1. An overview of pre-eclampsia (PE)
PE, a pregnancy-associated multisystem disorder, is
defined by hypertension, proteinuria and other systemic
disturbances in the last trimester of pregnancy, during labor, or
shortly following delivery, affecting ~3–8% of pregnancies
worldwide (1). The International
Society for the Study of Hypertension in Pregnancy (2) classifies PE as: i) mild PE, with a
maternal systolic blood pressure of ≥140 mmHg and/or diastolic
blood pressure of ≥90 mmHg on two occasions separated by 6 h, and
significant proteinuria (≥300 mg protein in a 24-h urine specimen,
or ≥1+ by dipstick) after 20 weeks of gestation; or ii) severe PE,
where either severe hypertension (systolic blood pressure of ≥160
mmHg and/or diastolic blood pressure of ≥110 mmHg on at least two
occasions 6 h apart) plus mild proteinuria or mild hypertension
plus severe proteinuria (≥2 g/24 h, or ≥2+ by dipstick). Several
modifiable and non-modifiable factors were documented for
increasing the risk of PE. These include first pregnancy,
pre-existing diabetes mellitus and insulin resistance, a high body
mass index prior to pregnancy, advanced maternal age, renal disease
and hypertension (3).
Molecular mechanisms and pathogenesis of
PE
PE is termed 'the disease of theories' due to its
elusive origin, which is considered to be multifactorial. The
clinical manifestations of PE, i.e. the defining lines of the
disease, represent the last stage of PE, which begins in the early
stages of gestation (4). In
general, vascular homeostasis, along with placental and fetal
development, is disturbed in PE. The placenta is a key organ
involved in pathogenesis, as its removal attenuates the clinical
manifestation of PE. Genetic and epigenetic pathways were reported
to induce alterations in the placental transcriptome in PE
(5). Observational data have
supported the association of PE with endothelial dysfunction,
including vasoconstriction and end-organ ischemia (4). For example, circulating and placental
levels of soluble fms-like tyrosine kinase-1 (sFlt-1) and soluble
endoglin (sEng) are raised in women with PE compared with women
experiencing a normal pregnancy (5). sFlt-1 and sEng are anti-angiogenic
proteins, which antagonize proangiogenic factors, including
vascular endothelial growth factor (VEGF) and placental growth
factor, thereby resulting in inadequate placental vascular
development (5).
In addition, hypoxia is considered a key determinant
of the risk of PE, highlighted by a significant rise in protein
levels of hypoxia-inducible factor (HIF) in the placenta of women
with PE (6). The restriction of
placental perfusion in several animal species has resulted in
PE-like illness (7), and oxidative
stress is central to stimulating the release of cytokines,
anti-angiogenic factors and related linkers. Systemic inflammatory
responses and endothelial dysfunction in a number of organ systems,
including the vasculature, kidney, liver, and so on, was eventually
observed in women with PE (5,6).
This suggested that a search for a diagnostic, as well as a
therapeutic, pathway for PE requires a thorough assessment of these
multiple pathways.
2. MicroRNAs (miRNAs)
MiRNAs are regulatory RNAs, 21–23 nucleotides long,
which were identified initially in Caenorhabditis elegans in
1993 (8). They are involved in the
transcriptional and post-transcriptional regulation of gene
expression, and reportedly exert an important role in biological
pathways, including cell development, cell differentiation,
regulation of cell cycle, metabolism and apoptosis (9). The human genome encodes >1,000
miRNA species, and appears to target 60% of the genes of humans and
other mammals (10).
Biogenesis
miRNAs represent a conserved group of nucleotide
regulatory RNAs that are involved in multiple pathways (11). miRNAs contribute to the regulatory
network by the complementary binding of their 'seed sequences' to
targets in the 3′ untranslated region (3′ UTR) of mRNAs (Fig 1). Most miRNAs are involved in
regulating the translation of a large number of different mRNAs,
with each mRNA possessing multiple binding sites for single or
several different miRNAs (12).
The most up-to-date comprehensive miRNA database, miRBase, lists
2,578 human mature miRNAs and 1,872 precursors (12).
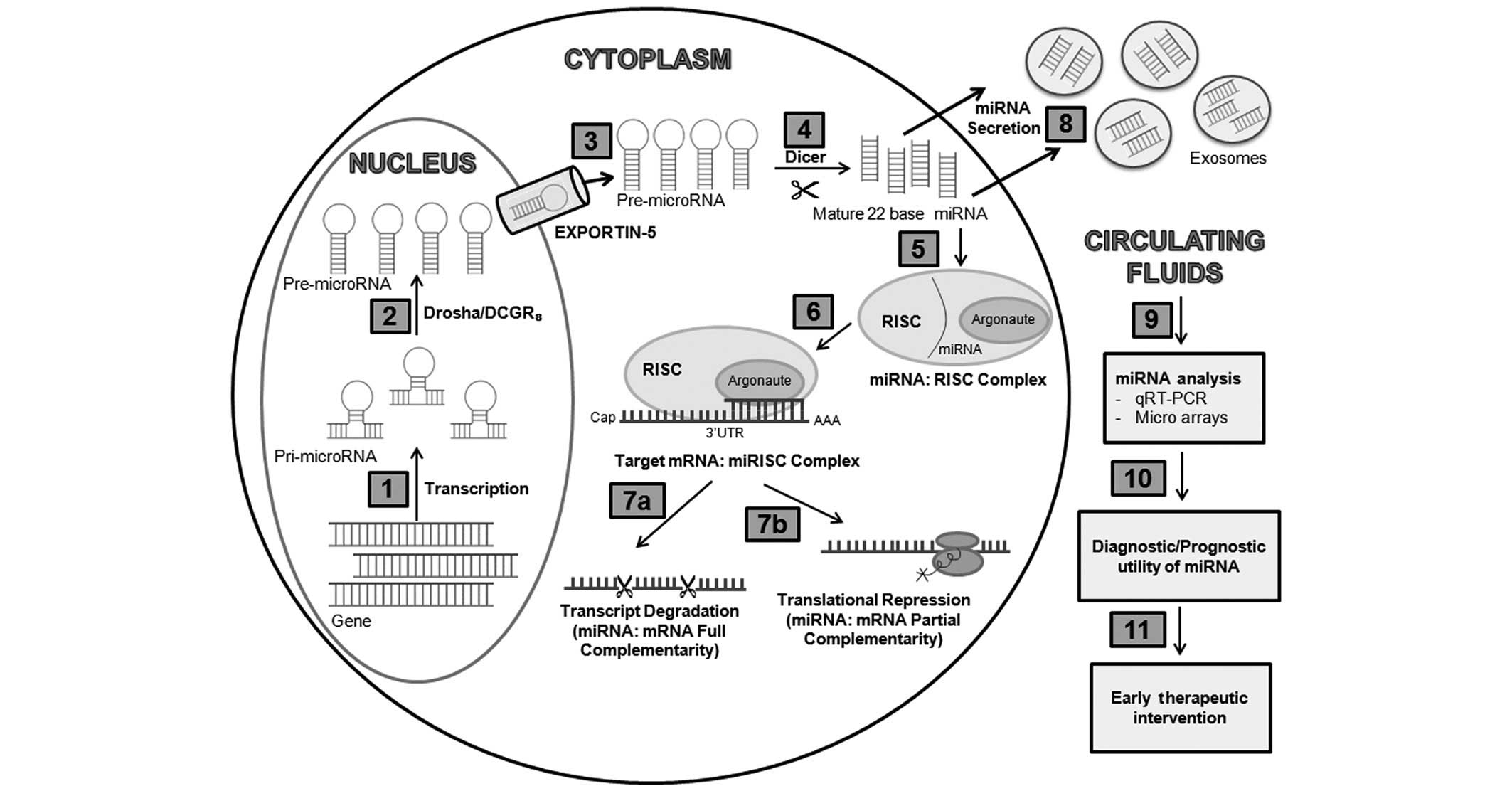 | Figure 1miRNA biogenesis and function. miRNAs
are transcribed by RNA polymerase II as Pri-miRNAs (1), which are cleaved by Drosha into
Pre-miRNAs almost 70 nucleotides in length (2), prior to being exported from the
nucleus into the cytoplasm via an exportin-5/Ran GTP complex
(3). Further processing by the
RNAse III, Dicer, generates mature miRNA-miRNA duplexes (4). One of the strands of mature miRNA is
incorporated into RISC (5), and
together they interact with the 3′UTR of the target mRNA (6), leading to either transcript
degradation (miRNA: mRNA full complementarity) (7a) or
translational repression (miRNA: mRNA partial complementarity)
(7b). Mature miRNA duplexes have the tendency to escape cells
enclosed in vesicles exosomes (8),
which move to the other cells or are present in the peripheral
fluids and can be easily detected by using advanced techniques
(9), increasing their scope in
diagnosis and allowing early therapeutic intervention (10,11).
miRNA, microRNA; qT-PCR, real-time quantitative polymerase chain
reaction; RISC, RNA-induced silencing complex; 3′UTR,
3′-untranslated. |
Role of miRNA in disease
Dysregulation of the expression of miRNAs is
associated with several diseases, including diabetes mellitus
(13), neurodegeneration,
rheumatoid arthritis, gastrointestinal diseases and cancer
(9). A burgeoning number of
studies have suggested a close link between the disrupted
expression of miRNAs and adverse pregnancy complications, including
early spontaneous miscarriage (14).
Biochemistry of miRNAs
Tissue- and disease-specific miRNA expression
profiles are more discriminatory and stable compared with total
mRNA profiles. The biological effectiveness of specific mRNAs
resides in their subsequent translation into specific proteins,
contrasting with miRNAs, which act as the key regulators of
multiple genes; therefore, miRNAs reflect more accurately the
altered physiology. Moreover, circulating miRNAs appear to be
resistant to endogenous ribonuclease activity, and are therefore
fairly stable (15). miRNAs have
been demonstrated to be ten times more stable compared with mRNA,
the half-life of which is ~10 h (16). The turnover trend of miRNAs, as
determined according to a mathematical model, yielded an average
miRNA half-life of 119 h, with certain miRNAs, for example,
miR-125b (half-life, 225 h), being more persistent compared with
others (16). This is likely to be
due to their intrinsic structural features, presented in
particulate form when associated with exosomes (Fig 1), or in the soluble form when
complexed with RNA-binding proteins, such as argonaute 2, or with
high-density lipoproteins (17).
Recent advances in polymerase chain reaction
(PCR)-based detection methods, such as singleplex real-time
quantitative (RT-q)PCR or high-throughput RT-qPCR platforms, has
ensured that the quantitative analysis of miRNAs from tissue
specimens and plasma/serum has become easier and more precise
(Fig. 1) (18). This highlights the ease of
exploiting miRNAs as biomarkers, for the development of practicable
detection methods for numerous diseases.
Despite the importance of miRNAs as regulators of
gene expression, studies which have implicated an altered miRNA
expression in obstetrics and gynecology have been limited in scope
and number, and the majority of published studies have focused on
an association with ovarian (19)
or uterine (20) cancers. At
present, the link between miRNAs and PE has been addressed by only
a few studies. The current review summarizes the state of knowledge
pertaining to the role of miRNAs in the pathophysiology of PE, and
the possible implications of this association, namely early
diagnosis, are discussed
3. Placenta-specific miRNAs
Concerning normal human placenta, the expression of
>600 miRNAs belonging to the clusters, chromosome 19 miRNA
cluster (C19MC), C14MC and miR-371-3, exhibiting gestational-age
dependent changes in their expression has been identified (21). A previous report has demonstrated
that the placental miRNome predominantly comprises miRNAs belonging
to C14MC in the first trimester, and miRNAs from C19MC in the third
trimester (22). In particular,
C14MC miRNAs are imprinted from the maternal chromosome and C19MC
miRNAs are imprinted from the paternal chromosome, both considered
to exert important roles in the regulation of cellular
differentiation and immunomodulation during pregnancy (22).
C14MC, the largest miRNA cluster comprising 52 miRNA
genes, is preserved without major structural changes and is
uniquely identified in placental mammals (23). This suggests an important role for
C14MC miRNAs in controlling neurogenesis, embryonic development,
transcriptional regulation and RNA metabolism (23).
The C19MC cluster, comprising 46 miRNA genes, is
implicated for its important role in human embryonic development,
similarly to genes such as insulin-like growth factor 2 (24). Notably, several of the miRNAs from
the C19MC cluster have been detected in the maternal circulation
throughout gestation, including miR-515-3p, miR-516-5p, miR-517a,
miR-517c, miR-518b, miR-520a*, miR-520 h, miR-526a and miR-526b
(25). Thus, maternal circulating
miRNAs may have the potential to be novel diagnostic tools for
pregnancy disorders.
The miR-371-3 cluster consists predominantly of
three miRNAs, hsa-miR-371-5p, hsa-miR-372 and hsa-miR-373-5p, and
is also located on chromosome 19. This cluster is predominantly
expressed in the placenta, and the levels of its miRNAs decrease
during development (10), and
therefore would appear to be essential for cell cycle maintenance
and regulation of proliferation and apoptosis. In addition, miRNAs
from the miR-17-92 cluster have also been observed to be expressed
in normotensive term placentas. These miRNAs are important for
angiogenesis, affecting the expression of numerous genes, namely
HIF1A, interleukin 8, tissue inhibitor of metalloproteinase 2,
matrix metallopeptidase 2, VEGFA, ephrin-B and ephrin receptor B4
(21).
The abundant expression of miRNAs (359 miRNAs) in
amniotic fluid has been documented, comprising predominantly those
involved in maintaining embryonic development and several
immunity-related processes, thereby protecting the fetus from
foreign pathogens and the maternal host immune response (26).
4. Differential expression of miRNAs in
PE
Recognition of the importance of miRNAs to the
development of PE is a relatively recent phenomenon, dating to
2007, when the first study on the possible association of altered
placental miRNA expression with PE was published (27). Subsequent studies have reported
detection of miRNA species, predominantly in placental tissues
(Table I) (28–32),
and more recently, in circulating fluids, such as serum/plasma
(Table II) (30,31,33–35).
These studies allowed the identification of miRNAs with a minimal
overlapping pattern. For example, certain miRNAs, including
miR-210, miR-155, miR-196, miR-195, miR-26 and miR-181a, were
upregulated (27,28,30,31,35–37),
whereas others, including miR-144 and miR-223, were downregulated
(30,33,37).
Contradictory data concerning the regulation of other miRNAs in PE
were also reported. Furthermore, the upregulation of miR-195 in
placentas from severe PE pregnancies (36), as well as downregulation of its
expression, were reported (31,37).
 | Table IDifferentially expressed miRNAs in
pre-eclamptic placentas. |
Table I
Differentially expressed miRNAs in
pre-eclamptic placentas.
| Study
population | Initial pool | Upregulated | Downregulated | Reference |
|---|
|
African-American | 157 | miR-210, miR-155,
miR-181b, miR-182, miR-200b, miR-154, miR-183 | - | (27) |
| Chinese | 677 | miR-16, miR-29b,
miR-195, miR-26b, miR-181, miR-335, miR-222, miR-210 | - | (36) |
| Caucasian
American | 180 | miR-181a, miR-584,
miR-30a-3p, miR-210, miR-152, miR-517, miR-518b, miR-519e, miR-638,
miR-296, miR-362 | miR-101, miR-10b,
miR-218, miR-590, miR-204, miR-32, miR-126, miR-18a, miR-19a,
miR-411, miR-377, miR-154, miR-625, miR-144, miR-195, miR-150,
miR-1, miR-18b, miR-363, miR-342 3p, miR-450, miR-223, miR-374 | (37) |
| Chinese | 7 | miR-155 | - | (32) |
| Caucasian
American | 611 | miR-210 | miR-328,
miR-miR-584, miR-139-5p, miR-500, miR-1247, miR-34c 5p, miR-1 | (28) |
| Caucasian
American | 820 | miR-493 miR-200c,
miR-296 | miR-15b, miR-181a,
miR-210, miR-483-5p, | (29) |
 | Table IIDifferentially expressed miRNAs in
pre-eclamptic circulating fluids. |
Table II
Differentially expressed miRNAs in
pre-eclamptic circulating fluids.
| Study
population | Source | Initial pool | Upregulated | Downregulated | Reference |
|---|
| Chinese | Plasma | 821 | miR-574 5p,
miR-26a, miR-151 3p, miR-130a, miR-181a, miR-130b, miR-30d,
miR-145, miR-103, miR-425, miR-221, miR-342 3p, miR-24,
miR-210 | miR-144,
miR-16 | (30) |
| Chinese | Plasma | - | miR-519d, miR-517b,
miR-19a, miR-10a, miR-517c, miR-18a, miR-210, miR-221, miR-101,
miR-26b, miR-521, miR-378, miR-519a, miR-520h, miR-125b,
miR-125a-5p, miR-114, miR-29a, miR-144*,
miR-15b*, miR-182, miR-29c, miR-30a, miR-518c, miR-27a,
miR-24, miR-519e, miR-130a, miR-515-3p, miR-299a-5p, miR-518b,
miR-23a, miR-23b, miR-34a, miR-424, miR-525-3p, miR-199a-5p,
miR-100, miR-29b, miR-99a, miR-21, miR-145, miR-512 5p,
miR-30b | miR-19a, miR-144,
miR-19b, miR-25, miR-451, miR-15b, miR-223, miR-320c, miR-185,
miR-107, Let-7f | (33) |
| Chinese | Plasma | 184 | miR-210 | miR-18a, miR-19b1,
miR-92a1 | (31) |
| Mixed
population | Sera | 63 | miR-143, miR-125b,
miR-192 | miR-126, miR-127,
miR-221, miR-942 | (34) |
| Caucasian | Sera | 754 | miR-1233, miR-650,
miR-520a, miR-215, miR-210, miR-25, miR-518b, miR-193a-3p, miR-32,
miR-204, miR-296-5p, miR-152 | miR-126, miR-335,
miR-44, miR-204, miR-668, miR-376a, miR-15b | (35) |
It is clear that miRNAs exert an essential role in
regulation, as alterations in their expression may result in the
dysregulation of several processes, including antiapoptotic
survival (27), innate/adaptive
immunity (27,28), cell cycle regulation, adhesion and
migration (28). This was
exemplified by the findings that overexpression of miR-182,
miR-182*, miR-155 and miR-210 was associated with
increased apoptosis in the placentas of patients with PE, an
abnormal immune response and angiogenesis (27). Mice with deficient miRNA exhibited
inappropriate angiogenesis, resulting in embryotoxicity (38). In addition, miRNAs may influence
signal transduction events (36),
vascular remodeling and angiogenesis (21), particularly in cancer metastasis
(39), calcium and lipid
metabolism (27) and organ/system
development (36).
Placental versus circulating miRNAs in
PE
Heterogeneity in the sample type, including the
placenta (Table I) (27,32),
serum (34,35) and plasma (Table II) (30,31),
yielded often contradictory findings. This was exemplified by a
previous study, which examined miRNA profiles in severe PE in
different placenta sites, and in maternal plasma obtained at 15–18
and 35–38 weeks of gestation (31). In placental specimens, a total of
seven miRNAs, comprising miR-210, miR-30a-3p, miR-518b, miR-524,
miR-17-3p, miR-151 and miR-193b were upregulated, whereas nine
miRNAs, comprising miR-195, miR-223, miR-218, miR-17, miR-18a,
miR-196b1, miR-92a1, miR-379 and miR-411, were downregulated
(31). However, circulating levels
of miR-18a, miR-92a1 and miR-92b1 were markedly lower, whereas that
of miR-210 was higher in patients with severe PE at 15–18
gestational weeks, and at term when the plasma was assayed
(31).
In addition, differential localization of miRNAs
throughout the placenta was reported, as miR-210, miR-518b,
miR-524, miR-151 and miR-519e-5p were detected in the basal plate
and miR-125, miR-92a1 and miR-379 were expressed in the chorionic
plate of placenta (31). This
suggests that differences in the sample source may result in
differential, and even opposite, results in miRNA profiling
(31). The aberrantly expressed
miRNAs in this study were linked with modulating trophoblast cell
invasion, namely transforming growth factor-2 signaling via the
repression of Smad2/Smad6/Smad7/Smad4/Smad5 expression, resulting
in abnormal placentation.
The placental tissues analyzed in the
above-mentioned studies were obtained post-delivery, thereby
questioning the exact status of miRNA expression throughout
pregnancy. In view of the shortcomings associated with placental
sampling, a shift to non-invasive sources of miRNA, including
circulating fluids, was suggested (31, 33–35).
Role of the severity of PE and onset
time
Differential miRNA expression appears to be affected
by PE severity, i.e. mild and severe. The differential expression
of 51 miRNA species were previously documented, of which 22 were
upregulated and five were downregulated in plasma from patients
with severe PE, compared with 33 upregulated and six downregulated
miRNAs in cases of mild PE (33).
Notably, miR-141 and miR-29a were markedly overexpressed in mild
PE, whereas miR-144 was underexpressed in mild and severe PE
(33). miR-141 was identified as a
placenta-specific miRNA (33),
miR-144 as a regulator of placental ischemia and hypoxia (40), and miR-29a as a tumor suppressor or
promoter (41) in different
tumors. In addition, the aberrant expression of miR-29a has
previously been observed in diabetes (13) and Alzheimer's disease (42).
The onset time of PE is also an important
contributor to miRNA activity. Previous studies reported the
dysregulation of 22 (43), 15
(30) and 51 (33) miRNAs in the serum/plasma of women
with PE pregnancies in the advanced stage of gestation (third
trimester). A more recently published study revealed modest
differences in the expression levels of miRNAs in subjects with PE
vs. normal pregnancies at the first trimester (34). Of the 754 miRNAs analyzed in pooled
sera, 63 were consistently detected in the sera from PE and control
subjects, of which 15 miRNAs were differentially expressed with a
small fold change (34). Open
array profiling confirmed the over-representation of miR-143,
miR-125b and miR-192, and the under-representation of miR-126,
miR-127, miR-221 and miR-942 in the serum.
Functionally, the miRNAs detected by Luque et
al (34) were multipurpose in
nature, are were implicated in angiogenesis (miR-125b, miR-143 and
miR-942), inflammation (miR-126, miR-127, miR-192 and miR-221)
(44), hypoxia/ischemia (miR-127)
and cell migration/remodeling (miR-125b, miR-143 and miR-127)
(30), rather than being more
pregnancy-specific. This suggested a lack of miRNA discriminatory
power during the early, preclinical phase of PE, which may be an
outcome of the own etiology of the disease, i.e. the appearance of
miRNAs in the circulation is a relatively late event in PE
development (34).
The negative correlation was noted between the
levels of miR-942 and maternal arterial pressure, and between the
levels of miR-143 and the uterine artery Doppler pulsatility index
(34). This was in agreement with
an earlier study (30), which
documented a role for miRNA-143 in the regulation of blood pressure
and vascular function, highlighting the utility of miRNAs as
prognostic markers.
Another study along similar lines in Caucasians
demonstrated the differential expression of miRNAs in serum samples
from an early gestation stage (12–14 weeks) of pregnant women who
later developed severe PE in the third trimester (35). A total of 19 mature miRNAs appeared
to be differentially expressed, including 12 upregulated (miR-1233,
miR-650, miR-520a, miR-215, miR-210, miR-25, miR-518b, miR-193a-3p,
miR-32, miR-204, miR-296-5p and miR-152) and seven downregulated
(miR-126, miR-335, miR-144, miR-204, miR-668, miR-376a and miR-15b)
miRNAs (35). RT-qPCR validated
four miRNAs (miR-1233, miR-520a, miR-210 and miR-144), revealing a
fold change of >3, with miR-1233 exhibiting a fold change of
>5 between a group of women who developed severe PE compared
with a group of women with normotensive pregnancies. Notably, the
differentially expressed miRNAs in that study were implicated in
different types of cancer, for example, miR-1233 has already been
detected in renal carcinoma (45),
miR-650 has been implicated in hepatocellular carcinoma (46), and miR-215 and miR-204 have been
implicated in metastatic renal cell carcinoma. This clearly
suggested a pro-malignancy-like signature of circulating miRNAs in
women who develop severe PE at a later stage.
Evidence of placental leakage of miRNAs into the
circulation during PE prompted speculation of whether miRNAs could
be reliable biomarkers in the timely diagnosis of PE (47). Insofar as the placenta and tumors
constitute the primary source of RNA-containing exosomes, the
placental miRNAs originating from villous trophoblasts present as
placenta-derived exosomes may be detected in the plasma (48). Previously, a research group
profiled 377 human miRNAs in placental tissue and the plasma, and
315 of them were observed to be expressed in the placenta, with 286
detectable in the plasma (11).
Effect of ethnic background and technical
aspects
Differential expression of miRNAs with a minimum
overlap within the studies reported above has been observed,
thereby expanding the spectrum of miRNAs implicated in the
pathogenesis of PE. Such discrepancies can be attributed to the
heterogeneity in the ethnic background of study subjects (Tables I and II), as the majority of the reported
studies were performed on Chinese populations (30–33,36),
whereas the others involved African-American (27), Caucasian American (29), other Caucasian (35) and mixed population (34) subjects.
Variations in the techniques used in miRNA profiling
are also considered to be a factor contributing towards
inconsistencies in the PE-specific expression of miRNAs. Where
certain studies have been performed using RT-qPCR (34), others have utilized high-throughput
techniques, such as microarrays (30), and more sophisticated techniques,
such as next-generation sequencing (43). A major variability between studies
was also observed in statistical methods used for detecting
differentially expressed genes. In certain of the studies, an
inadequacy in the statistical procedures used was observed, whereas
others were not using stringent enough statistical criteria.
Whereas certain studies reported only miRNAs with a marked
differential expression of a >2-fold change (27,36),
others exercised a greater leniency, including in their analyses
miRNAs with a marked expression of only 1.5-fold changes (28,31).
Other factors which may contribute include the difference in the
study sizes, which have been relatively small in the majority of
cases, or different definitions of PE used for patient inclusion
criteria.
5. Prognostic value of miRNA in PE
It is interesting to observe that in PE, thus far,
almost 120 miRNAs have been reported to be dysregulated, and none
of the two independent studies discussed above have revealed a
complete overlapping of the miRNA panel. It is only miR-210 that
has been observed in the majority of the studies taken into
consideration in the present review, which indicates its importance
in normal placentation. miRNA-210 constitutes one of the
hypoxia-associated miRNAs (ʻhypoxamiRsʼ) upregulated by hypoxia
(6), and its consistently aberrant
expression in PE makes it a potential serum biomarker for PE
(31).
miR-210 has been associated with events integral to
the pathogenesis of PE, including the endothelial cell response to
hypoxia, formation of capillary-like structures, VEGF-driven cell
migration, cell differentiation and survival (28). Upregulation of miR-210 is
correlated with the inhibition of migration and the invasive
capability of trophoblasts, and is also linked to induction of the
activity of several intracellular transcription factors, including
nuclear factor-κB p50 (49). More
recently, a study on placentas from healthy pregnant individuals
and patients with PE has demonstrated that the aberrant expression
of miR-210 may contribute to the occurrence of PE by regulating
trophoblast cell invasion via targeting potassium channel
modulatory factor-1 mediated signaling in the human placenta
(50). Along with miR-210, the
aberrant expression of two more miRNAs has been observed in the
majority of the studies on PE, and has been suggested to have
prognostic value, namely miR-518c (51) and miR-155 (32).
In silico and in vivo studies have
demonstrated that the upregulated expression of miR-518c, along
with that of miR-210, target and dysregulate the post-transcription
of 17-β hydroxysteroid dehydrogenase 1 (HSD17B1), a placental
steriodogenetic enzyme (51). High
plasma levels of HSD17B1 observed in women with PE are
advantageous, as the enzyme is expected to be derived almost
exclusively from the placenta. In vitro studies on BeWo and
JEG-3 cells demonstrated upregulation of miR-518c and miR-210
compared with decreased mRNA levels of HSD17B1 following
exposure to hypoxia, confirming the miRNA-mediated dysregulation of
HSD17B1 expression (51).
Similarly, overexpression of miR-155 was reported to
downregulate cysteine-rich angiogenic inducer 61 (CYR61), a stress
gene expressed by vascular cells and trophoblasts and implicated in
cell migration, proliferation, differentiation and adhesion in the
placenta of women with PE (32).
CYR61 is an important early angiogenic regulating factor during
pregnancy, which induces the expression of VEGF, and its 3′-UTR was
validated as the target of miR-155 (32). miR-155 appears to control the
stability of CYR61 mRNA, and consequently its level of expression
thus directly links local ischemia and oxidative stress.
6. An hypothesis bridging miRNAs with
PE
At present, a large number of studies have
documented compelling evidence of aberrantly expressed miRNAs as
characteristic phenomena of established PE. A general hypothesis
may be drawn that the dysregulated expression of miRNAs observed in
PE has a reciprocal effect on their potential target genes, thus
curbing or elevating their normal function. Therefore, all the
events that occur normally in placental development, as a result of
the aberrant expression of miRNAs, now occur abnormally, resulting
in impaired cytotrophoblast differentiation and apoptosis,
incomplete spiral artery invasion and decreased blood flow to and
from the placenta. Trophoblast necrosis releases cell fragments
into the maternal bloodstream, which ultimately triggers a systemic
immunological response and oxidative stress in the placenta
(52).
Furthermore, certain of the miRNAs among the
population of dysregulated ones recorded in PE are not only
specific for the placenta, but are also part of a number of other
organ systems, including the liver, brain, immune system and, most
importantly, the kidney. Therefore, any dysregulation of these
miRNAs may affect the normal function of target genes in the
placenta, as well as in other systems. This explains why PE is
considered to be a multisystem disorder. The stepwise mechanism of
action that links miRNAs with PE has yet to be fully elucidated,
and thus it is necessary to identify the mechanisms of action for
all the miRNAs that account for the occurrence of PE, in order to
have a clear and detailed understanding of the pathogenesis of the
disease.
7. Conclusion
The exact pathogenesis of PE remains incompletely
understood. The identification of miRNAs that are reproducibly
detected in tissues and circulating fluids suggest their utility as
biomarkers. The majority of the differential expression profiles of
miRNAs identified in the placenta in cases of PE correlates with
those from maternal plasma/sera, demonstrating their prognostic
value in PE. However, larger and prospective cohort studies on
varied populations are required for the thorough assessment of the
contribution of miRNAs to PE. Functional studies, including in
vitro and in vivo approaches, are warranted to
comprehensively elucidate the role of the PE-associated miRNAs in
the onset and/or progression of PE. Additionally, it is necessary
to delineate the pathways by means of which certain dysregulated
miRNAs are able to detrimentally affect a given biological process,
and to investigate such pathways with a view to the opportunities
they afford for therapeutic intervention in PE.
References
|
1
|
Duley L: The global impact of
pre-eclampsia and eclampsia. Semin Perinatol. 33:130–137. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
ACOG Committee on Practice
Bulletin-Obstetrics: ACOG practice bulletin. Diagnosis and
management of preeclampsia and eclampsia Number 33, January 2002.
Obstet Gynecol. 99:159–167. 2002.
|
|
3
|
Duckitt K and Harrington D: Risk factors
for pre-eclampsia at antenatal booking: Systematic review of
controlled studies. BMJ. 330:565–570. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Redman CW and Sargent IL: Latest advances
in understanding preeclampsia. Science. 308:1592–1594. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Steegers EA, von Dadelszen P, Duvekot JJ
and Pijnenborg R: Pre-eclampsia. Lancet. 376:631–644. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chan YC, Banerjee J, Choi SY and Sen CK:
miR-210: The master hypoxamir. Microcirculation. 19:215–223. 2012.
View Article : Google Scholar :
|
|
7
|
Khalil RA and Granger JP: Vascular
mechanisms of increased arterial pressure in preeclampsia: Lessons
from animal models. Am J Physiol Regul Integr Comp Physiol.
283:R29–R45. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nat Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Devor EC, Santillan DA and Santillan MK:
Preeclampsia and MicroRNAs. Proceed Obstet Gynecol. 3:1–10.
2013.
|
|
12
|
Pillar N, Yoffe L, Hod M and Shorman N:
The possible involvement of microRNAs in preeclampsia and
gestational diabetes mellitus. Best Pract Res Clin Obstet Gynaecol.
29:176–182. 2015. View Article : Google Scholar
|
|
13
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P
and Stoffel M: A Pancreatic islet-specific microRNA regulates
insulin secretion. Nature. 432:226–230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ventura W, Koide K, Hori K, Yotsumoto J,
Sekizawa A, Saito H and Okai T: Placental expression of microRNA-17
and -19b is down-regulated in early pregnancy loss. Eur J Obstet
Gynecol Reprod Biol. 169:28–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Pogosova-Agadjanyan EL, Peterson
A, Noteboom J, O'Briant KC, et al: Circulating microRNAs as stable
blood-based markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gantier MP, McCoy CE, Rusinova I, Saulep
D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F and
Williams BR: Analysis of microRNA turnover in mammalian cells
following Dicer1 ablation. Nucleic Acids Res. 39:5692–5703. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK,
Pritchard CC, Gibson DF, Mitchell PS, Bennett CF,
Pogosova-Agadjanyan EL, Stirewalt DL, et al: Argonaute2 complexes
carry a population of circulating microRNAs independent of vesicles
in human plasma. Proc Natl Acad Sci USA. 108:5003–5008. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kroh EM, Parkin RK, Mitchell PS and Tewari
M: Analysis of circulating microRNA biomarkers in plasma and serum
using quantitative reverse transcription-PCR (qRT-PCR). Methods.
50:298–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot
CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, et al: Integrated
analyses identify a master microRNA regulatory network for the
mesenchymal subtype in serous ovarian cancer. Cancer Cell.
23:186–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Devor EJ, Mik JN, Ramachandran S,
Goodheart MJ and Leslie KK: Global dysregulation of the chromosome
14q32 imprinted region in uterine carcinosarcoma. Exp Ther Med.
3:677–682. 2012.PubMed/NCBI
|
|
21
|
Wang W, Feng L, Zhang H, Hachy S, Satohisa
S, Laurent LC, Parast M, Zheng J and Chen DB: Preeclampsia
up-regulates angiogenesis-associated microRNA (ie, miR-17,-20a and
20b) that target ephrin-B2 and EPHB4 in human placenta. J Clin
Endocrinol Metab. 97:1051–1059. 2012. View Article : Google Scholar
|
|
22
|
Morales-Prieto DM, Ospina-Prieto S,
Chaiwangyen W, Schoenleben M and Markert UR: Pregnancy-associated
miRNA-clusters. J Reprod Immunol. 97:51–61. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Glazov EA, McWilliam S, Barris WC and
Dalrymple BP: Origin, evolution and biological role of miRNA
cluster in DLK-DIO3 genomic region in placental mammals. Mol Biol
Evol. 25:939–948. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Noguer-Dance M, Abu-Amero S, Al-Khtib M,
Lefèvre A, Coullin P, Moore GE and Cavaillé J: The primate-specific
microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol
Genet. 19:3566–3582. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kotlabova K, Doucha J and Hromadnikova I:
Placental-specific microRNA in maternal circulation-identification
of appropriate pregnancy-associated microRNAs with diagnostic
potential. J Reprod Immunol. 89:185–191. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pineles BL, Romero R, Montenegro D, Tarca
AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P,
et al: Distinct subsets of microRNAs are expressed differentially
in the human placentas of patients with preeclampsia. Am J Obstet
Gynecol. 196:261–266. 2007.PubMed/NCBI
|
|
28
|
Enquobahrie DA, Abetew DF, Sorensen TK,
Willoughby D, Chidambaram K and Williams MA: Placental microRNA
expression in pregnancies complicated by preeclampsia. Am J Obstet
Gynecol. 204:12–21. 2011.
|
|
29
|
Mayor-Lynn K, Toloubeydokhti T, Cruz AC
and Chegini N: Expression profile of microRNAs and mRNAs in human
placentas from pregnancies complicated by preeclampsia and preterm
labor. Reprod Sci. 18:46–56. 2011. View Article : Google Scholar
|
|
30
|
Wu L, Zhou H, Lin H, Qi J, Zhu C, Gao Z
and Wang H: Circulating microRNAs are elevated in plasma from
severe preeclamptic pregnancies. Reproduction. 143:389–397. 2012.
View Article : Google Scholar
|
|
31
|
Xu P, Zhao Y, Liu M, Wang Y, Wang H, Li
YX, Zhu X, Yao Y, Wang H, Qiao J, et al: Variations of microRNAs in
human placentas and plasma from preeclamptic pregnancy.
Hypertension. 63:1276–1284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Y, Diao Z, Su L, Sun H, Li R, Cui H
and Hu Y: MicroRNA-155 contributes to preeclampsia by
down-regulating CYR61. Am J Obstet Gynecol. 202:466–472.
2010.PubMed/NCBI
|
|
33
|
Li H, Ge Q, Guo L and Lu Z: Maternal
Plasma miRNAs expression in Preeclamptic Pregnancies. Biomed Res
Int. 2013:9702652013.PubMed/NCBI
|
|
34
|
Luque A, Farwati A, Crovetto F, Crispi F,
Figueras F, Gratacos E and Aran JM: Usefulness of circulating
microRNAs for the prediction of early preeclampsia at
first-trimester of pregnancy. Sci Rep. 4:48822014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ura B, Feriotto G, Monasta L, Bilel S,
Zweyer M and Celeghini C: Potential role of circulating microRNAs
as early markers of preeclampsia. Taiwan J Obstet Gynecol.
53:232–234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu Y, Li P, Hao S, Liu L, Zhao J and Hou
Y: Differential expression of microRNAs in the placentae of Chinese
patients with severe pre-eclampsia. Clin Chem Lab Med. 47:923–929.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu XM, Han T, Sargent IL, Yin GW and Yao
YQ: Differential expression profile of microRNAs in human placentas
from preeclamptic pregnancies vs normal pregnancies. Am J Obstet
Gynecol. 200:661–667. 2009.PubMed/NCBI
|
|
38
|
Yang WJ, Yang DD, Na S, Sandusky GE, Zhang
Q and Zhao G: Dicer is required for embryonic angiogenesis during
mouse development. J Biol Chem. 280:9330–9335. 2005. View Article : Google Scholar
|
|
39
|
Su X, Chakravarti D, Cho MS, Liu L, Gi YJ,
Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, et al:
Tap63 suppresses metastasis through coordinate regulation of Dicer
and miRNAs. Nature. 467:986–990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vitoratos N, Hassiakos D and Lavazzo C:
Molecular mechanisms of preeclampsia. J Pregnancy.
2012:2983432012.PubMed/NCBI
|
|
41
|
Muniyappa MK, Dowling P, Henry M, Meleady
P, Doolan P, Gammell P, Clynes M and Barron N: MiRNA-29a regulates
the expression of numerous proteins and reduces the invasiveness
and proliferation of human carcinoma cell lines. Eur J Cancer.
45:3104–3118. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shioya M, Obayashi S, Tabunoki H, Arima K,
Saito Y, Ishida T and Satoh J: Aberrant microRNA expression in the
brains of neurodegenerative diseases: MiR-29a decreased in
Alzheimer disease brains targets neurone navigator 3. Neuropathol
Appl Neurobiol. 36:320–330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang Q, Lu J, Wang S, Li H, Ge Q and Lu Z:
Application of next-generation sequencing technology to profile the
circulating microRNAs in the serum of preeclampsia versus normal
pregnant women. Clin Chim Acta. 412:2167–2173. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xie T, Liang J, Liu N, Wang Q, Li Y, Noble
PW and Jiang D: MicroRNA-127 inhibits lung inflammation by
targeting IgG Fcγ receptor I. J Immunol. 188:2437–2444. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wulfken LM, Moritz R, Ohlmann C,
Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brünagel
G, von Ruecker A, Müller SC and Ellinger J: MicroRNAs in renal cell
carcinoma: Diagnostic implications of serum miR-1233 levels. PLoS
One. 6:e257872011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zeng ZL, Li FJ, Gao F, Sun DS and Yao L:
Upregulation of miR-650 is correlated with down-regulation of ING4
and progression of hepatocellular carcinoma. J Surg Oncol.
107:105–110. 2013. View Article : Google Scholar
|
|
47
|
Chim SS, Shing TK, Hung EC, Leung TY, Lau
TK, Chiu RW and Lo YM: Detection and characterization of placental
microRNAs in maternal plasma. Clin Chem. 54:482–490. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mincheva-Nilsson L and Baranov V: The role
of placental exosomes in reproduction. Am J Reprod Immunol.
63:520–533. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang Y, Fei M, Xue G, Zhou Q, Jia Y, Li
L, Xin H and Sun S: Elevated levels of hypoxia-inducible
microRNA-210 in pre-eclampsia: New insights into molecular
mechanisms for the disease. J Cell Mol Med. 16:249–259. 2012.
View Article : Google Scholar
|
|
50
|
Luo R, Shao X, Xu P, Liu Y, Wang Y, Zhao
Y, Liu M, Ji L, Li YX, Chang C, et al: MicroRNA-210 contributes to
preeclampsia by downregulating potassium channel modulatory factor
1. Hypertension. 64:839–845. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ishibashi O, Ohkuchi A, Ali MM, Kurashina
R, Luo S, Ishikawa T, Takizawa T, Hirashima C, Takahashi K, Migita
M, et al: Hydroxysteroid (17-β) dehydrogenase 1 is dysregulated by
miR-210 and miR-518c that are aberrantly expressed in preeclamptic
placentas: A novel marker for predicting preeclampsia.
Hypertension. 59:265–273. 2012. View Article : Google Scholar
|
|
52
|
Duley L, Henderson-Smart DJ, Meher S and
King JF: Antiplatelet agents for preventing pre-eclampsia and its
complications. Cochrane Database Syst Rev. CD0046592007.PubMed/NCBI
|















