Introduction
Colorectal cancer (CRC) is the third most common
cancer type and the fourth leading cause of cancer-associated
mortality worldwide, with an estimated incidence of 1.2 million
cases and a mortality of >600,000 annually (8% of all
cancer-associated mortalities), making this a global public health
problem. The incidence and mortality rates of CRC are increasing
rapidly in Asian countries (1).
Early-stage diagnosis of CRC potentially reduces the mortality of
this disease. Although colonoscopic screening for CRC is currently
the most reliable diagnostic tool, its cost and invasive nature
limit its use. Therefore, there is a pressing requirement for
developing novel, non-invasive, highly sensitive effective
therapeutic methods (2).
MicroRNAs (miRs) belong to a class of highly
conserved, ~22-nucleotide, single-stranded RNAs, which
epigenetically regulate protein translation by binding to the
3′-untranslated region (UTR) of the target messenger RNA (mRNA) and
mediating either mRNA degradation or translational repression
(3). miRs function as potential
oncogenes or tumor suppressors in cancer development. A global
impairment of miR has been described in various human cancer types,
including CRC (4).
miR-1246 is a p53 transcriptional target, which is
involved in the regulation of the known p53 functions, including
cell cycle, apoptosis and senescence (5). The levels of serum miR-1246 can
predict lymph node metastasis in cervical squamous cell carcinoma,
which induces cervical squamous cell carcinoma SiHa cell
proliferation, invasion and migration by targeting THBS2 (6,7) and
it may be a prognostic biomarker for oesophageal squamous cell
carcinoma (8). miR-1246 is
important in differentiation, invasion and metastasis of esophageal
squamous cell carcinoma and hepatocellular carcinoma (9–11).
miR-1246 was reported to be one of the driving miRs in CRC
(12) since miR-1246 was
significantly higher in tumor tissues compared with stromal
tissues, and is present in higher serum exosomal levels in patients
with CRC (13,14). miR-1246 is considered to be
responsible for proangiogenic function by activating Smad1/5/8
signaling in CRC (15). However,
other biofunctions remain to be elucidated.
CCNG2 encodes an unconventional cyclin homolog,
cyclin G2 (CycG2), associated with growth inhibition. The
expression of CycG2 is repressed by mitogens, however, is
upregulated during cell cycle arrest responses to antiproliferative
signals (16). The expression of
CycG2 decreased in thyroid cancer, esophageal, prostate, kidney and
gastric cancer tissues and cells, and is significantly correlated
with lymph node metastasis, clinical stage, histological grade and
poor overall survival, suggesting that CCNG2 may be important as a
negative regulator in thyroid cancer K1 cells by promoting the
degradation of CDK2 (17–21). In addition, abnormal expression of
CycG2 was observed in nasopharyngeal carcinoma cells and was
closely associated with tumor differentiation, origin and
progression (22). In colorectal
carcinoma, the protein expression level of CycG2 was significantly
lower in CRC tissue compared with normal tissues, and biological
function revealed that SW480 cells transfected with CCNG2 exhibited
a lower survival fraction, higher percentage of the G0/G1 phases
and reduced protein expression of CDK2 (21). The underlying molecular mechanism
of the downregulation of CycG2 remains to be fully elucidated. In
laryngeal squamous cell carcinoma, CCNG2 was confirmed as a direct
target of miR-93 (23).
Notably, the expression of miR-1246 is associated
with chemoresistance and cancer stem cell-like properties via
CCNG2, and may predict a poorer prognosis in patients with
pancreatic cancer (24). These
results suggested that upregulation of miR-1246 may be one of the
molecular mechanism of the regression of CCNG2 in pancreatic cancer
cells. However, whether this is also the case in CRC cells remains
to be elucidated.
The present study constructed cell models with
gain-of-and loss-of-function of miR-1246 to assess its biofunction
and direct target in CRC cell lines. These results may support
CCNG2 and miR-1246 as novel diagnostic or therapeutic target for
CRC.
Materials and methods
Ethics statement
Prior written informed consent was obtained from all
patients and the study was approved by the Protection of Human
Subjects Committee of the Second XiangYa Hospital of Central South
University (Hunan, China).
Tumor tissue sample preparation
A total of 10 patients, including six males and four
females aged 45–76 years (mean, 63.2 years), diagnosed with CRC at
The Second XiangYa Hospital of Central South University were
recruited. The experimental protocols were approved by the Ethics
Committee of The Second XiangYa Hospital of Central South
University. RNA or tissue samples were prepared from the tumor
tissues and their matched adjacent non-tumor tissues, and were then
subjected to reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) or western blotting.
Tissue microarray
immunohistochemistry
A CRC tissue microarray (TC0128) was purchased from
Auragene Biotechnology (Changsha, China). Tissue microarray blocks
were incubated with anti-CCNG2 antibody (cat. no. ab5502; Abcam,
Cambridge, MA, USA) or normal rabbit immunoglobulin G as a negative
control. Immunostaining was performed using the Moticcam3000 system
(Motic Group Co., Ltd., Xiamen, China) with diaminobenzidine
(Zhongshan Jinqiao, Beijing, China). Image-Pro-Plus software (Media
Cybernetics, Inc., Rockville, MD, USA) was used to quantify the
mean density of CCNG2 staining, according to the manufacturer's
protocols.
Cell culture
The SW620, SW480, HCT116, HT29 and LOVO CRC cell
lines, and intestinal epithelial cells (IECs) were purchased from
China Center for Type Culture Collection (Wuhan, China). All cell
lines were cultured in Dulbecco's modified Eagle's medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented
with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc.), 100 IU/ml penicillin and 100 µg/ml streptomycin
sulfate (Beyotime Institiute of Biotechnology, Shanghai, China) at
37°C in a humidified incubator, containing 5% CO2.
Transfection
For the miR-1246 functional analyses, the
pre-miR-1246, pre-con, anti-1246 or anti-con (GeneCopoeia,
Guangdong, China) were transfected into HCT-116 and LOVO cell lines
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocols.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocols. The relative expression level of
miR-1246 and CCNG2 were determined by RT-qPCR using mirVana™
qRT-PCR microRNA detection kit (Ambion, Applied Biosystems, Austin,
TX, USA), according to the manufacturer's protocols. The specific
primer settings for miR-1246 (cat. no. HmiRQP0078) and U6 (cat. no.
HmiRQP9003) were obtained from GeneCopoeia. The mRNA expression of
CCNG2 was detected by RT-qPCR using the standard SYBR Green RT-PCR
kit (Takara Bio, Inc., Otsu, Japan), according to the manufacture's
protocols. The specific primer pairs were as follows: CCNG2, sense:
5′-AAGAAGAGAGATTCCAACC-3′ and antisense: 5′-CCAGCAAAAAAGAACAGAC-3′;
β-actin, sense: 5′-CCCATCTATGAGGGTTACGC-3′ and anti-sense:
5′-TTTAATGTCACGCACGATTTC-3′. The mRNA expression levels were
normalized against that of β-actin. The relative mRNA expression
levels of CCNG2 or miR-1246 was quantified using the GraphPad Prism
4.0 software (GraphPad Software, San Diego, CA, USA) and the
2−ΔΔCq method (25).
Cell proliferation assay
Cells in the exponential growth phase were plated at
a final concentration of 2×103 cells/well into 96-well
culture plates. The viability of the cells was determined by MTT
assay 24, 48 and 72 h following seeding. The optical density at 570
nm of each well was measured using an ELISA reader (ELX-800;
Bio-Tek, Winooski, VT, USA).
Colony-formation assay
For all groups, 3 ml complete medium, containing 150
cells were added into each well of a 6-well plate. The plates were
incubated at 37°C with 5% CO2 for 14 days. Following
incubation, the cells were gently washed and stained with Giems
(Solabrio, Beijing, China). Colonies containing at least 50 cells
(0.3–1.0 mm in diameter) were counted using a microscope (AE31;
Motic Group Co., Ltd.).
Cell apoptosis assay
The cells were transfected with 80 nM pre-miR-1246
or anti-miR-1246, and 48 h after transfection, the cells were
subjected to an apoptosis assay. Apoptosis was detected by annexin
V/propidium iodide staining using an apoptosis detection kit (BD
Pharmingen, San Diego, CA, USA). Briefly, 106 treated
cells were incubated with annexin V/propidium iodide for 20 min at
25°C and were subsequently analyzed by flow cytometry (FACSCalibur;
Beckman Coulter, Brea, CA, USA).
Cell invasion assay
The invasive ability of CRC cells was determined in
24-well Transwell chambers (Chemicon, Danvers, MA, USA), containing
a layer of matrigel. For all groups, 200 µl cell suspension
(1×106 cells/ml) was added in triplicate wells.
Following an incubation for 24 h, the dye on the membrane was
dissolved with 10% acetic acid, dispensed into 96-well plates (150
µl/well) and the optical density at 570 nm of each well was
measured using an ELISA reader (ELX-800; BioTek).
Cell migration assay
The cell migratory capability was estimated using a
wound healing assay, as described previously (26). Briefly, the cells were cultured to
confluence. Wounds of ~1 mm width were created using a pipette tip
of 1 ml, and the cells were washed and incubated in a serum-free
medium. Following incubation for 24 h, the cells were incubated in
a medium, containing 10% fetal bovine serum. Cultures at 0 and 48 h
were fixed and observed under a microscope (AE31; Motic).
Dual luciferase reporter assay
The 3′-UTR of CCNG2 (NM_004354.2), containing the
miR-1246 binding sites and its corresponding mutated sequence, were
cloned into the psi-CHECK2 luciferase reporter vector (Promega,
Madison, WI, USA) downstream of Renilla luciferase, termed
CCNG2-3′-UTR and CCNG2-Mut 3′-UTR, respectively. Using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
the HCT-116 and LOVO cells were co-transfected with the recombinant
reporter constructs and miR-1246 mimics, miR-1246 inhibitor,
negative control (NC) or the negative control inhibitor
(GeneCopopia, Rockville, MD, USA). Luciferase activity was
determined after 48 h using the Dual-Glo substrate system (Promega)
and an LD400 luminometer (Beckman Coulter). The data are presented
as the ratio of experimental (Renilla) luciferase against
control (Firefly) luciferase.
Western blotting
The tissues or cells were solubilized in cold
radioimmunoprecipitation lysis buffer (Auragene, Changsha, China).
Following lysis, a Bicinchoninic Acid Assay kit was used (Auragene)
to determine protein concentration. The proteins (2
µg/µl) were separated on 10% SDS-PAGE gels (Amresco
LLC, Solon, OH, USA) and were subsequently transferred onto a
polyvinylidene membrane (EMD Millipore, Bedford, MA, USA). The
membranes were blocked in 10% non-fat dried milk in
phosphate-buffered saline containing 0.05% Tween-20 for 3 h and
were subsequently incubated overnight at 4°C with rabbit anti-human
CCNG2 polyclonal antibody (cat. no. ab5502; 1:1,000; Abcam), with
β-actin (rabbit anti-human polyclonal antibody; 1:2,000; cat. no.
189073; Abcam) used as a control. Following primary antibody
incubation, the membrane was incubated with the
goat-anti-rabbit-horseradish peroxidase secondary antibody (cat.
no. 111-035-003; Jackson ImmunoResearch Laboratories, West Grove,
PA, USA) at room temperature for 1 h, and the immune complexes were
detected using an enhanced chemiluminescence kit (Auragene).
Statistical analysis
The data are expressed as the mean ± standard
deviation from at least three separate experiments. Statistical
analysis was performed using SPSS 16.0 software (SPSS, Inc.,
Chicago, IL, USA). The difference between two groups was analyzed
by Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
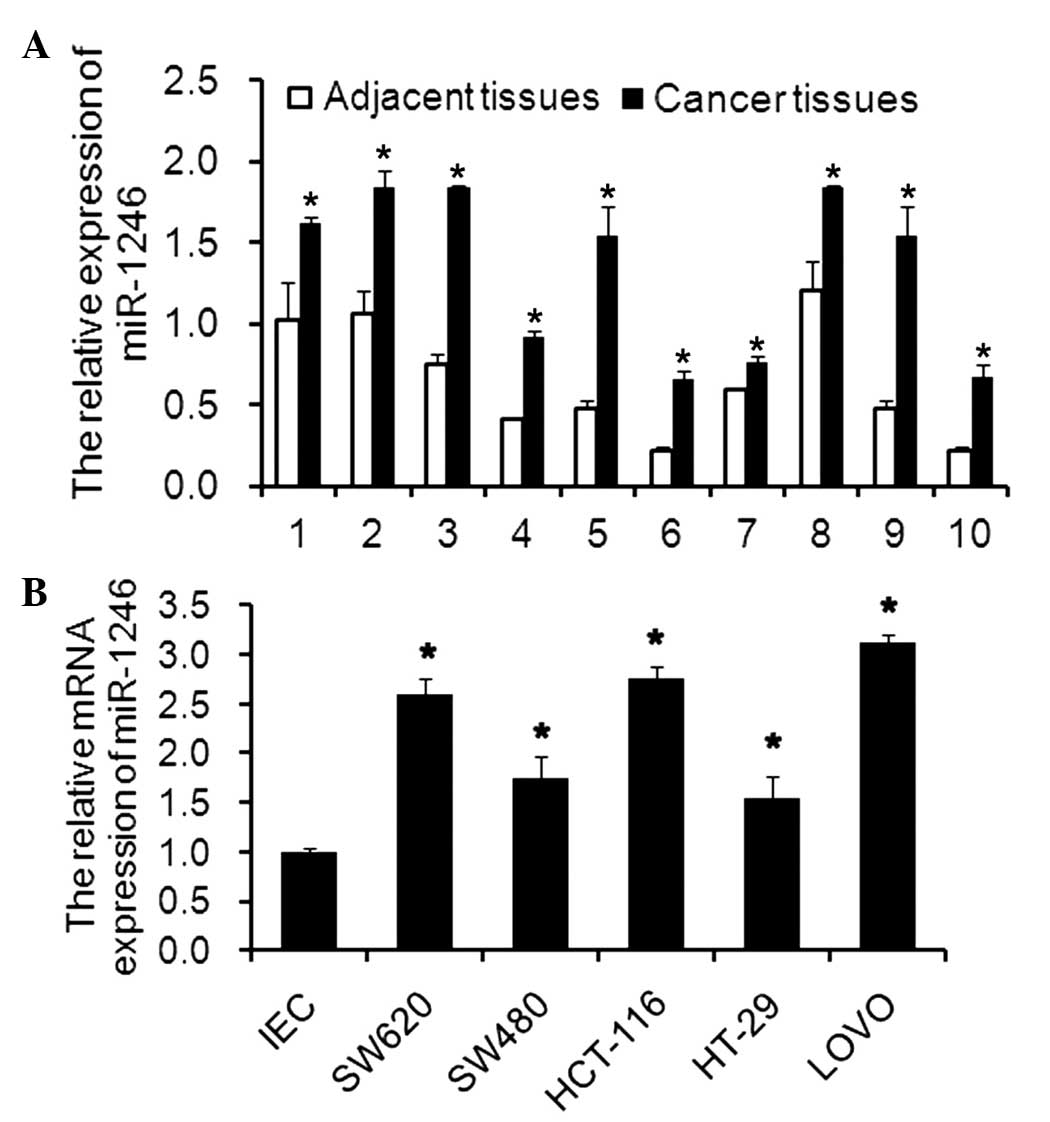
Expression of miR-1246 in CRC tissues and
cell lines
The mRNA expression levels of miR-1246 in clinical
tissues or in CRC cell lines were determined using RT-qPCR.
Compared with the paired adjacent tissues, the mRNA expression
levels of miR-1246 in the tumor tissues were significantly higher
(Fig. 1A). Additionally, the mRNA
expression levels of miR-1246 in SW620, SW480, HCT116, HT29 and
LOVO cell lines were also upregulated compared with the levels in
the IECs (Fig. 1B). The results
suggested that miR-1246 may be an onco-miR involved in the
progression and development of CRC.
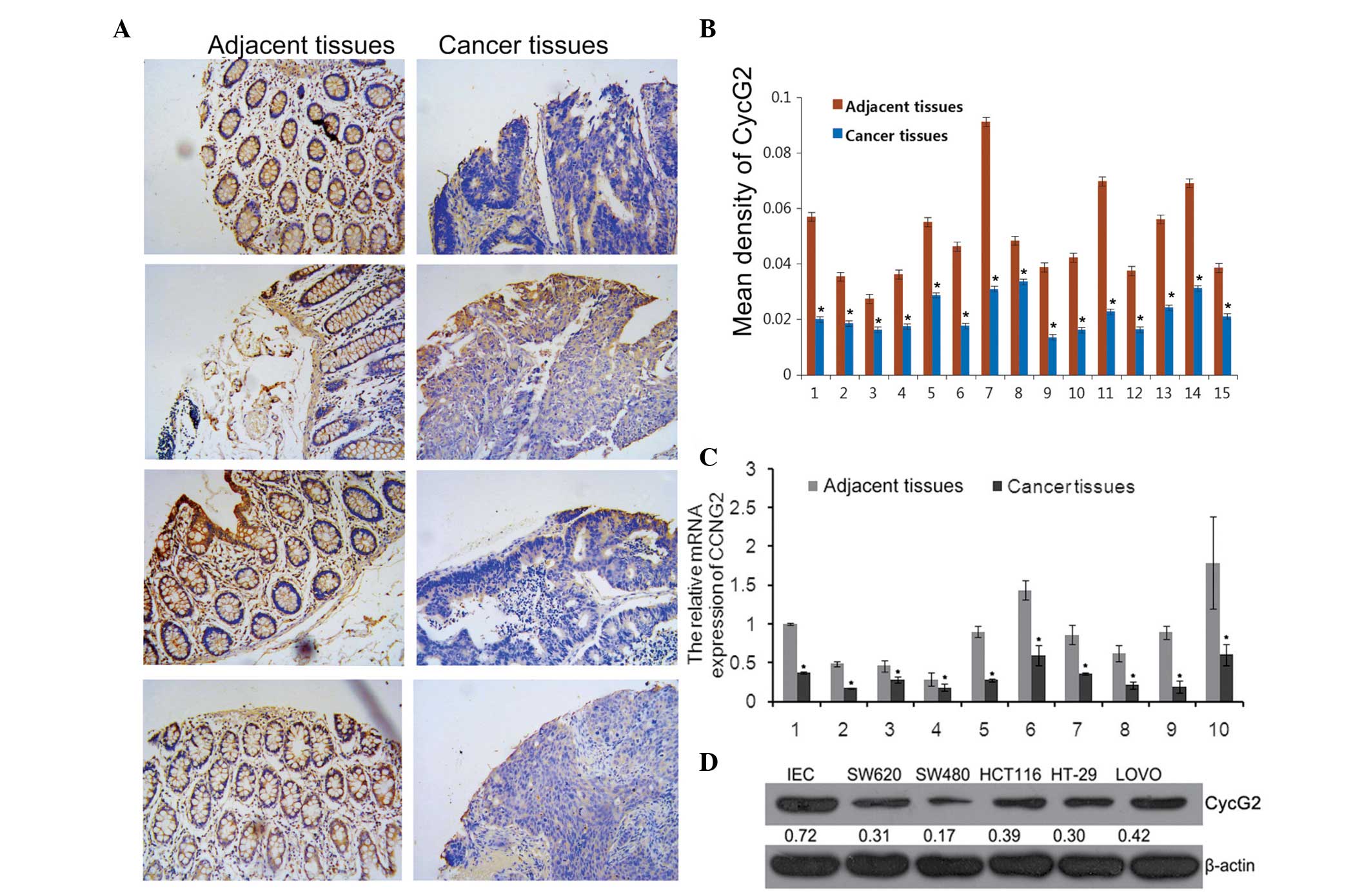
Expression of CycG2 in CRC tissues and
cell lines
CCNG2 was a predicted underlying target gene of
miR-1246. The tissue microarray analysis results demonstrated that
the CycG2 mean density was significantly lower in tumor tissues
compared with paired adjacent tissues (Fig. 2A and B).
The expression levels of CycG2 in collected CRC
tissues and cell lines were assessed using RT-qPCR and western
blotting. The results revealed that compared with the paired
adjacent tissues, the mRNA expression levels of CCNG2 in tumor
tissues were significantly lower (Fig.
2C) and the protein expression levels of CycG2 in the SW620,
SW480, HCT116, HT29, LOVO cell lines were also downregulated
compared with the IECs (Fig. 2D),
which were consistent with the TMA results.
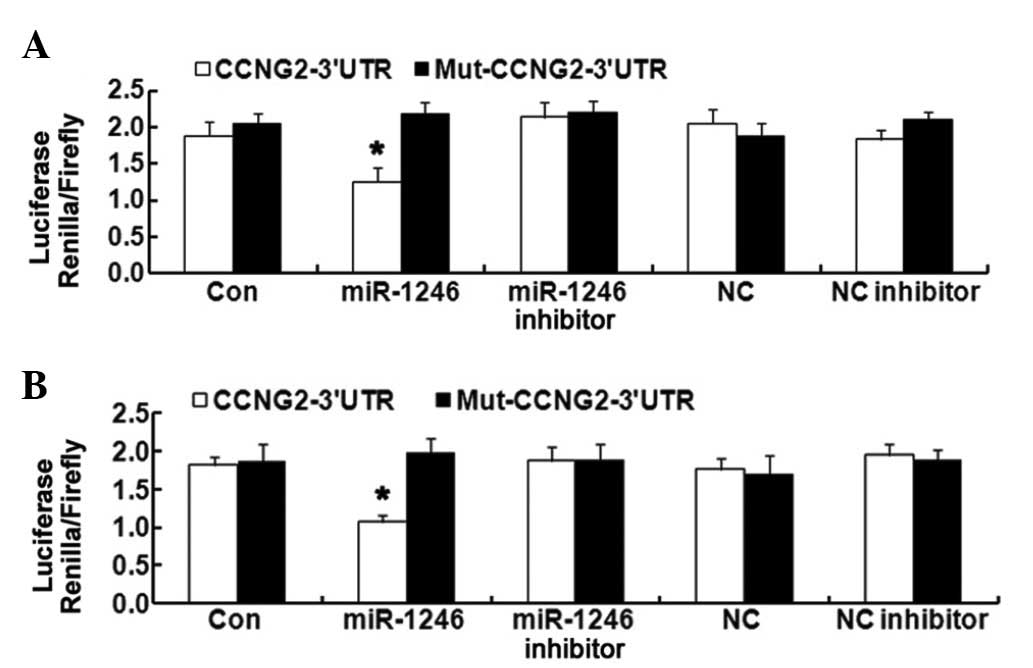
CCNG2 is a direct target of miR-1246 in
the HCT-116 and LOVO cell lines
The coexistence of miR-1246 upregulation and CCNG2
downregulation in CRC tissues and cells implies a potential
regulatory correlation between them. To confirm the direct
targeting association between them, a luciferase reporter assay was
used. The CCNG2 3′-UTR fragment, containing the miR-1246 binding
site and its mutated sequence, were cloned into psi-CHECK2 dual
luciferase reporter vectors. miR-1246 significantly inhibited the
luciferase activity in HCT-116 and LOVO cells transfected with the
CCNG2-3′-UTR. However, miR-1246 mimics revealed no suppression on
the luciferase activity levels in HCT-116 and LOVO cells
transfected with Mut-CCNG2-3′-UTR (Fig. 3). These results confirmed that
CCNG2 is a direct target of miR-1246 in HCT-116 and LOVO cells.
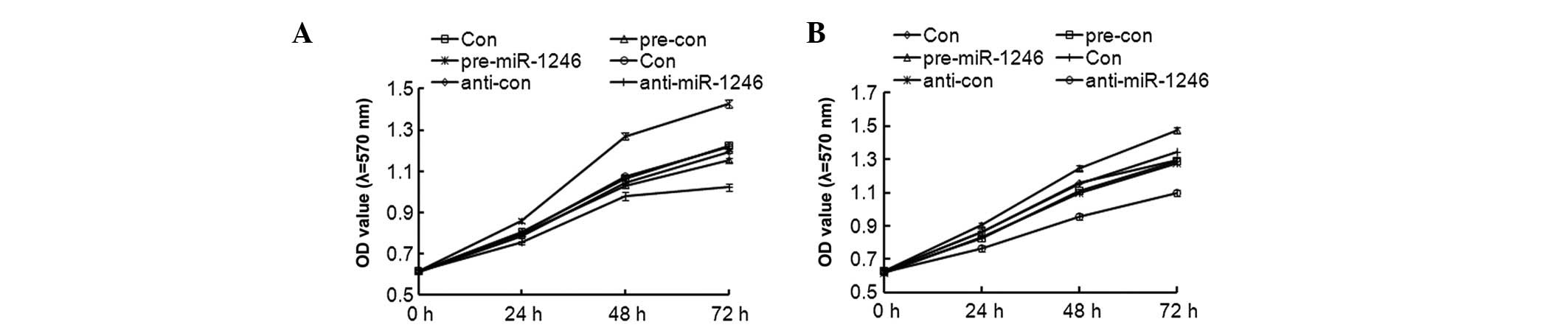
Effects of miR-1246 on CRC
proliferation
To further investigate the role of miR-1246 in CRC
in vitro, a cell proliferation assay was performed to
determine the effects of miR-1246 overexpression and knockdown on
cell proliferation in HCT-116 and LOVO cells. Following
transfection with pre-miR-1246, anti-miR-1246 and their negative
control (pre-con and anti-con), RT-qPCR data revealed that the mRNA
expression of miR-1246 was successfully altered. The expression of
miR-1246 increased by almost 12 times in the HCT116 cell line, and
15 times in the LOVO cells, compared with the pre-con groups,
following transfection with pre-miR-1246 transfection (data not
shown). As shown in Fig. 4, the
cell proliferation rate was even higher in the
miR-1246-overexpressed HCT-116 and LOVO cells when compared with
their parental or control groups; however, in miR-1246-knockdown
HCT-116 and LOVO cells, the cell proliferation was reduced. These
findings suggested that miR-1246 has a positive role in HCT-116 and
LOVO cell proliferation.
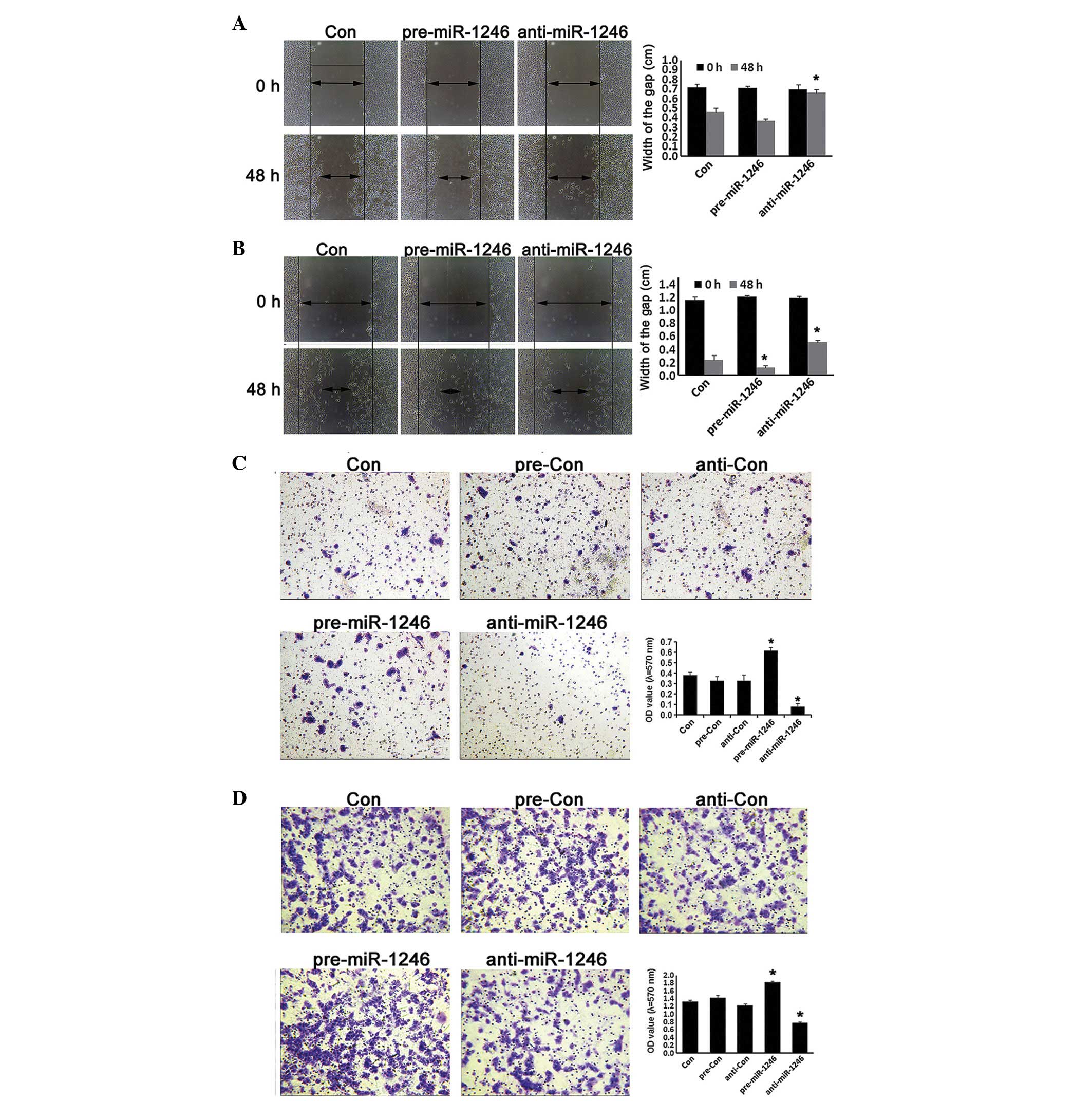
Effects of miR-1246 on CRC cell invasion
and migration
The alternations of human CRC invasion were further
investigated in miR-1246 gain- and loss-of-function HCT-116 and
LOVO cells. As shown in Fig. 5,
miR-1246-overexpression cells exhibited a higher invasive and
migration capacity compared with cells in the control groups, while
miR-1246-reduced cells exhibited a lower invasive phenotype and
migration capacity compared with the relative controls (P<0.05;
Fig. 5). These data indicated that
miR-1246 promoted the invasive ability and migration capacity in
human CRC.
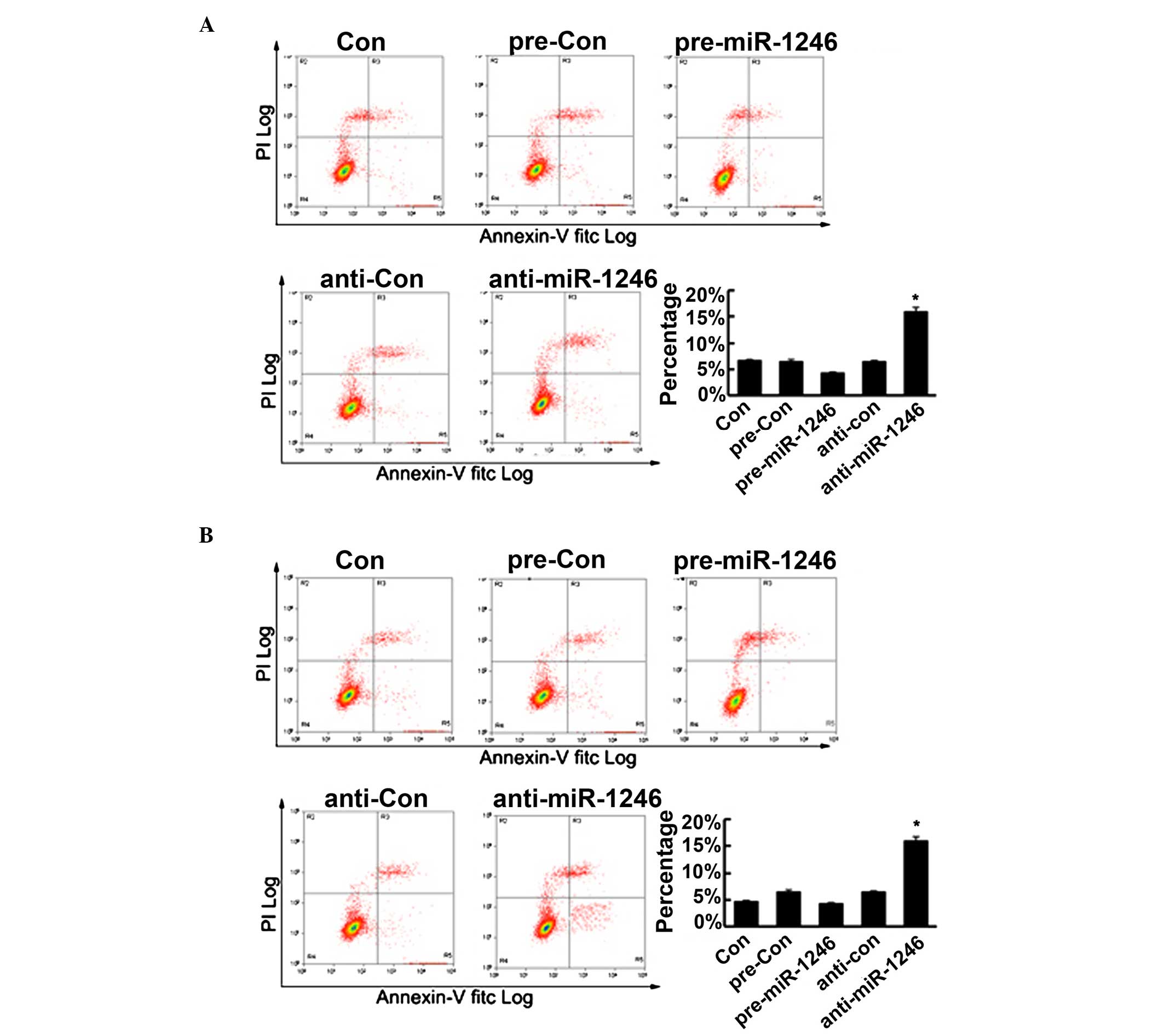
miR-1246 regulates cell apoptosis in CRC
cells
To further determine the role of miR-1246 in CRC
cells, cell apoptosis of the HCT-116 and LOVO cells was determined,
subsequent to upregulating or downregulating miR-1246, using flow
cytometry. As shown in Fig. 6, the
upregulation of miR-1246 reduced cell apoptosis, however, this was
not statistically significant. Conversely, the downregulation of
miR-1246 significantly promoted cell apoptosis. Therefore, these
findings confirmed that miR-1246 mediated the cell apoptosis in CRC
cells in vitro.
Discussion
miR-1246 is known to be important in the progression
of diverse cancer types. In the present study, it was revealed that
miR-1246 was upregulated in the examined 10 tissues samples, which
is consistent with previous reports (5,8,27).
These finding further illustrated that miR-1246 serves as an
oncogene in CRC and its abnormal upregulation facilitates the
progression of the tumor. It is well known that miRNAs function
predominantly via special binding to the 3′-UTR of its target
genes. Theoretical analysis suggests that the 3′-UTR of CCNG2
includes the binding sites of miR-1246. To confirm this, the
HCT-116 and LOVO CRC cells were transfected with miR-1246 mimics or
inhibitor. The results revealed that the protein expression levels
of CycG2 were accordingly decreased or increased, suggesting that
miR-1246 negatively regulated the expression of CCNG2 in CRC cells
and that CCNG2 is the potential target gene of miR-1246.
Subsequently, the dual luciferase reporter assay revealed that only
miR-1246 mimics inhibited luciferase activity in HCT-116 and LOVO
CRC cells transfected with 3′-UTR CCNG2 reporter vector. Therefore,
for the first time, to the best of our knowledge, the present study
demonstrated that CCNG2 is a target gene of miR-1246 in CRC
cells.
The present study next investigated the potential
effects of the miR-1246/CCNG2 pathway on HCT-116 and LOVO cells.
The results revealed that the upregulation of miR-1246, which led
to a decreased expression of CycG2, promoted the proliferation,
colony formation, invasion and migration, and inhibited the
apoptosis of CRC cells. However, downregulation of miR-1246 by its
inhibitor, which led to increased expression of CycG2, inhibited
these processes. These results suggested that miR-1246 upregulation
may be critical in the malignant progression of CRC and its
mechanism of action is partly involved in CCNG2.
The mechanisms underlying the function of miR-1246
as an inducer in CRC are implicated in promoting proliferation,
invasion, migration, apoptosis and differentiation (for example, by
suppressing THBS2 and CADM1) (7,10).
In the present study, certain of these molecules or others were
possibly involved in the process of miR-1246 affecting growth and
metastasis of HCT-116 and LOVO CRC cells. However, the present
study hypothesized that CCNG2, as target of miR-1246, may also be
involved in miR-1246 by promoting growth and metastasis of CRC
cells. Previous studies indicated that CCNG2 is important for tumor
cell proliferation. Downregulation of CycG2 can lead to growth
enhancement in various cancer types. It was previously reported
that downregulation of CCNG2 led to a reduced percentage of cells
in the G0/G1 phases (P<0.05) and higher CDK2 protein expression
in several types of tumor cell (17,18,20).
CCNG2 is also associated with invasion and lymph node metastasis,
as confirmed in prostate cancer cells (19). These results confirmed that
miR-1246 was a negative regulator of CCNG2 expression in CRC cells.
Although the target gene of miR-1246 is not singular, it is logical
to deduce that the upregulation of miR-1246, promoting
proliferation, invasion, migration and inhibiting apoptosis in CRC
cells is, at least partly, due to the downregulation of CCNG2.
miR-1246 was reported to be involved in stem cell
characteristics, including chemoresistance and cancer stem
cell-like properties via CCNG2 in pancreatic cancer (24), which may be an area of future work,
to determine if miR-1246 influences the stemness of CRC stem cells
and chemosensitivity of CRC cells.
In conclusion, miR-1246 was upregulated in CRC and
negatively regulates the expression of CCNG2 in HCT-116 and LOVO
CRC cells. miR-1246 upregulation mediated the malignant progression
of CRC, partly via the inhibition of CCNG2 expression. These
findings provided a molecular basis for the role of miR-1246/CCNG2
in the progression of human CRC cells and suggested a novel target
for the treatment of CRC.
References
|
1
|
Dong Y, Yu J and Ng SS: MicroRNA
dysregulation as a prognostic biomarker in colorectal cancer.
Cancer Manag Res. 6:405–422. 2014.PubMed/NCBI
|
|
2
|
Yang Y, Gu X, Zhou M, Xiang J and Chen Z:
Serum microRNAs: A new diagnostic method for colorectal cancer.
Biomed Rep. 1:495–498. 2013.
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liao JM, Zhou X, Zhang Y and Lu H:
MiR-1246: A new link of the p53 family with cancer and Down
syndrome. Cell Cycle. 11:2624–2630. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen J, Yao D, Li Y, Chen H, He C, Ding N,
Lu Y, Ou T, Zhao S, Li L and Long F: Serum microRNA expression
levels can predict lymph node metastasis in patients with
early-stage cervical squamous cell carcinoma. Int J Mol Med.
32:557–567. 2013.PubMed/NCBI
|
|
7
|
Chen J, Yao D, Zhao S, He C, Ding N, Li L
and Long F: MiR-1246 promotes SiHa cervical cancer cell
proliferation, invasion and migration through suppression of its
target gene thrombospondin 2. Arch Gynecol Obstet. 290:725–732.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takeshita N, Hoshino I, Mori M, Akutsu Y,
Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, et
al: Serum microRNA expression profile: miR-1246 as a novel
diagnostic and prognostic biomarker for oesophageal squamous cell
carcinoma. Br J Cancer. 108:644–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fu HL, Wu de P, Wang XF, Wang JG, Jiao F,
Song LL, Xie H, Wen XY, Shan HS, Du YX and Zhao YP: Altered miRNA
expression is associated with differentiation, invasion and
metastasis of esophageal squamous cell carcinoma (ESCC) in patients
from Huaian, China. Cell Biochem Biophys. 67:657–668. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun Z, Meng C, Wang S, Zhou N, Guan M, Bai
C, Lu S, Han Q and Zhao RC: MicroRNA-1246 enhances migration and
invasion through CADM1 in hepatocellular carcinoma. BMC Cancer.
14:6162014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan H, Wang S, Yu H, Zhu J and Chen C:
Molecular pathways and functional analysis of miRNA expression
associated with paclitaxel-induced apoptosis in hepatocellular
carcinoma cells. Pharmacology. 92:167–174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Piepoli A, Tavano F, Copetti M, Mazza T,
Palumbo O, Panza A, di Mola FF, Pazienza V, Mazzoccoli G, Biscaglia
G, et al: Mirna expression profiles identify drivers in colorectal
and pancreatic cancers. PloS one. 7:e336632012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Della Vittoria Scarpati G, Calura E, Di
Marino M, Romualdi C, Beltrame L, Malapelle U, Troncone G, De
Stefano A, Pepe S, De Placido S, et al: Analysis of differential
miRNA expression in primary tumor and stroma of colorectal cancer
patients. Biomed Res Int. 2014:8409212014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ogata-Kawata H, Izumiya M, Kurioka D,
Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H,
et al: Circulating exosomal microRNAs as biomarkers of colon
cancer. PloS One. 9:e929212014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamada N, Tsujimura N, Kumazaki M,
Shinohara H, Taniguchi K, Nakagawa Y, Naoe T and Akao Y: Colorectal
cancer cell-derived microvesicles containing microRNA-1246 promote
angiogenesis by activating Smad 1/5/8 signaling elicited by PML
down-regulation in endothelial cells. Biochim Biophys Acta.
1839:1256–1272. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zimmermann M, Arachchige-Don AS, Donaldson
MS, Dallapiazza RF, Cowan CE and Horne MC: Elevated cyclin G2
expression intersects with DNA damage checkpoint signaling and is
required for a potent G2/M checkpoint arrest response to
doxorubicin. J Biol Chem. 287:22838–22853. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen JQ, Liu CJ, Wen HX, Shi CL, Zhang HS,
Li M and Sun GG: Changes in the expression of cyclin G2 in
esophageal cancer cell and its significance. Tumour Biol.
35:3355–3362. 2014. View Article : Google Scholar
|
|
18
|
Li WJ, Liu GL, Yu F, Xiang XX, Lu YF, Xiao
HZ and Shi YP: CCNG2 suppressor biological effects on thyroid
cancer cell through promotion of CDK2 degradation. Asian Pac J
Cancer Prev. 14:6165–6171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cui DW, Cheng YJ, Jing SW and Sun GG:
Effect of cyclin G2 on proliferative ability of prostate cancer
PC-3 cell. Tumour Biol. 35:3017–3024. 2014. View Article : Google Scholar
|
|
20
|
Cui DW, Sun GG and Cheng YJ: Change in
expression of cyclin G2 in kidney cancer cell and its significance.
Tumour Biol. 35:3177–3183. 2014. View Article : Google Scholar
|
|
21
|
Sun GG, Zhang J and Hu WN: CCNG2
expression is down-regulated in colorectal carcinoma and its
clinical significance. Tumour Biol. 35:3339–3346. 2014. View Article : Google Scholar
|
|
22
|
Ye XX, Liu CB, Chen JY, Tao BH and Zhi-Yi
C: The expression of cyclin G in nasopharyngeal carcinoma and its
significance. Clin Exp Med. 12:21–24. 2012. View Article : Google Scholar
|
|
23
|
Xiao X, Zhou L, Cao P, Gong H and Zhang Y:
MicroRNA-93 regulates cyclin G2 expression and plays an oncogenic
role in laryngeal squamous cell carcinoma. Int J Oncol.
46:161–1741. 2015.
|
|
24
|
Hasegawa S, Eguchi H, Nagano H, Konno M,
Tomimaru Y, Wada H, Hama N, Kawamoto K, Kobayashi S, Nishida N, et
al: MicroRNA-1246 expression associated with CCNG2-mediated
chemoresistance and stemness in pancreatic cancer. Br J Cancer.
111:1572–1580. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Arocho A, Chen B, Ladanyi M and Pan Q:
Validation of the 2-DeltaDeltaCt calculation as an alternate method
of data analysis for quantitative PCR of BCR-ABL P210 transcripts.
Diagn Mol Pathol. 15:56–61. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saadoun S, Papadopoulos MC, Hara-Chikuma M
and Verkman AS: Impairment of angiogenesis and cell migration by
targeted aquaporin-1 gene disruption. Nature. 434:786–792. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pigati L, Yaddanapudi SC, Iyengar R, Kim
DJ, Hearn SA, Danforth D, Hastings ML and Duelli DM: Selective
release of microRNA species from normal and malignant mammary
epithelial cells. PloS one. 5:e135152010. View Article : Google Scholar : PubMed/NCBI
|