Introduction
The mammalian genome contains 23 fibroblast growth
factors (FGFs) (1), which have
essential roles in metabolism and development. FGFs have been
identified to be involved in processes of embryogenesis, including
gastrulation, somitogenesis, body plan formation and organogenesis,
as well as in skin wound healing (2–7).
Initially, 1, a member of the FGF family, was reported to be
preferentially expressed in the liver (8). However, recent studies have
identified that FGF21 is inducible by starvation or certain drugs,
and have reported on its diverse functions in glucose homeostasis
as well as hepato- and cardioprotection (9–11).
FGF19, FGF21 and FGF23 belong to the FGF19 sub-family. Among them,
FGF21 primarily activates FGF receptor (FGFR)1c, for which
co-receptor β-klotho is required (12,13).
Previous efforts to produce recombinant human FGF21
(rhFGF21) using an Escherichia (E.) coli
system resulted in low expression of soluble protein, indicating
that the majority of recombinant protein localized in inclusion
bodies. Although the small ubiquitin-like modifier (SUMO) fusion
system has been shown to facilitate the soluble expression and
enhance the production of bioactive rhFGF21 (14), additional steps are required for
the removal of tags from the protein expressed in vitro. At
present, Pichia (P.) pastoris (yeast) is among
the most successful eukaryotic protein expression systems. Compared
with mammalian cells, culture of P. pastoris requires simple
and cheap growth media and conditions, and P. pastoris is
known to secrete proteins via signaling peptide-mediated secretion
pathways. These features render the P. pastoris system
suitable and efficient for recombinant protein expression (15).
In the present study rhFGF21 was expressed in P.
pastoris cells and isolated from the culture media.
Furthermore, examination of the expression of rhFGF21 in a variety
of tissue types revealed elevated expression of FGF21 in skin
compared with other tissue types, including that in the liver.
Further experiments showed that FGF21 was induced by wounding and
that exogenous treatment with FGF21 promoted cell migration, a key
step of the wound healing process. The present study provided a
novel method to express FGF21, which was not only more efficient
than previous systems, but also provided a basis for FGF production
in general. In addition, FGF21 accelerated fibroblast-cell
migration, which suggested its applicability in the treatment of
skin wounds.
Materials and methods
Creation of skin wounds on mice
Twelve male C57/BL6J mice, aged three months old and
weighing 28–35 g, were obtained from the Laboratory Animal Centre
of Wenzhou Medical University (Wenzhou, China), were divided into
two equal groups. All mice were anesthetized via intraperitoneal
injection of 3% sodium pentobarbital (45 mg/kg) and their dorsal
areas were completely depilated using Na2S (8.0%; w/v)
(both Sigma-Aldrich, St. Louis, MO, USA). Heart, liver, kidney and
skin tissues were harvested from one group to analyze FGF21 tissue
distribution; whereas skin wounds (length, 1 cm; depth, 0.3 cm)
were created on the backs of the mice in the remaining group using
a sharp laser blade (Swann-Morton, Sheffield, UK) to perforate the
hypodermis. At 0 and 3 h following wound creation, 1 cm2
of the dorsal area surrounding the wound was collected in order to
analyze the expression patterns of FGF21 and β-koltho. No mice were
sacrificed before wounding. Following experimentation, mice were
anesthetized using sodium pentobarbital and sacrificed via cervical
dislocation.
Total RNA extraction, complementary DNA
synthesis and polymerase chain reaction (PCR)
All frozen liver, heart, kidney and skin tissues
were powdered using a mortar and pestle in liquid nitrogen and were
homogenized for 30–45 sec in 1 ml TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Waltham, MA, USA) at increasing speed to
the maximum setting on a IKA T-10 Basic Ultra Turrax homogenizer
(IKA Werke GmbH & Co. KG, Staufen, Germany). Following total
RNA extraction with TRIzol reagent, RNA was quantified using a
Nanodrop 2000 photometer (Thermo Fisher Scientific, Inc.). A total
of 2 µg total RNA was reverse-transcribed using a GoScript
Reverse Transcription kit (Reverse Transcription System; Promega
Corp., Madison, WI, USA) following the manufacturer's instructions.
PCR was performed using a PCR Mastermix kit (Takara Biotechnology
Co., Ltd., Dalian, China) on a T100 thermal cycler (Bio-Rad
Laboratories, Inc., Hercules, CA, USA), as follows: 95°C for 5 min,
followed by 35 cycles of 94°C for 30 sec and 58°C for 30 sec, 72°C
for 30 sec, elongation at 72°C for 5 min and holding at 10°C. Gene
expression levels were quantified as described previously (16). mRNA levels were normalized against
those of GAPDH (cat. no. ab9482; 1:2500 dilution; Abcam, Cambridge,
UK) using Image J software (National Institutes of Health,
Bethesda, MD, USA) and the 2−∆∆Cq method (17). Gene-specific primer sequences were
synthesized by Sangon Biotech Co., Ltd., (Shanghai, China), as
follows: FGF21 forward, 5′-GATGACGACCAAGACACTG-3′ and reverse,
5′-CGGCCCTGTAAAGGCTCT-3′; β-klotho forward,
5′-ACGACCCGACGAGGGCTGTT-3′ and reverse,
5′-GGAGGAGACCGTAAACTCGGGCTTA-3′ and GAPDH forward,
5′-GCACAGTCAAGGCCGAGAAT-3′ and reverse, 5′-GCCTTCTCCATGGTGGTGAA-3′.
For the examination of PCR products size, a DL 2000 DNA ladder
(Takara Biotechnology Co., Ltd.) was used in the present study.
Construction of recombinant expression
plasmids
Based on the sequences of the mature human
polypeptide of FGF21, which were obtained from NCBI, a FGF21 coding
sequence was designed by replacing the non-preferred codon of P.
pastoris with the corresponding preferred one (Table I) using Vector NTI Advance 10.0
software (Thermo Fisher Scientific, Inc.). Primers were synthesized
by Sangon Biotech Co., Ltd., which also confirmed the correct
insertion and reading frame of FGF21 by DNA sequencing. The
sequences for the synthesis of the optimized-codon human FGF21 gene
are stated in Table II. Pyrobest
DNA polymerase (0.05 U/µl; Takara Biotechnology Co., Ltd.)
was used for polymerase chain reaction (PCR) and codon-optimized
FGF21 open reading frame sequences were obtained using seven pairs
of 55–59-nt primers with seven rounds of PCR. The first round of
PCR was performed using the P1/RP1 primer pair and P2/RP2 was used
for the second round, with the previous round of PCR product used
as template, and so forth. PCR thermal cycling was completed as
follows: 94°C for 4 min, followed by 32 cycles for 94°C for 30 sec,
56°C for 30 sec, 72°C at 30 sec, and extension ay 72°C for 5 min.
An XhoI restriction site was introduced during primer
synthesis to allow for in-frame cloning into the a-factor secretion
signal sequences containing expression vector pPIC9K (Invitrogen)
in order to express the native N-terminus of FGF21. Furthermore, a
nucleotide sequence encoding the KEX2 cleavage site was placed
ahead of FGF21. At the C-terminus, a stop codon was introduced
along with an EcoRI site. Synthesized FGF21 was cloned into
XhoI and EcoRI sites (0.3 U/µl) in the pPIC9K
vector.
 | Table IUnpreferred and preferred codons
based on the codon bias of Pichia pastoris. |
Table I
Unpreferred and preferred codons
based on the codon bias of Pichia pastoris.
| Amino acid | Leucine | Proline | Glutamic acid | Glutamine | Serine | Alanine | Glycine | Leucine | Threonine |
|---|
| Unpreferred | CTG | CCG | GAG | CAG | AGC | GCC | GGC | CTG | ACG |
| Preferred | TTA | CCA | GAA | CAA | TCA | GCA | GGT | TTG | ACT |
 | Table IIPrimer sequences. |
Table II
Primer sequences.
| Primer | Sequence
(5′-3′) |
|---|
| P1 |
TGCCAACGCCCAGACGGGGCTCTCTATGGCAGTCTTCACTTCGATCCGGAAGCCTGTTC |
| P2 |
GTGTGATTCAGATACTTGGCGTAAAAACCTCCCGTTTCTTATGCCAACGCCCAGACGGG |
| P3 |
TCACCCGAATCTCTCTTGCAACTAAAAGCCTTGAAGCCTGGTGTGATTCAGATACTTG |
| P4 |
GATTCGAGAAGATGGAACTGTTGGTGGAGCCGCAGACCAGTCACCCGAATCTCTCTTGC |
| P5 |
CTGTATACAGATGACGCACAGCAAACGGAGGCACATCTCGAGATTCGAGAAGATGGAA |
| P6 |
CCCACTACTTCAATTTGGGGGGCAGGTGAGGCAACGATACCTGTATACAGATGACGCAC |
| P7 |
CCGCTCGAGAAAAGACATCCCATACCTGATAGCTCCCCACTACTTCAATTTGG |
| RP1 |
ATACGTTGTATCCATCTTCCAAGAGAAGCTCACGAAATGAACAGGCTTCCGGATCG |
| RP2 |
GCCAGGCAGGTGCAGTGGTAAACCATGCGCCTCACTCTGATATACGTTGTATCCATCTT |
| RP3 |
GCAGGACCCCTAGGTGCTGGGTCCCGGTGTGGGCTCTTATTGCCAGGCAGGTGCAGTG |
| RP4 |
CGGCAAAGCGGGAGGTAGGCCCGGTAATGGTAAAAATCTCGCAGGACCCCTAGGTGCTG |
| RP5 |
TCCGACGTCGGGCGGCTGTGGAGCTAGGATTCCAGGGGGTTCCGGCAAAGCGGGAGGTA |
| RP6 |
TGAGAGGGCCCAACCATCGACAGCGGATCACTAGATCCGACGTCGGGCGGCTGT |
| RP7 |
GGAATTCCTTAGCTCGCGTATGACGGCGATCTACCTTGAGAGGGCCCAACCATCG |
Yeast transfection, protein expression
and purification
The plasmid pPIC9K-FGF21 DNA was linearized by 0.3
U/µl SalI and then electroporated into the P.
pastoris strain SMD1168 (Invitrogen) at 1.5 KV, 25 µF
and 200 Ω using a Gene Pulser apparatus (Bio-Rad Laboratories,
Inc.). After electroporation, 1 ml of ice-cold 1 M sorbitol was
immediately added to the cells. The cells were incubated for 2 h at
30°C and transfected cells were selected on minimal dextrose (MD)
plates [1.34% yeast nitrogen base medium (YNB), 4×10−5%
biotin, 1.5% agar (all Difco; BD Biosciences, Franklin Lakes, NJ,
USA) and 2% dextrose (Sigma-Aldrich)] at 30°C for 2–3 days. The
methyl utilization plus (Mut+) and methyl utilization
slow (Muts) phenotypes of the transfected cells were
screened by replica-plating them onto minimal medium (MM) plates
(1.34% YNB, 4×10−5% biotin, 0.5% methanol
(Sigma-Aldrich) as the primary carbon source and 1.5% agar) and MD
plates to determine the methanol-utilizing phenotypes. Verification
by PCR was performed using alcohol oxidase (AOX)1 universal primers
(Sangon Biotech Co., Ltd.).
The Mut+ and Muts strains were
subjected to shake-flask cultivation. Six positive colonies were
selected and cultured in 100 ml BMGYH [2% peptone, 1% yeast
extract, 1.34% YNB, 100 mM potassium phosphate (pH 6.0), 1%
glycerol, 0.004% histidine and 1.61 µM biotin] in a 500-ml
shaking flask until OD600=5–6 at 30°C. Following
centrifugation at 1,000 × g for 5 min, the pellet was re-suspended
to achieve an OD600 of 1.0 in 300 ml BMMYH (BMGYH with
1% glycerol replaced with 0.8% methanol) to induce expression.
Subsequently, cells were cultured for 72 h with addition of 0.8%
methanol every 24 h. Proteins were separated by 15% sodium dodecyl
sulfate polyacrylamide gel electrophoresis (SDS-PAGE; Beyotime
Institute of Biotechnology, Shanghai, China) and western blot
analysis was performed using mouse anti-FGF21 monoclonal antibody
(1:1,000; MAB25371; R&D Systems, Inc., Minneapolis, MN, USA)
(18).
Following centrifugation of the yeast cells, the
supernatant (300 ml) was subjected to sequential precipitation by
slow addition of 142.8 g ammonium sulfate (Sigma-Aldrich) to a
final concentration of 70% saturation over 2 h. Following agitation
overnight at 4°C, the suspension was centrifuged at 12,000 × g for
20 min to harvest the protein precipitate, 100 ml of which was
re-suspended in 20 mM phosphate buffer (PB; pH 8). The solution was
dialyzed in 11 PB (20mM) overnight at 4°C using a dialysis bag
(pore size, 3kD; Beijing Dingguo Changsheng Biotechnology Co.,
Ltd., Beijing, China) with three changes of PB. Subsequently, the
solution was loaded, at a flow rate of 5 ml/min, onto a 2.5×15 cm
diethylaminoethanol (DEAE)-Sepharose column (bed volume, 75 ml; GE
Healthcare Life Sciences, Chalfont, UK), which had been
equilibrated with five bed volumes of starting buffer containing
0.05 M NaCl and 20 mM PB (pH 8.0). Following washing with starting
buffer, the bound proteins were eluted with a PB buffer gradient
supplemented with 0.15, 0.3, 0.5 and 1.0 M NaCl. All fractions were
collected and examined using 15% SDS-PAGE, and the eluted fractions
at 0.3 M NaCl containing FGF21 were further purified by a 2×80 cm
Sephadex G-50 column (bed volume, 250 ml; GE Healthcare Life
Sciences). The fraction containing FGF21 was concentrated to 5 ml
with polyethylene 2000. A total of 100 µl of the
concentrated filtrate was subjected to semi-preparative
reverse-phase high-performance liquid chromatography (HPLC) using
an Agilent Technologies 1200 series equipped with an Agilent 1260
Infinite Diode Array Detector (G4212B; ID, 2.1×100 mm; particle
size, 2.5 µm; Agilent Technologies, Inc., NC, USA) on a C18
column pre-equilibrated with 0.1% trifluoroacetic acid (Merck
Millipore, Darmstadt, Germany). Bound protein was eluted using a
linear gradient of acetonitrile (10–90%; Merck Millipore) in 0.1%
TFA at a flow rate of 1.35 ml/min and monitored at 280 nm using a
diode-array. The purity and integrity of FGF21 purified by
reverse-phase HPLC was analyzed by determining the percentage of
the major peak in the total area and electrospray ionization mass
spectrometry (Bruker Reflex III MALDI-TOF MS, Bremen, Germany) with
nitrogen at 337 nm (19). The
fraction containing FGF21 at a purity of >90% was subsequently
lyophilized and the powder was stored at −70°C for subsequent
use.
Human foreskin fibroblast cell
culture
Fibroblast culture was performed as previously
described (20). Briefly, human
foreskin samples (2.1–3.6 cm2) were collected from six
individuals aged between 14 and 26 years at the Department of
Dermatology at The Second Affiliated Hospital, Wenzhou Medical
University between January and February 2014. The protocol of the
present study was approved by the Institutional Ethical Committee
of the Second Affiliated Hospital, Wenzhou Medical University
(Wenzhou, China). Written informed consent was obtained from the
patient. Following the removal of all fat, the tissue was cut into
3-mm/2-mm (length/width) strips and incubated in Dulbecco's
modified Eagle's medium (DMEM; Hyclone, Logan, UT, USA)
supplemented with 10% fetal bovine serum (FBS; Hyclone), 1%
penicillin-streptomycin (Gibco; Thermo Fisher Scientific) and 0.05%
dispase (Sigma-Aldrich) at 4°C overnight. Subsequently, the
epidermis was removed and the dermis was harvested in
25-cm2 flasks pre-treated with FBS, which were stored
horizontally for 1 h and vertically for 2 h in a culture chamber
containing 5% CO2 at 37°C. The tissues were cultured in
DMEM, containing 5.5 mM glucose, 10% FBS and 1%
penicillin-streptomycin, and the medium was changed every three
days. Cultured cells were digested and passaged with 0.25% trypsin
(Gibco) once cell confluence reached ~80%. Cells at passage 4–5
were used in the subsequent experiments.
Wound healing assay
The effects of FGF21 on the migratory capacity of
fibroblasts was determined using the wound healing scratch assay,
as previously described (20).
Primary fibroblast cells were seeded onto 6-well plates at 80–90%
confluence and incubated overnight in DMEM containing 0.5% FBS and
5 µg/ml mitomycin-C (Sigma-Aldrich). Linear scratch wounds
were subsequently created in the confluent fibroblast monolayer
using a sterile 200 µl pipette tip (MFLab, Ningbo, China).
The medium was immediately replaced with prewarmed (37°C) fresh
DMEM containing 0.5 % FBS and 5 µg/ml mitomycin-C. Following
6 h culture, 100 ng/ml FGF21 protein was added to the culture
medium and gently shaken. At 0 and 24 h after wounding, images were
captured using a microscope (IX70; Olympus, Tokyo, Japan) equipped
with a CCD camera (CoolSNAP HQ; Nippon Roper, Chiba, Japan), which
were controlled by MetaMorph 7.1 software (Molecular Devices, LLC,
Sunnyvale, CA, USA). To quantify cell migration, 20 cells on the
border of the wound area were randomly selected from each well and
the migration distance was measured using ImageJ 14.8 software at
the indicated time points (21).
Western blot analysis
Following the indicated treatments, cells were lysed
in an ice-cold lysis solution (2 M thiourea, 7 M urea, 2%
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 40 mM
dithiothreitol, 40 mM Tris base and 1% protease inhibitor;
Sigma-Aldrich). Following centrifugation at 15,000 × g for 15 min,
the concentration of total protein in the supernatant was
determined using the Bradfold protein assay (Bio-Rad Laboratories,
Inc.). A total of 25 µg protein was separated using 15%
SDS-PAGE, followed by electrotransfer onto Immobilon-P transfer
membranes (Millipore, Billerica, MA, USA). Subsequent to blocking
in Tris-buffered saline containing 5% skimmed milk and 0.05%
Tween-20 for 1 h, membranes were blotted with the following primary
rabbit monoclonal antibodies at 4°C overnight: Anti-phospho
(p)-stress-activated protein kinase/c-Jun N-terminal kinase (JNK)
(Thr183/Tyr185) (cat. no. 4668; 1:1,000
dilution; Cell Signaling Technology, Inc., Danvers, MA, USA),
anti-JNK (cat. no. ab179461; 1:1,2000) and anti-FGF21 (cat. no.
ab171941; 1:2,000 dilution; both Abcam). Following three washes
with TBST, the membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody (cat. no.
7074; 1:2,000 dilution; Cell Signaling Technology, Inc.) for 1 h at
room temperature. Antigen-antibody complexes were then visualized
using an enhanced chemiluminescence kit (GE Healthcare Life
Sciences). Images of the western blot and X-ray film were captured
using an ImageQuant LAS 4000 Mini (GE Healthcare Life Sciences) and
an Epson Perfection V700 photo scanner (Epson Corporation, Nagano,
Japan). Protein levels were normalized against those of total total
c-Jun N-terminal kinase (t-JNK) using Image J software. A protein
molecular weight marker (Takara Biotechnology Co., Ltd.) was used
to analyze protein size.
Statistical analysis
Statistical analysis was performed using two-way
analysis of variance and Bonferroni post-hoc analysis on Prism 5
software (GraphPad, Inc., La Jolla, CA, USA). Comparison between
two groups was performed using an unpaired Student's t-test. Values
are expressed as the mean ± standard error. P<0.05 was
considered to indicate a statistically significant difference
between values.
Results
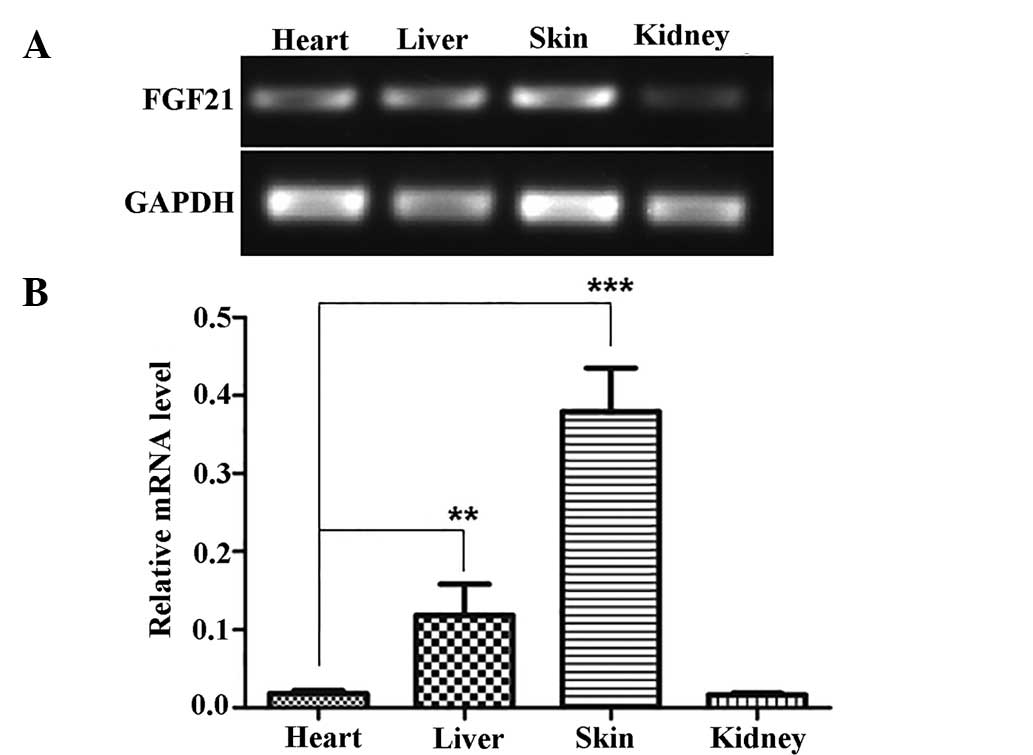
FGF21 is highly expressed in skin tissues
and is upregulated following wounding
Initially, FGF21 has been reported to be
preferentially expressed in the liver (8). To determine FGF21 expression patterns
in other tissue types, the present study subjected liver, heart,
kidney and skin samples to RT-PCR analysis of FGF21. The results
showed that FGF21 is highly expressed in the liver; however, an
even higher expression (~3-fold) was detected in the skin (Fig. 1). Historical identifi-cation of the
FGF family via the analysis of numerous tissues demonstrated that
basic fibroblast growth factor (bFGF) was a major member of the
fibroblast growth factor in the brain, fibroblasts and other
tissues (22–24). Given that skin wound healing is a
process regulated by various proteins and that bFGF is a member of
the FGF gene family, which is widely known for its implication in
wound healing (7,21), the present study investigated the
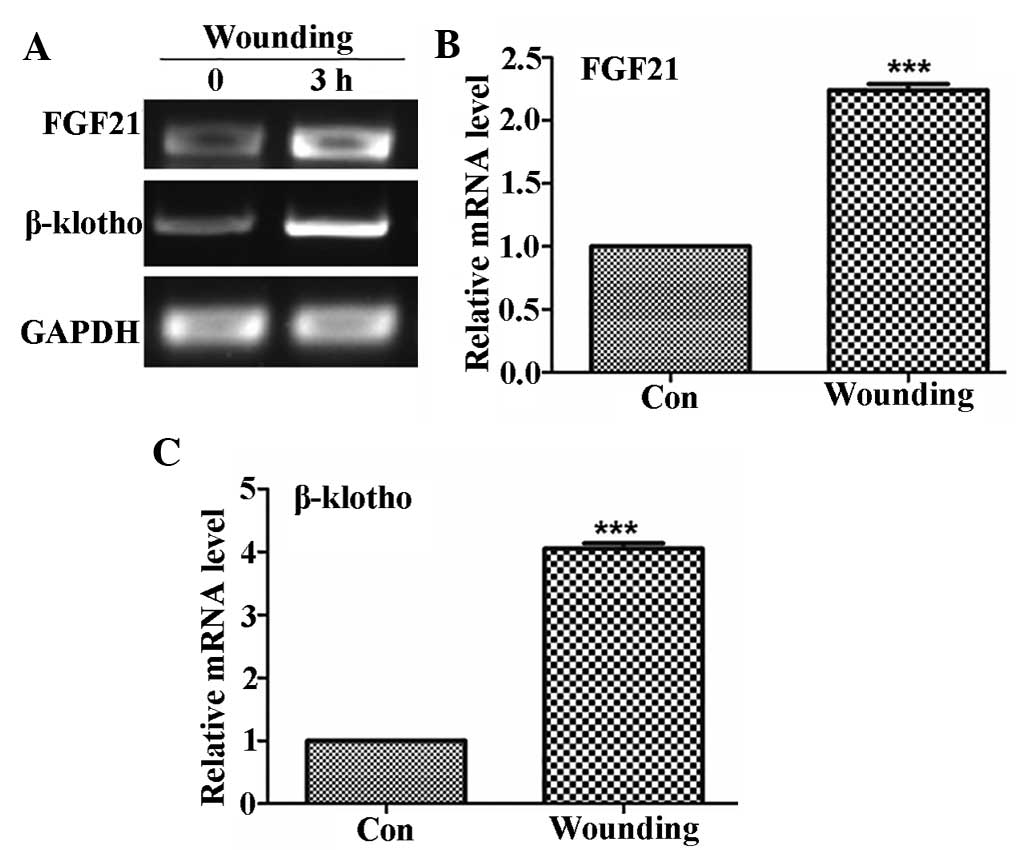
role of FGF21 in wound healing. Expression patterns of FGF21 and
FGFR co-receptor β-klotho in the skin of mice were analyzed prior
to and 3 h following wounding. As shown in Fig. 2, FGF21 expression in the skin of
mice was ~2.2-fold increased following wounding, while β-klotho was
induced by ~4-fold after wounding. These results indicated that
FGF21 was highly expressed in the skin and induced by skin
wounding.
Construction of expression vector
Previous studies have reported that expression of
FGF21 in E. coli is challenging and inefficient (25,26).
To establish an alternative eukaryotic system for FGF expression
with higher efficiency, a yeast system was utilized. Compared to
bacteria, yeast is an eukaryotic organism with higher genetic
similarity to humans; furthermore, yeast cells secrete proteins
into the culture medium, allowing for soluble proteins to be
harvested without the necessity of cell lysis. Translationally
preferred codons are selected for accurate and efficient
translation in bacteria, yeast, worm, fly, and mammals (27); therefore, the present study
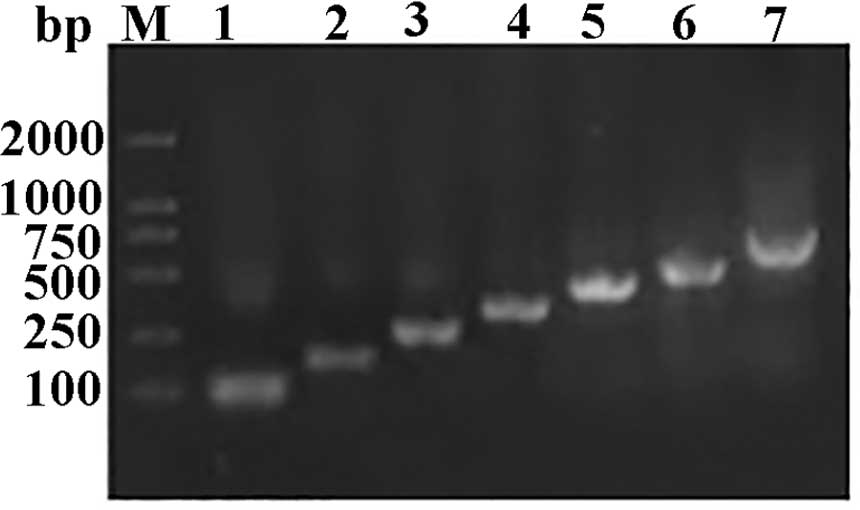
optimized the codon usage of the wild-type FGF21 gene based on
codon bias of P. pastoris without changing the amino acid
composition (Table I) and designed
seven pairs of primers (Table II)
to synthesize FGF21 open reading frame. The full length of FGF21
was obtained by employing seven PCR cycles (Fig. 3) and was cloned into the
XhoI/EcoRI sites of the pPIC9K expression vector to
yield recombinant FGF21. Correct insertion and reading frame of
FGF21 were confirmed by DNA sequencing.
Phenotype screening
After 2–3 days following transfection, the
Mut+ phenotypes grew normally on MM as well as on MD
plates. Although Muts phenotypes grew as well as
Mut+ on MD plates, the colony sizes of Muts
phenotype cells were smaller on MM plates, as compared with the MD
plates at the same time point (data not shown). The Mut+
cells expressed the AOX1 gene, which encodes alcohol oxidase,
enabling recombinants to rapidly grow with methanol as the sole
carbon source. However, due to a recombination at the AOX1 locus to
result in loss of the AOX1 gene in the Muts phenotype,
the cells' ability to metabolize methanol and consequently their
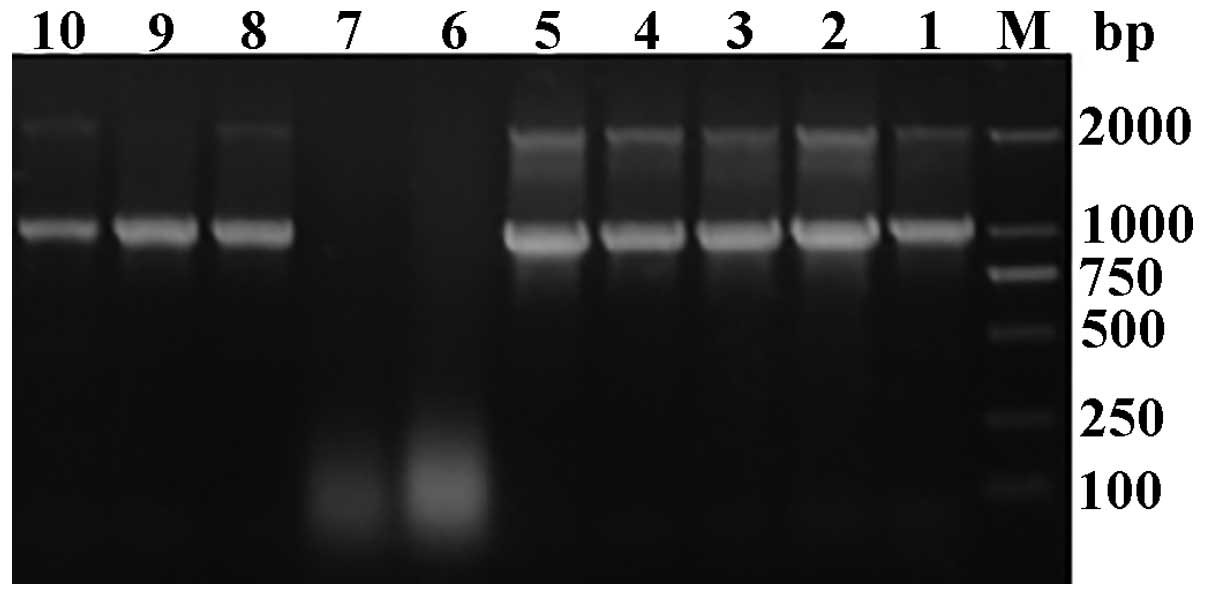
growth was reduced. The genomic DNA of the transformants was
extracted and used as the template for PCR analysis. There were two
fragments of 1.0-kb product (containing the insert gene sequence of
the 546- and 492-bp from the vector) and a 2.2-kb product (AOX1
gene) which was amplified with the AOX1 universal primers for
positive yeast transformants, whereas the negative recombi-nants
exhibited only one fragment of 2.2 kb, demonstrating that the
recombinant expression vector had been successfully transfected
into the yeast genome, while no inserted fragment was detected for
negative yeast recombinants (Fig.
4).
Expression and purification of FGF21 in
P. pastoris
The Mut+ (SMD1168) strains transfected
with the FGF21 expression vector were incubated in flasks with
agitation, and their FGF21 production was assessed. After 96 h of
induction with 0.8% methanol, 1 l cell culture supernatant
containing FGF21 was collected and purified by ammonium sulfate
precipitation, Sephadex G-50 gel filtration and DEAE-Sepharose
ion-exchange chromatography. The putative FGF21 protein was
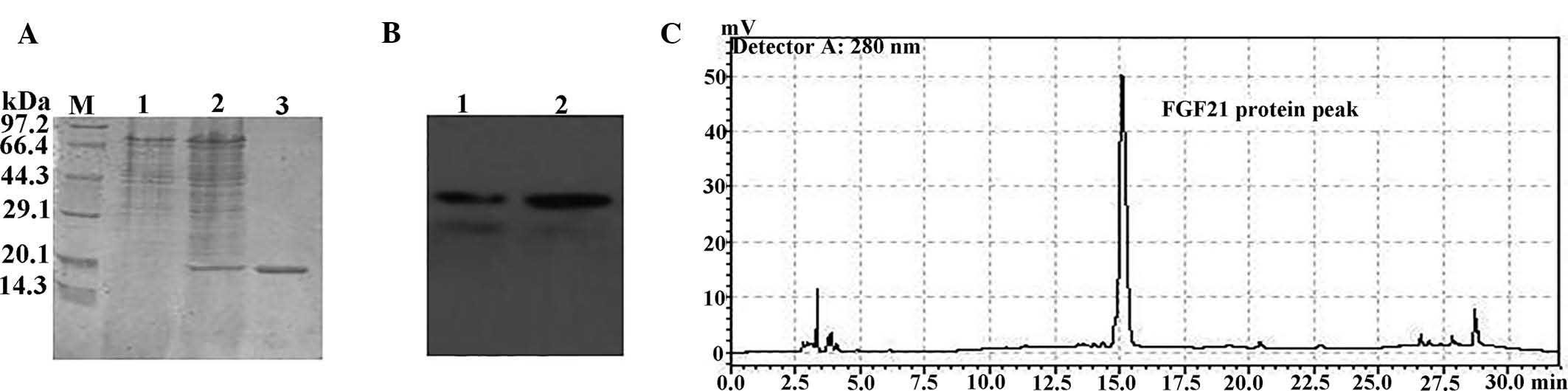
analyzed by 15% SDS-PAGE, revealing a ~20-kDa product (Fig. 5A). Purified recombinant FGF21 was
further confirmed by western blot analysis using an FGF21-specific
antibody (Fig. 5B). Analytical
HPLC was further employed to reveal that the FGF21 protein was
obtained with >90% purity (Fig.
5C). The yield of the recombinant protein (~15 mg/l) was
relatively low in P. pastoris, as compared with that of
E. coli (213±17 mg/l) (28). These results demonstrated that
FGF21 was successfully expressed in P. pastoris.
FGF21 promotes fibroblast migration
The present study further investigated whether the
produced and purified FGF21 exerted any effects on cell migration,
which may be beneficial in wound healing. As FGF21 was highly
expressed in the skin of mice and induced after wounding, its
function in wound healing was further examined. The wound healing
process comprises multiple steps, including proliferation and
migration of fibroblasts, which are regulated by various proteins,
including FGFs. To test effects of FGF21 on the cell migration
process, human primary foreskin fibroblast cells were grown in
low-glucose DMEM containing 5 µg/ml mitomycin-C to prevent
cell proliferation for one day, following which the cell monolayers
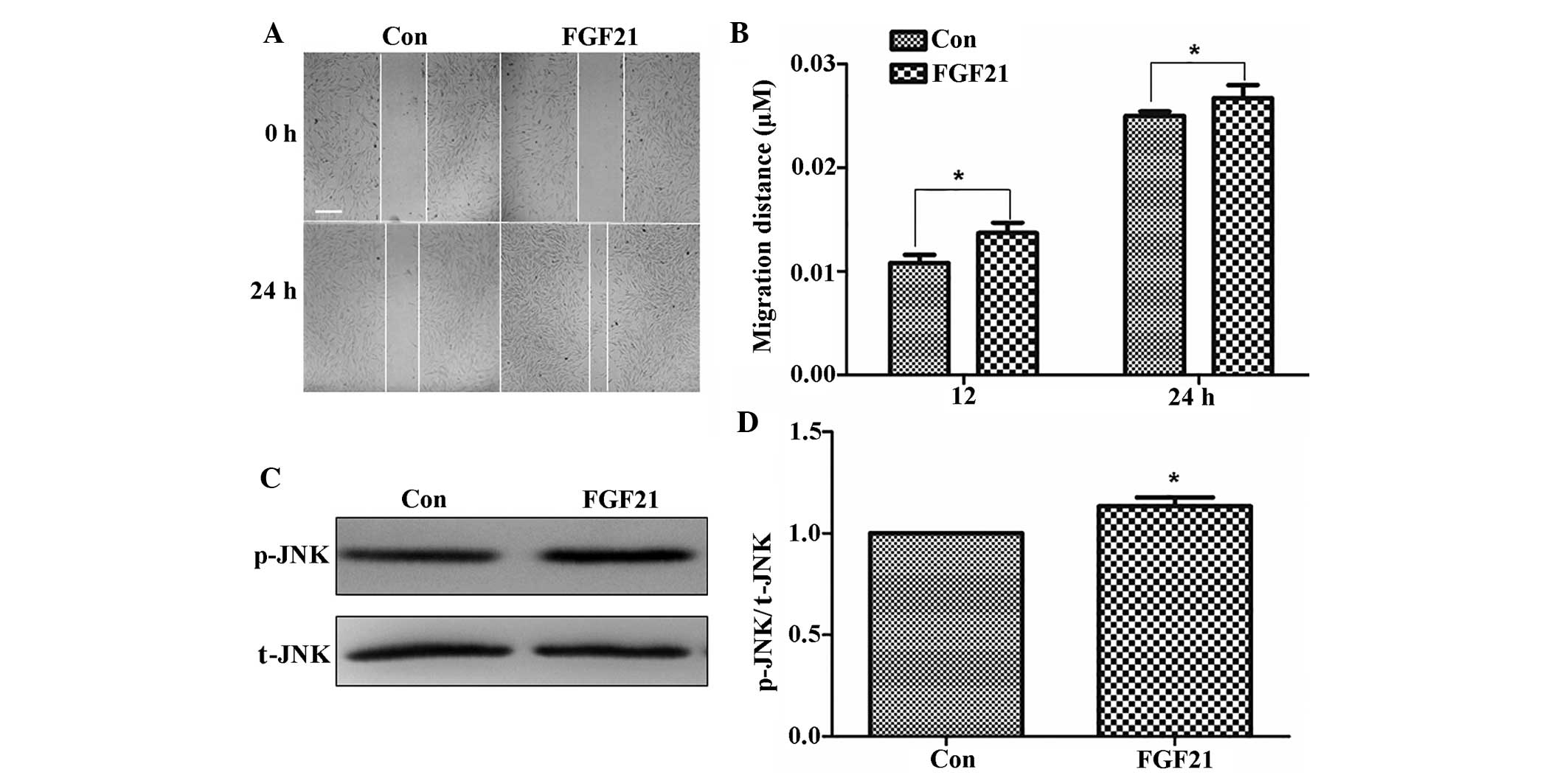
were wounded and incubated with 100 ng/ml FGF21. As shown in
Fig. 6A and B, the cells treated
with FGF21 displayed enhanced cell migration at 24 h following
wound generation.
FGF21 activates the JNK pathway in
fibroblasts
JNK activation is associated with its
phosphorylation and has an important role during cell migration
during wound healing (20).
Therefore, JNK phosphorylation in fibroblasts was examined
following FGF21 treatment. FGF21 treatment activated p-JNK compared
to that in the control group (Fig. 6C
and D). FGF21 increased JNK activation, which may potentially
represent the underlying mechanism by which it accelerates
fibroblast-cell migration.
Discussion
Skin wound healing is a complex process that
requires the actions of keratinocytes, fibroblasts, endothelial
cells, macrophages and platelets. These cells undergo multiple
steps, including proliferation and migration, to rebuild the skin
(29). In this process,
fibroblasts cause wound contraction, and fibroblast-cell migration
is considered to be a fundamental step towards wound healing. Cell
migration is a multi-step cyclic process, including extension of a
protrusion, stable attachment to a site proximal to the leading
edge of the protrusion, forward movement of the cell body, release
of adhesions and retraction at the cell rear (30). bFGF is another member of the FGF
family, whose efficacy in promoting fibroblast-cell migration has
been previously described (7). The
functions of FGF21 in glucose homeostasis as well as in hepato- and
cardioprotection have been reported (9–11).
While it has been reported that FGF21 is highly expressed in the
liver (8), the present study
revealed that its expression was even higher in the injured skin of
mice. Following wounding, the expression of FGF21 and β-klotho,
co-receptor of FGF21 necessary for activation of FGF signaling, was
enhanced in the skin of mice, suggesting that FGF21 is involved in
the wound healing process. Furthermore, recombinant FGF21 protein
was identified to enhance the migration of human fibroblast cells,
therefore exerting a similar function to that of bFGF (7). bFGF was shown to accelerate
fibroblast migration via activation of the phosphoinositide-3
kinase/RacI/JNK pathway (7). In
the present study, FGF21 application activated JNK phosphorylation
in fibroblasts, which may be the underlying mechanism of its
enhancing effects on cell migration. These findings led to the
hypothesis that FGF21 promotes wound repair, and that the
underlying mechanism include the activation of the JNK signaling
pathway.
E. coli, the most widely used system for
heterologous protein expression, has been previously used for FGF21
production (14,25). This type of engineering uses
polyethylene glycol or SUMO protein tags to increase the production
and stability of soluble recombinant protein; however, this
technique requires additional steps to remove the tags. In the
present study, P. pastoris was used to express recombinant
FGF21, which offers a simple technique for obtaining soluble
proteins due to their secretion into the culture media. In the
present study, only a small amount of recombinant FGF21 protein was
isolated from the media, implying that FGF21 expression in this
yeast system may still be low.
Of note, the FGF21 produced in the present study was
demonstrated to promote wound healing in vitro. Furthermore,
mechanistic analysis showed that recombinant FGF21 triggered the
activation of JNK, a downstream regulator of FGFR. These findings
suggested that FGF21 expressed by P. pastoris was
biologically active.
Methods provided by previous studies may be utilized
for designing strains for the production of specific proteins,
including transcription and translation factors (31) or chaperones (32). Modification of yeast strains will
enhance their capacity for protein production. Recently, a study on
Saccharomyces cerevisiae indicated that the choice of
auxotrophic marker was shown to be crucial for developing a yeast
expression system with stable heterologous protein production
(33). The present study was a
first attempt to produce FGF21 using a yeast strain, and further
engineering of yeast strains as well as additional optimizations
will provide a platform for the efficient expression of proteins of
interest, such as FGF21.
In conclusion, the present study demonstrated that
FGF21 expression in skin tissue was higher compared to that in
other tissues. Furthermore, FGF21 expression was identified to be
induced following skin wounding. Exogenous treatment of fibroblasts
with FGF21 produced by an engineered P. pastoris yeast
strain promoted cell migration and activated JNK phosphorylation, a
key regulator of wound healing. The present study therefore
provided a novel eukaryotic system for the expression of FGFs and
indicated that FGF21 may aid in accelerating skin wound
healing.
Abbreviations:
|
FGF21
|
fibroblast growth factor 21
|
|
JNK
|
c-Jun N-terminal kinase
|
|
SUMO
|
small ubiquitin-like modifier
|
|
MD
|
minimal dextrose
|
|
MM
|
minimal medium
|
|
Muts
|
methanol utilization slow
|
|
Mut+
|
methanol utilization plus
|
|
FGFR
|
fibroblast growth factor receptor
|
|
DEAE
|
diethylaminoethyl
|
Acknowledgments
The present study was supported by the Research
Development Fund of Wenzhou Medical University (grant no.
QTJ14029).
References
|
1
|
Mohammadi M, Olsen SK and Ibrahimi OA:
Structural basis for fibroblast growth factor receptor activation.
Cytokine Growth Factor Rev. 16:107–137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feldman B, Poueymirou W, Papaioannou VE,
DeChiara TM and Goldfarb M: Requirement of FGF-4 for
postimplantation mouse development. Science. 267:246–249. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dubrulle J and Pourquié O: Fgf8 mRNA decay
establishes a gradient that couples axial elongation to patterning
in the vertebrate embryo. Nature. 427:419–422. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun X, Meyers EN, Lewandoski M and Martin
GR: Targeted disruption of Fgf8 causes failure of cell migration in
the gastrulating mouse embryo. Genes Dev. 13:1834–1846. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martin GR: The roles of FGFs in the early
development of vertebrate limbs. Genes Dev. 12:1571–1586. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goldfarb M: Functions of fibroblast growth
factors in vertebrate development. Cytokine Growth Factor Rev.
7:311–325. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kanazawa S, Fujiwara T, Matsuzaki S,
Shingaki K, Taniguchi M, Miyata S, Tohyama M, Sakai Y, Yano K,
Hosokawa K and Kubo T: BFGF regulates PI3-kinase-Rac1-JNK pathway
and promotes fibroblast migration in wound healing. PLoS One.
5:e122282010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishimura T, Nakatake Y, Konishi M and
Itoh N: Identification of a novel FGF, FGF-21, preferentially
expressed in the liver. Biochim Biophys Acta. 1492:203–206. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang Q, Zhong L, Zhang J, Wang Y,
Bornstein SR, Triggle CR, Ding H, Lam KS and Xu A: FGF21 maintains
glucose homeostasis by mediating the cross talk between liver and
brain during prolonged fasting. Diabetes. 63:4064–4075. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin Z, Tian H, Lam KS, Lin S, Hoo RC,
Konishi M, Itoh N, Wang Y, Bornstein SR, Xu A and Li X: Adiponectin
mediates the metabolic effects of FGF21 on glucose homeostasis and
insulin sensitivity in mice. Cell Metab. 17:779–789. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin Z, Wu F, Lin S, Pan X, Jin L, Lu T,
Shi L, Wang Y, Xu A and Li X: Adiponectin protects against
acetaminophen-induced mitochondrial dysfunction and acute liver
injury by promoting autophagy in mice. J Hepatol. 61:825–831. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yie J, Wang W, Deng L, Tam LT, Stevens J,
Chen MM, Li Y, Xu J, Lindberg R, Hecht R, et al: Understanding the
physical interactions in the FGF21/FGFR/β-Klotho complex:
Structural requirements and implications in FGF21 signaling. Chem
Biol Drug Des. 79:398–410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Belov AA and Mohammadi M: Molecular
mechanisms of fibroblast growth factor signaling in physiology and
pathology. Cold Spring Harb Perspect Biol. 5:a0159582013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Xiao Y, Fu L, Zhao H, Zhang Y, Wan
X, Qin Y, Huang Y, Gao H and Li X: High-level expression and
purification of soluble recombinant FGF21 protein by SUMO fusion in
Escherichia coli. BMC Biotechnol. 10:142010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Daly R and Hearn MT: Expression of
heterologous proteins in Pichia pastoris: A useful experimental
tool in protein engineering and production. J Mol Recognit.
18:119–138. 2005. View
Article : Google Scholar
|
|
16
|
Zittermann SI and Issekutz AC: Basic
fibroblast growth factor (bFGF, FGF-2) potentiates leukocyte
recruitment to inflammation by enhancing endothelial adhesion
molecule expression. Am J Pathol. 168:835–846. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
18
|
Jin F, Xu X, Zhang W and Gu D: Expression
and characterization of a housefly cecropin gene in the
methylotrophic yeast, Pichia pastoris. Protein Expr Purif.
49:39–46. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin FL, Xu XX, Yu XQ and Ren SX:
High-level expression of active recombinant ubiquitin
carboxyl-terminal hydrolase of Drosophila melanogaster in Pichia
pastoris. Protein Expr Purif. 65:115–121. 2009. View Article : Google Scholar
|
|
20
|
Xuan YH, Huang BB, Tian HS, Chi LS, Duan
YM, Wang X, Zhu ZX, Cai WH, Zhu YT, Wei TM, et al: High-glucose
inhibits human fibroblast cell migration in wound healing via
repression of bFGF-regulating JNK phosphorylation. PLoS One.
9:e1081822014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iyer VR, Eisen MB, Ross DT, Schuler G,
Moore T, Lee JC, Trent JM, Staudt LM, Hudson J Jr, Boguski MS, et
al: The transcriptional program in the response of human
fibroblasts to serum. Science. 283:83–87. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Burgess WH and Maciag T: The
heparin-binding (fibroblast) growth factor family of proteins. Annu
Rev Biochemistry. 58:575–602. 1989. View Article : Google Scholar
|
|
23
|
Courty J, Dauchel MC, Caruelle D,
Perderiset M and Barritault D: Mitogenic properties of a new
endothelial cell growth factor related to pleiotrophin. Biochem
Biophys Res Commun. 180:145–151. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fernig DG and Gallagher JT: Fibroblast
growth factors and their receptors: An information network
controlling tissue growth, morphogenesis and repair. Prog Growth
Factor Res. 5:353–377. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang Z, Wang H, Lu M, Sun C, Wu X, Tan Y,
Ye C, Zhu G, Wang X, Cai L and Li X: A better anti-diabetic
recombinant human fibroblast growth factor 21 (rhFGF21) modified
with polyethylene glycol. PLoS One. 6:e206692011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kharitonenkov A, Shiyanova TL, Koester A,
Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers
JS, Owens RA, et al: FGF-21 as a novel metabolic regulator. J Clin
Invest. 115:1627–1635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gu W, Zhou T and Wilke CO: A universal
trend of reduced mRNA stability near the translation-initiation
site in prokaryotes and eukaryotes. PLoS Comput Biol. 6:e1000664.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang M, Jiang X, Su Z, Lin J, Xiang Q,
Yang Z, Huang Y and Li X: Large-scale expression, purification, and
glucose uptake activity of recombinant human FGF21 in Escherichia
coli. Appl Microbiol Biotechnol. 93:613–621. 2012. View Article : Google Scholar
|
|
29
|
Martin P: Wound healing-aiming for perfect
skin regeneration. Science. 276:75–81. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cao C, Sun Y, Healey S, Bi Z, Hu G, Wan S,
Kouttab N, Chu W and Wan Y: EGFR-mediated expression of aquaporin-3
is involved in human skin fibroblast migration. Biochem J.
400:225–234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dever TE: Using GCN4 as a reporter of eIF2
alpha phosphorylation and translational regulation in yeast.
Methods. 11:403–417. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Payne T, Finnis C, Evans LR, Mead DJ,
Avery SV, Archer DB and Sleep D: Modulation of chaperone gene
expression in mutagenized Saccharomyces cerevisiae strains
developed for recombinant human albumin production results in
increased production of multiple heterologous proteins. Appl
Environ Microbiol. 74:7759–7766. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kazemi Seresht A, Nørgaard P, Palmqvist
EA, Andersen AS and Olsson L: Modulating heterologous protein
production in yeast: The applicability of truncated auxotrophic
markers. Appl Microbiol Biotechnol. 97:3939–3948. 2013. View Article : Google Scholar
|