and all staff, students and parents
of Chongqing Yongchuan Special Education School (Chongqing, China)
for their support and help.
The present study was supported by the Science and
Technology Research Project of Chongqing Municipal Education
Commission (grant no. KJ110326) and the Hospital Project of
Yongchuan Hospital, Chongqing Medical University (grant no.
YJYB201025).
References
|
1
|
Morton NE: Genetic epidemiology of hearing
impairment. Ann N Y Acad Sci. 630:16–31. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen G, Wang X and Fu S: Prevalence of
A1555G mitochondrial mutation in Chinese new borns and the
correlation with neonatal hearing screening. Int J Pediatr
Otorhinolaryngol. 75:532–534. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Géléoc GS and Holt JR: Sound strategies
for hearing restoration. Science. 344:12410622014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Taneja MK: Preimplantation genetic
diagnosis: Its role in prevention of deafness. Indian J Otolaryngol
Head Neck Surg. 66:1–3. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dai P, Yu F, Han B, Yuan Y, Li Q, Wang G,
Liu X, He J, Huang D, Kang D, et al: The prevalence of the 235delC
GJB2 mutation in a Chinese deaf population. Genet Med. 9:283–289.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan Y, You Y, Huang D, Cui J, Wang Y,
Wang Q, Yu F, Kang D, Yuan H, Han D, et al: Comprehensive molecular
etiology analysis of nonsyndromic hearing impairment from typical
areas in China. J Transl Med. 7:792009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Du W, Wang Q, Zhu Y, Wang Y and Guo Y:
Associations between GJB2, mitochondrial 12S rRNA, SLC26A4
mutations and hearing loss among three ethnicities. Biomed Res Int.
2014:7468382014. View Article : Google Scholar
|
|
8
|
Yin A, Liu C, Zhang Y, Wu J, Mai M, Ding
H, Yang J and Zhang X: Genetic counseling and prenatal diagnosis
for hereditary hearing loss in high-risk families. Int J Pediatr
Otorhinolaryngol. 78:1356–1359. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang XH, Wang JB and Kong WJ: Practice of
Otorhinolaryngology-Head and Neck Surgery. 2nd Edition. People's
Medical Publishing House; Beijing: pp. p10062007, In Chinese.
|
|
10
|
De Keulenaer S, Hellemans J, Lefever S,
Renard JP, De Schrijver J, Van de Voorde H, Tabatabaiefar MA, Van
Nieuwerburgh F, Flamez D, Pattyn F, et al: Molecular diagnostics
for congenital hearing loss including 15 deafness genes using a
next generation sequencing platform. BMC Med Genomics. 5:172012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu F, Han DY, Dai P, Kang DY, Zhang X, Liu
X, Zhu QW, Yuan YY, Sun Q, Xue DD, et al: Mutation of GJB2 gene in
nonsyndromic hearing impairment patients: Analysis of 1190 cases.
(Article in Chinese). Zhonghua Yi Xue Za Zhi. 87:2814–2819. 2007.In
Chinese.
|
|
12
|
Kenneson A, Van Naarden Braun K and Boyle
C: GJB2 (connexin 26) variants and nonsyndromic sensorineural
hearing loss: A HuGE review. Genet Med. 4:258–274. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen T, Liu Q, Jiang L, Liu C and Ou Q:
Mitochondrial COX2 G7598A mutation may have a modifying role in the
phenotypic manifestation of aminoglycoside antibiotic-induced
deafness associated with 12S rRNA A1555G mutation in a Han Chinese
pedigree. Genet Test Mol Biomarkers. 17:122–130. 2013. View Article : Google Scholar :
|
|
14
|
Prak HJ, Shaukat S, Liu XZ, Hahn SH, Naz
S, Ghosh M, Kim HN, Moon SK, Abe S, Tukamoto K, et al: Origins and
frequencies of SLC26A4 (PDS) mutations in east and south Asians:
Global implications for the epidemiology of deafness. J Med Genet.
40:242–248. 2003. View Article : Google Scholar
|
|
15
|
Hutchin T, Coy N, Conlon H, Telford E,
Bromelow K, Blaydon D, Taylor G, Coghill E, Brown S, Trembath R, et
al: Assessment of the genetic causes of recessive childhood
non-syndromic hearing loss in the UK-implications for genetic
testing. Clin Genet. 68:506–512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bizanova A and Kopp P: Genetics and
phenomics of pendred syndrome. Mol Cell Endocrinol. 322:83–90.
2010. View Article : Google Scholar
|
|
17
|
Dai P, Li Q, Huang D, Yuan Y, Kang D,
Miller DT, Shao H, Zhu Q, He J, Yu F, et al: SLC26A4c.919–2A>G
varies among Chinese ethnic groups as a cause of hearing loss.
Genet Med. 10:586–592. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang H, Chen J, Shan XJ, Li Y, He JG and
Yang BB: Prevalence and range of GJB2 and SLC26A4 mutations in
patients with autosomal recessive non-syndromic hearing loss. Mol
Med Rep. 10:379–386. 2014.PubMed/NCBI
|
|
19
|
Yuan Y, Guo W, Tang J, Zhang G, Wang G,
Han M, Zhang X, Yang S, He DZ and Dai P: Molecular epidemiology and
functional assessment of novel allelic variants of SLC26A4 in
non-syndromic hearing loss patients with enlarged vestibular
aqueduct in China. PLoS One. 7:e499842012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu YM, Guo YF, Liu XW, Wang Y, Xu B, Ji
Y, Li J, Li Z and Wang Q: The hereditary etiology analysis of deaf
students from Shaanxi province. J Audiol Speech Pathol. 18:225–228.
2010.
|
|
21
|
Alford RL, Arnos KS, Fox M, Lin JW, Palmer
CG, Pandya A, Rehm HL, Robin NH, Scott DA, Yoshinaga-Itano C, et
al: American college of medical genetics and genomics guideline for
the clinical evaluation and etiologic diagnosis of hearing loss.
Genet Med. 16:347–355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Levenson D: New testing guidelines for
hearing loss support next-generation sequencing: Testing method may
help determine genetic causes of hearing loss among patients whose
phenotypes are not easily distinguished clinically. Am J Med Genet
A. 164:vii–viii. 2014.
|