Introduction
Hepatocellular carcinoma is one of the most common
types of cancer worldwide, and is the third most common cause of
cancer-associated mortality (1).
Patients with hepatocellular carcinoma have a poor prognosis, with
a 5-year survival rate of only 3–5% (2). Radiation has been used in cancer
therapy for several decades, and has been integrated into one of
the treatments strategies for hepatocellular carcinoma (3). Despite initial promising clinical
responses, there is increasing evidence that hepatocellular
carcinoma can acquire resistance to radiation (4,5).
Although the mechanism underlying radioresistance remains to be
fully elucidated, several factors affecting the sensitivity of
radioresistant cancer cells have been identified, and
investigations are continuing to examine combination treatments
using novel strategies, which may allow for decreased dosage and
enhanced sensitivity to radiation (5).
MicroRNA (miRNA; miR) is non-coding single-stranded
RNA of ~22 nucleotides in length (6). Although miRNAs are generally
downregulated in cancer, several studies have shown that aberrant
miRNA expression is correlated with the development and progression
of cancer (7–9). It has been reported that miRNAs
affect numerous cancer-associated processes, including
proliferation, cell cycle control, apoptosis, differentiation,
migration and metabolism (8–10).
Therefore, miRNAs may be used as biomarkers for diagnosis, and may
be vital targets for cancer molecular therapies.
The miR-34 family consists of three closely
associated miRNAs; miR-34a, miR-34b and miR-34c (11). The miR-34 members have been
reported to be important downstream effectors of the p53 tumor
suppressor (11). In addition,
canonical p53 target sites are present in the miR-34 promoters
(12). miR-34a has been
investigated as a key regulator of tumor suppression (13). The reported miR-34a targets include
B cell lymphoma 2, Notch1, cyclin D1, cyclin E2, cyclin-dependent
kinase 4, MET and sirtuin 1 (14),
suggesting that miR-34a may serve as a cancer therapeutic
target.
Lactate dehydrogenase-A (LDHA) is one of the
predominant isoforms of LDH, regulating the conversion of pyruvate
to lactate of the cellular glycolytic process (15). Previous studies have demonstrated
that the expression of LDHA in cancer cells is associated with
radiotherapy sensitivity (16). In
addition, LDHA has been demonstrated to contribute to paclitaxel
resistance in breast cancer (17),
indicating that LDHA may be a therapeutic target for overcoming
drug resistance or radioresistance. In the present study, the roles
of LDHA in radiation resistance were investigated in order to
identify novel mechanisms underlying miR-34a-mediated
radioresistance in human hepatocellular cancer.
Materials and methods
Cell lines and irradiation treatment
HepG2 and Huh7 human hepatocellular carcinoma cell
lines, and normal CL-48 hepatocytes were obtained from American
Type Culture Collection (Manassas, VA, USA). The cell cultures were
maintained in Dulbecco's modified Eagle's medium, supplemented with
100 µM non-essential amino acids, 100 µM two-fold
vitamin solution (all obtained from Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 2 mM L-glutamine (Sigma-Aldrich, St. Louis, MO,
USA), 1 mM sodium pyruvate, 10% fetal bovine serum, 50 U/ml
penicillin and 50 µg/ml streptomycin (Flow Labs, Rockville,
MD, USA). The cells were cultured at 37°C in a humidified
atmosphere containing 5% CO2 and 95% air. The cells were
irradiated at 0, 2, 4, 8, 16 or 20 Gy with 6 MV X-rays, generated
using a Siemens accelerator (KD-2; Siemens AG, Munich, Germany) at
an average dose rate of 200 cGy/min. The cells were trypsinized
(Sigma-Aldrich) and re-plated at a density of 1×106
cells/well in the cell culture dish in the incubator for 16 h at
37°C, followed by the downstream analysis. Cells were counted using
a TC20™ Automated Cell Counter (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) according to the manufacturer's protocols.
Establishment of radioresistant cell
lines
A radioresistant hepatocellular carcinoma cell line
was generated using HepG2 parental cells, by exposing the cells to
repeated irradiation for a duration of 3 months. Cells were counted
and passaged for culture overnight. Cells were treated with 10 Gy
X-ray generated using a Siemens accelerator (KD-2). Cells were
cultured in medium prior to the next passage on day 5. This was
repeated every 2 weeks until radioresistant cell sublines were
established. Numerous radioresistant cell clones were developed and
pooled for the following experiments.
Reagents and antibodies
The antibodies used in the present study were rabbit
anti-human polyclonal LDHA (cat. no. 2012; Cell Signaling
Technology, Inc., Danvers, MA, USA) and rabbit anti-human
polyclonal β-actin (cat. no. 4967; Cell Signaling Technology,
Inc.). The transfection reagent kit containing Invitrogen
Lipofectamine® 2000 transfection reagent, and Applied
Biosystems miRNA precursor (pre-miRNA-34a) and control miRNAs
(control-miRNAs) were purchased from Thermo Fisher Scientific,
Inc.
Pre-miRNA or anti-miRNA transfection
The miRNA precursors (pre-miRNAs) and miRNA control
(control-miRNA) were transfected into the cells using
Lipofectamine® 2000. At 48 h post-transfection, the
expression levels of miR-34a were quantified using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR), and
the protein expression levels of LDHA, a target of miR-34a, were
determined using western blotting.
Quantification of the expression levels
of miR-34a
For the analysis of miRNA expression levels, RTq-PCR
was performed. RNA was isolated from cultured cells using the
RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) with an on-column
DNase digestion step, according to the manufacturer's protocols.
The quality and quantity of total RNA samples was checked by
agarose gel electrophoresis and using the Bioanalyzer RNA 6000 Nano
assay (Agilent Technologies GmbH, Waldbronn, Germany). The RNA was
polyadenylated by poly(A) polymerase (New England BioLabs, Inc.,
Ipswich, MA, USA) and reverse transcribed into cDNA with a Promega
reverse transcription kit (Promega Corporation, Madison, WI, USA)
according to the manufacturer's instructions. qPCR was performed in
a 20-µl reaction containing 1 µl TaqMan®
Small RNA Assay (20X; Thermo Fisher Scientific, Inc.), 1 µl
cDNA, 0.5 µl miRNA primers (20 mmol/l; Invitrogen; Thermo
Fisher Scientific, Inc.), 10 µl TaqMan® Universal
PCR Master Mix II (2X; Thermo Fisher Scientific, Inc.) and 7.5
µl nuclease-free water. The reaction mixtures were incubated
at 95°C for 15 min, followed by 40 cycles at 95°C for 10 sec, at
60°C for 30 sec and at 72°C for 30 sec, running on a Mx3000P™
thermocycler (Agilent Technologies, Inc., Santa Clara, CA, USA).
The miRNA primer sequences were as follows: Sense, 5′-CGC TTC GGC
AGC ACA TAT ACTA-3′ and antisense, 5′-CGC TTC ACG AAT TTG CGT
GTCA-3′ for U6 small nuclear RNA (snRNA); and sense, 5′-TTT AAG CTT
ATG CGC CCT GCC-3′ and antisense, 5′-TTT CTC GAG AGA GCT TCC GAA
GTC CTGG-3′ for miR-34a. Quantitative miRNA expression level data
were analyzed by MxPro software (Agilent Technologies, Inc.) and
the normalization was performed with U6 snRNA. The miRNA was
quantified using the formula: ΔCqmiRNA = CqmiRNA - CqU6 (12,17).
All reactions were performed in triplicate.
Plasmid DNA and small interfering (si)RNA
transfection
Transfection was performed using
Lipofectamine® 2000 transfection reagent, according to
the manufacturer's protocol. Overexpression vectors containing
wild-type LDHA (cat. no. RC209378) were purchased from Origene
Technologies, Inc. (Rockville, MD, USA). The Invitrogen siLDHA and
negative control siRNA were purchased from Thermo Fisher
Scientific, Inc. At 48 h post-transfection, the cells were
collected and the whole-cell lysates were prepared using the
Mammalian Protein Extraction reagent (Pierce Biotechnology, Inc.,
Rockford, IL, USA) for further analysis.
Glucose uptake and lactate product
assays
Glucose levels were determined using a Glucose Assay
kit (BioVision, Inc., Milpitas, CA, USA), and glucose consumption
was calculated as the difference in glucose concentration between
the original media and the media from the cell cultures. Absorbance
was measured at 563 nm using a Spectra Max M5 plate reader
(Molecular Devices, LLC, Sunnyvale, CA, USA). Extracellular lactate
levels were measured in the cell culture media using a Lactate
Assay kit (BioVision, Inc.), according to the manufacturer's
protocol. The results were normalized to the quantity of total
protein in the control cells.
Cell viability assay
Following irradiation treatments, 5×104
cells were added to each well of the 96-well plates and incubated
for 24 h at 37°C. The present study used irradiation treatment
doses of 0, 2, 4, 8, 16 or 20 Gys. Sensitivities to these radiation
treatments were determined using an MTT assay. The incubation
medium was removed, and 50 µl MTT solution (1 mg/ml;
Sigma-Aldrich) was added to each well prior to incubation for a
further 4 h. Following incubation at 37°C, 200 µl dimethyl
sulfoxide (Sigma-Aldrich) was added to each well. The optical
density values were measured at 570 nm using a ELx808™ Absorbance
Microplate Reader (BioTek Instruments, Inc., Winooski, VT, USA).
All experiments were repeated at least twice.
Western blot analysis
Total protein was extracted using Mammalian Protein
Extraction reagent and the protein concentration was measured using
the Bradford assay (Bio-Rad Laboratories, Inc.). The protein
concentrations were measured by reading the absorbance at a
wavelength of 595 nm on the ELx808™ Absorbance Microplate Reader.
An equal quantity (50 µg) of protein from each cell line was
loaded per lane and separated on 10% SDS-PAGE (Bio-Rad
Laboratories, Inc.). The gels were electroblotted onto
nitrocellulose membranes (Novex, San Diego, CA, USA) and blocked
overnight at room temperature with 1X Tris-buffered saline with
0.1% Tween (Bio-Rad Laboratories, Inc.) and 4% non-fat dry milk.
The membranes were then probed with primary antibodies against LDHA
or β-actin at 1:1,000 dilution at 37°C. The bound antibodies were
then detected using goat anti-rabbit polyclonal peroxidase-coupled
secondary antibodies (1:3,000; Cell Signaling Technology, Inc.;
cat. no. 7074) for 1 h at 37°C prior to detection with a Super
Signal Enhanced Chemiluminescence kit (Pierce Biotechnology,
Inc.).
Prediction of miR-34a targets
The putative targets of miR-34a were predicted from
the public miRNA database, TargetScan (www.targetscan.org) by searching the 3′-untranslated
region of mRNA, which contains a highly conserved binding site for
miR-34a.
Statistical analysis
Statistical evaluation for data analysis was
determined using an unpaired Student's t-test. Student's t-test was
calculated using GraphPad Prism 5.0 software (GraphPad Software,
Inc., La Jolla, CA, USA) to assess for statistical differences. All
data are presented as the mean ± standard error of the mean.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-34a is downregulated in human
hepatocellular carcinoma cells and correlates with radiation
sensitivity
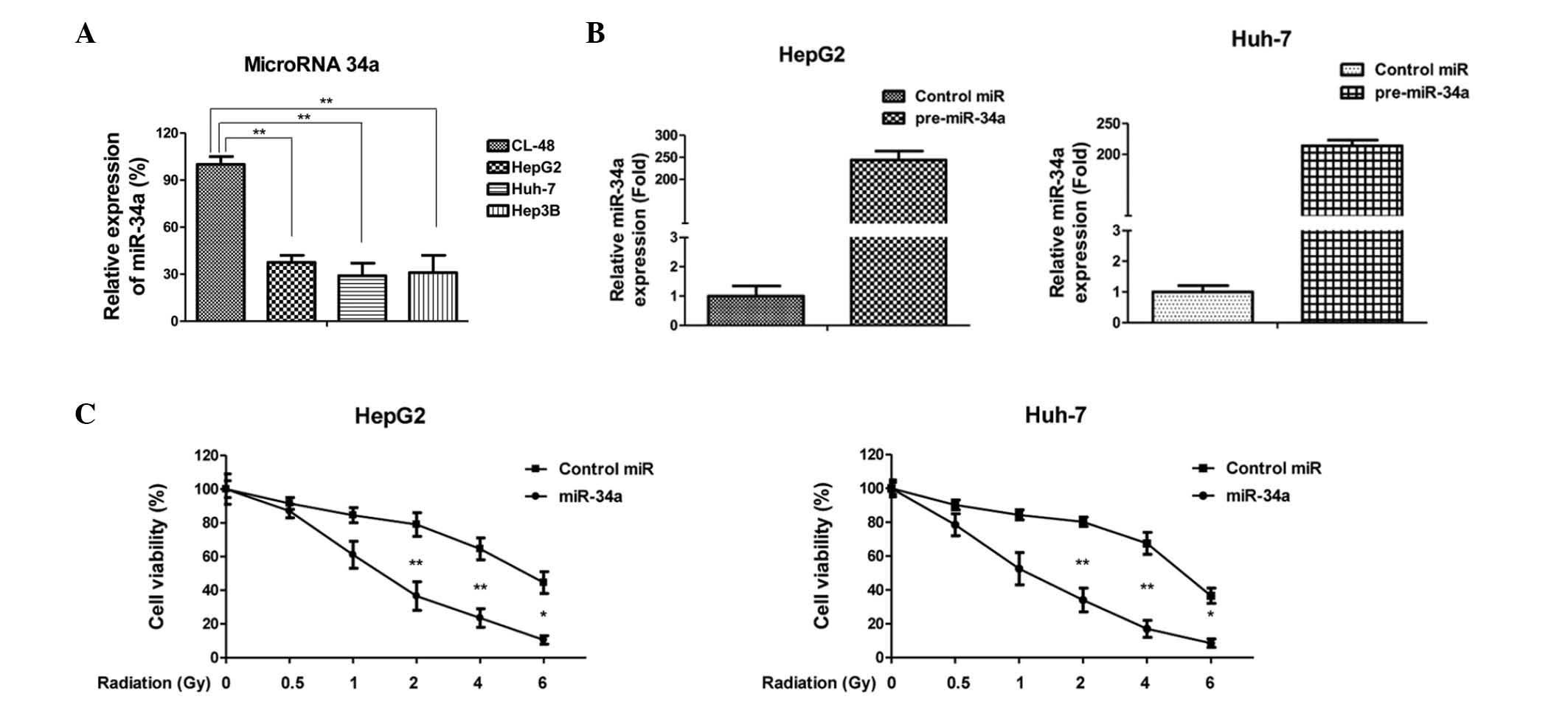
As previous studies demonstrated that miR-34a acts
as a tumor suppressor in several types of tumor (11–14),
the present study examined the expression levels of miR-34a in
human hepatocellular carcinoma cells, which were then compared with
normal CL-48 hepatocyte cells. As expected, the expression levels
of miR-34a were significantly downregulated in the hepatocellular
carcinoma cell lines, compared with the normal hepatocyte cell line
(Fig. 1A), suggesting miR-34a may
act as a tumor suppressor in hepatocellular carcinoma cells. The
roles of miR-34a in radiation sensitivity were also investigated.
The cells were transfected with pre-miR-34a for 48 h, followed by
exposure to radiation treatments between 0 and 6 Gy. Overexpression
of miR-34a in the HepG2 and Huh-7 cells (Fig. 1B) significantly decreased cell
viability, compared with the control miRNA (Fig. 1C), indicating the potential for
miR-34a to be applied clinically to enhance the radiosensitivity of
hepatocellular carcinoma.
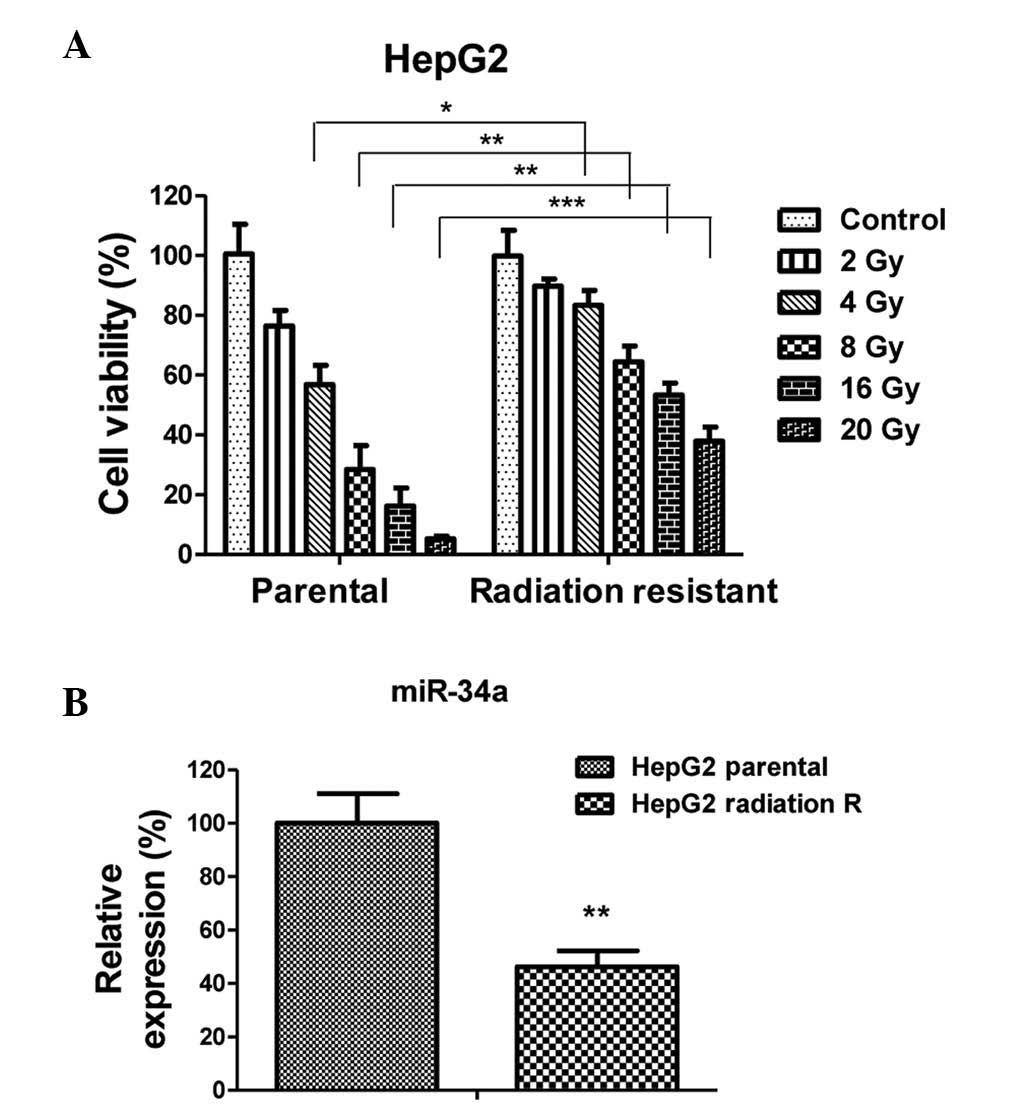
Radioresistant hepatocellular carcinoma
cells exhibit lower expression levels of miR-34a
The correlation between the expression of miR-34a
and radioresistance in cancer cells remains to be fully elucidated.
To investigate the mechanisms underlying miR-34a-mediated radiation
sensitivity, a radioresistant hepatocellular carcinoma cell line
was generated using HepG2 parental cells, by exposing the cells to
repeated irradiation for a duration of 3 months. Numerous
radioresistant cell clones were developed and pooled for the
following experiments. To verify radioresistance, the parental
cells and radioresistant cells were treated with various doses of
radiation(2–20 Gy). Subsequent cell viability assays demonstrated
that the radioresistant HepG2 cells tolerated treatment with higher
doses of radiation, compared with the parental (sensitive) cells,
which exhibited a significant inhibition in viability between 4 and
20 Gy (Fig. 2A). As expected, the
expression levels of miR-34a in the HepG2 radioresistant cells were
significantly downregulated (Fig.
2B), compared with the parental cells, indicating that miR-34a
may serve as a therapeutic agent for selection in the treatment of
radioresistant cancer patients.
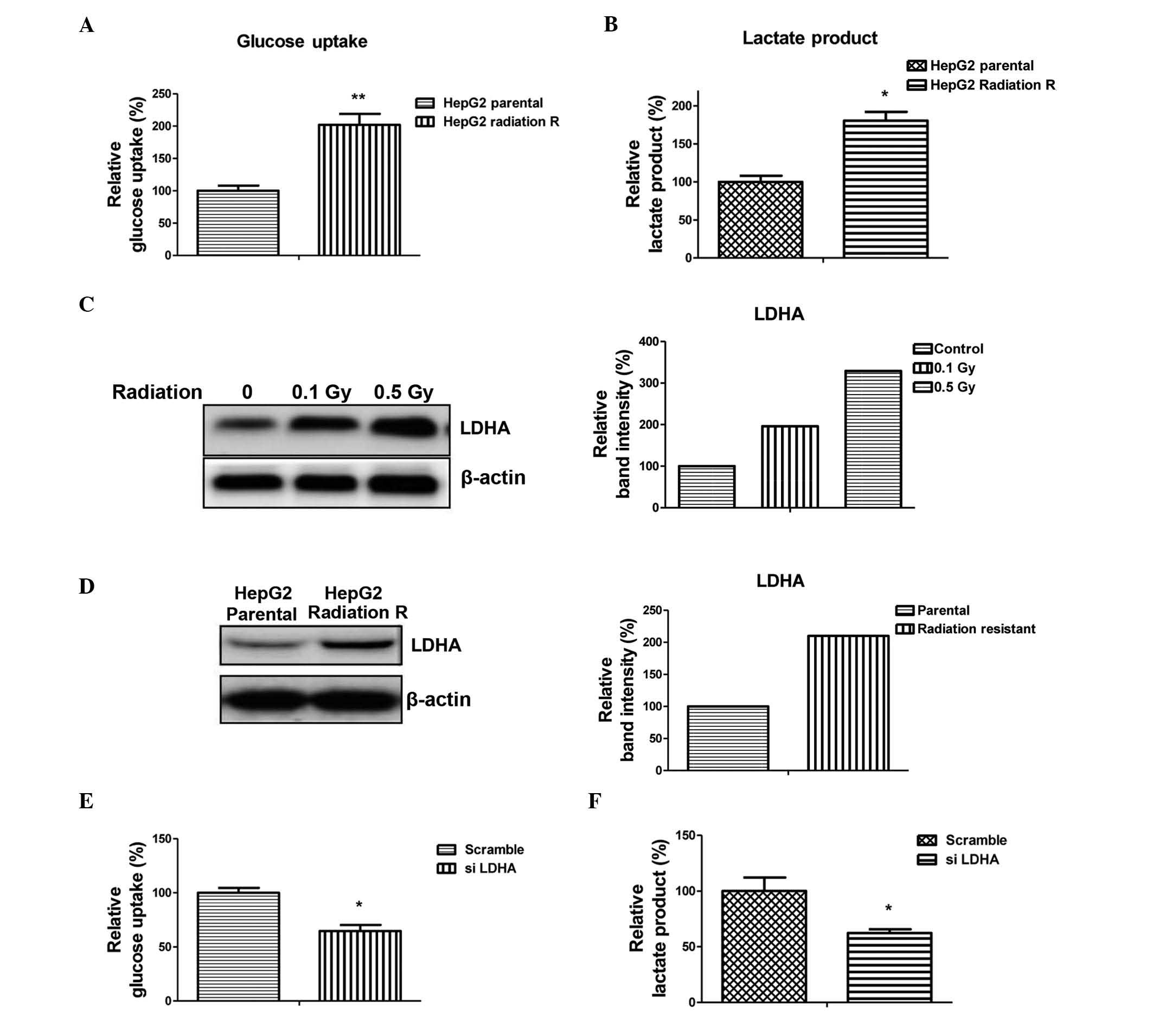
Glucose metabolism is upregulated in
radioresistant HepG2 cells through the induction of the expression
of LDHA
It has previously been reported that enhanced
aerobic glycolysis causes acquired radioresistance in human tumor
cells (18), indicating that
dysregulated glucose metabolism may contribute to radioresistance.
In the present study, glucose uptake and lactate production were
measured, which reflected the activity of glycolysis. As shown in
Fig. 3A and B, glucose uptake and
lactate production in the HepG2 radioresistant cells were
upregulated, compared with the parental cells. LDHA catalyzes the
interconversion between pyruvate and L-lactate, which is further
secreted from the cells (15).
Therefore, the upregulation of LDHA may serve as a marker for
increased glycolysis in cancer cells (15). Treatments with radiation at low
doses (0.1 and 0.5 Gy) induced the expression of LDHA in the HepG2
cells (Fig. 3C). Concordantly, the
expression of LDHA was upregulated in the radioresistant HepG2
cells (Fig. 3D), indicating that
LDHA was involved in the radioresistance of hepatocellular
carcinoma cells. To investigate whether the increase in glycolysis
in the radioresistant HepG2 cells occurred via the increased
expression levels of LDHA, the expression of LDHA was knocked down
by siRNA to monitor the rate of glycolysis. As expected, the levels
of glucose uptake and lactate production were lower in the
LDHA-knockdown radioresistant HepG2 cells (Fig. 3E and F).
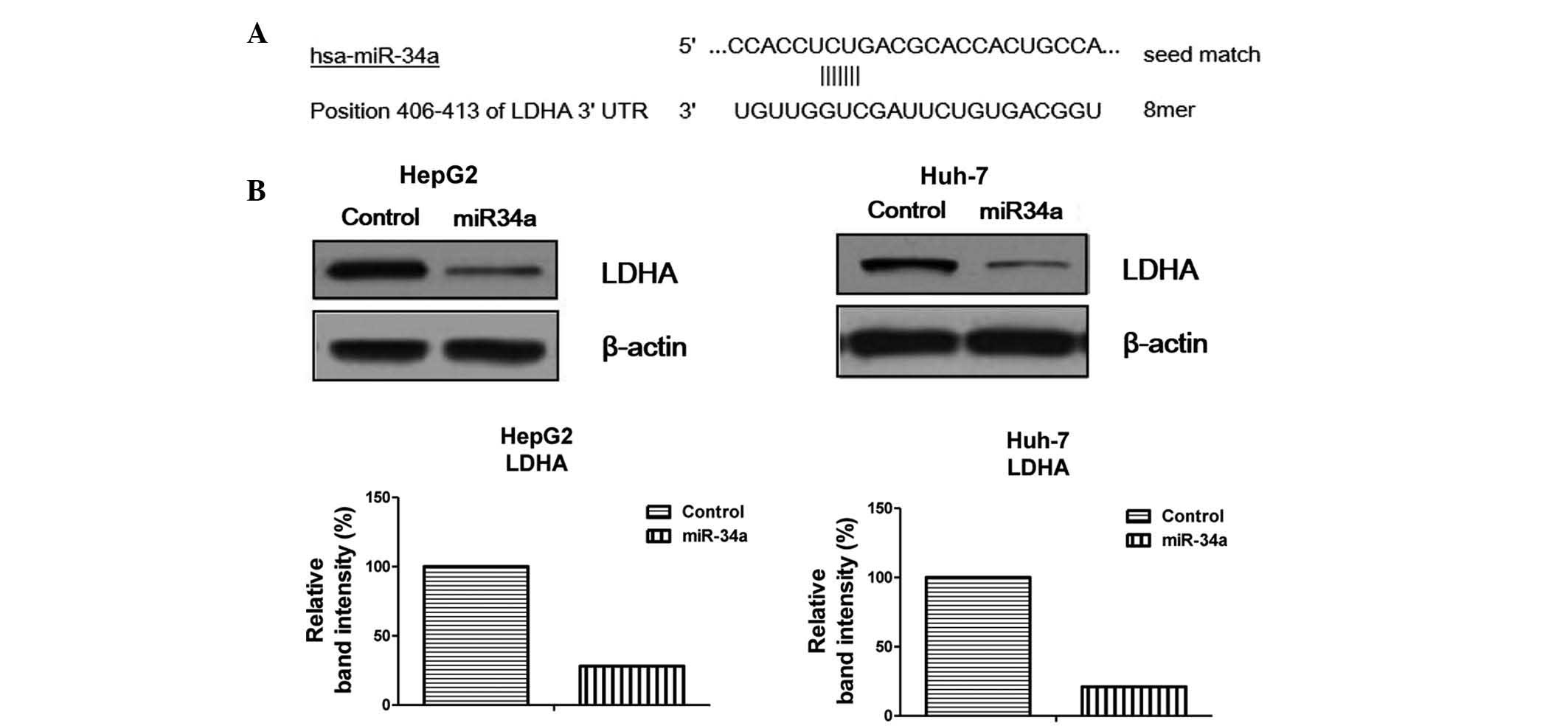
LDHA is a target of miR-34a in
hepatocellular carcinoma cells
To further examine the potential association between
dysregulated expression levels of miR-34a and increased glycolysis
in radioresistant hepatocellular carcinoma cells, an miRNA database
was accessed to identify potential miR-34a targets that may
contribute to radiation resistance. The public miRNA database,
TargetScan, predicted that LDHA may be a target of miR-34a, and
that the 3′-untranslated region of LDHA contains a highly conserved
binding site for miR-34a (Fig.
4A). To determine whether miR-34a targets LDHA in human
hepatocellular carcinoma cells, pre-miR-34a was transfected into
HepG2 and Huh-7 cells. The overexpression of miR-34a markedly
downregulated the protein expression levels of LDHA in the two cell
lines (Fig. 4B), indicating that
miR-34a-mediated radiosensitivity may act through the inhibition of
glycolysis by targeting LDHA.
Overexpression of miR-34a resensitizes
radioresistant HepG2 cells to radiation through the inhibition of
LDHA
The data described above revealed a potential role
for miR-34a in radioresistance and the regulation of glycolysis.
Therefore, the present study hypothesized that the downregulation
of LDHA by the overexpression of miR-34a in hepatocellular
carcinoma cells may resensitize radioresistant cells to radiation
treatment. To confirm this hypothesis, the HepG2 radioresistant
cells were transfected with pre-miR-34a and, after 48 h, the cells
were subjected to radiation treatment at various concentrations for
24 h. As shown in Fig. 5A, the
overexpression of miR-34a significantly resensitized the resistant
cells to radiation treatment. In addition, glucose uptake (Fig. 5B left) and lactate production
(Fig. 5B right) were decreased
following transfection of the HepG2 radioresistant cells with
miR-34a. To further support these results, the expression of LDHA
was recovered by transfecting the miR-34a pre-transfected
radioresistant cells with an LDHA expression vector (Fig. 5C). The recovery of LDHA increased
the resistance of the HepG2 cells to radiation (Fig. 5D), indicating that miR-34a-mediated
radiosensitivity occurred through the inhibition of LDHA.
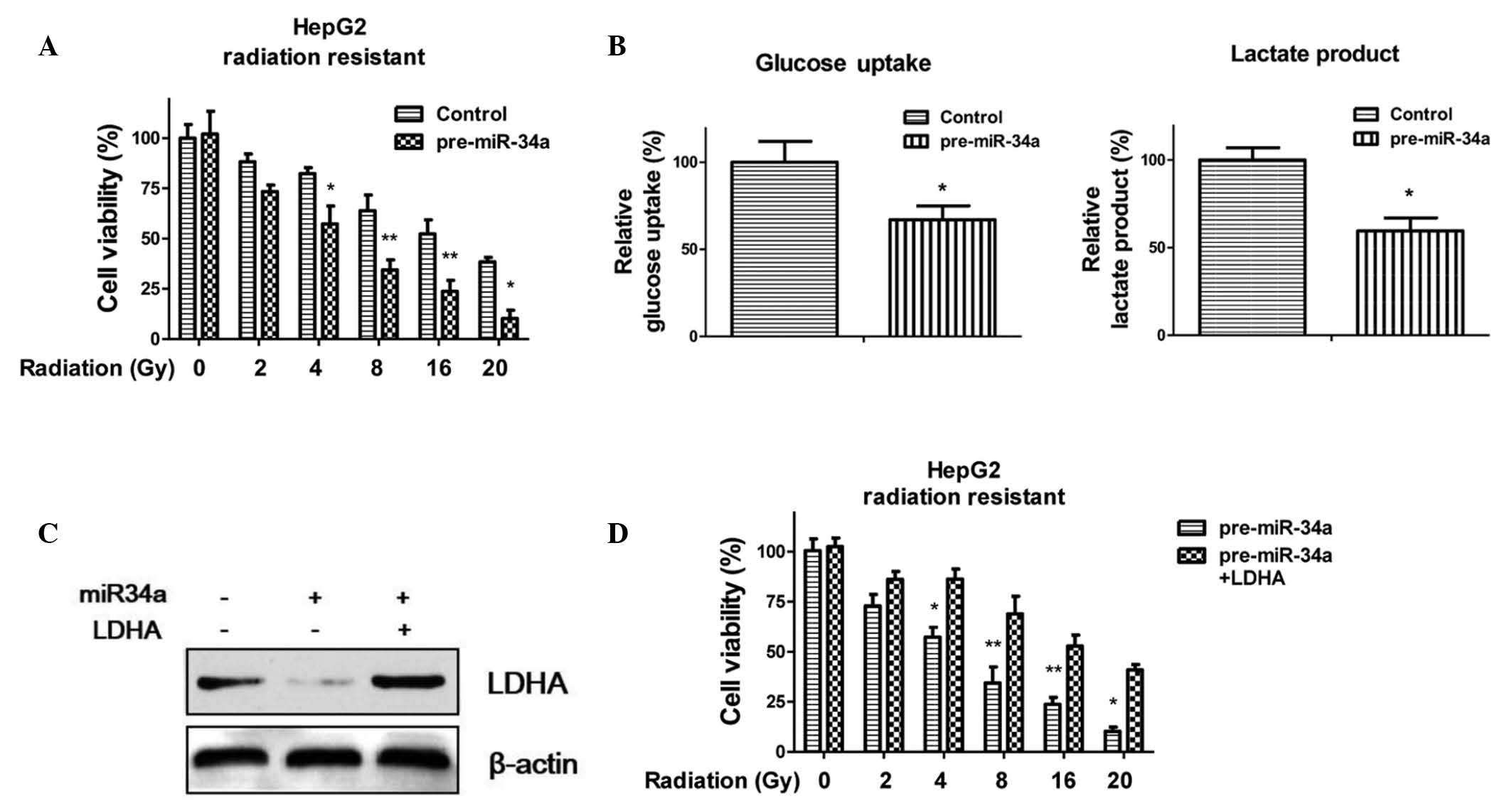 | Figure 5Overexpression of miR-34a in HepG2
radioresistant cells enhances radiosensitivity. (A) HepG2
radioresistant cells were transfected with scramble control siRNA
or per-miR-34a for 48 h, and the cells were then exposed to
radiation (0, 2, 4, 8, 16 and 20 Gys) prior to cell viability
analysis. (B) Overexpression of miR-34a in the radioresistant HepG2
cells inhibited glucose metabolism. (C) Radioresistant HepG2 cells
were transfected with pre-miR-34a or control siRNA for 48 h,
following which the miR-34a-overexpressing cells were transfected
with a control vector or an overexpression vector containing LDHA
for 24 h. The cells were then subjected to western blot analysis.
β-actin was used as a loading control. *P<0.05 and
**P<0.01 vs. the control. (D) Cells were transfected
with pre-miR-34a for 48 h, and transfected with a control vector or
an overexpression vector containing LDHA for 48 h. Cells were
treated with radiation (0, 2, 4, 8, 16 and 20 Gys), prior to cell
viability analysis. *P<0.05 and
**P<0.01 vs. the pre-miR-34a + LDHA of each
treatment. Columns represent the mean of three independent
experiments. Data are presented as the mean ± standard error of the
mean. siRNA, small interfering RNA; miR, microRNA; LDHA, lactate
dehydrogenase A. |
Discussion
miR-34a has been recognized as a key regulator of
tumor suppression (11–14). Furthermore, several studies have
reported that miR-34a modulates anticancer drug responses in
multiple types of cancer. It has been reported that the ectopic
overexpression of miR-34a increases sensitivity to doxorubicin
(19), 5-fluorouracil (20), Erlotinib (21) and Taxol (22) treatment. In the present study,
miR-34a was found to be involved in the resistance of human
hepatocyte cancer cells to radiation. The expression of miR-34a was
downregulated in human hepatocyte cancer cell lines, compared with
normal hepatocytes. In addition, the overexpression of miR-34a
contributed to radioresistance, which was concordant with previous
reports demonstrating that miR-34a acts as a tumor suppressor and
confers chemosensitivity (11–14).
The majority of cancer cells rely on aerobic
glycolysis to generate the energy required for cellular processes.
By contrast, normal differentiated cells rely primarily on
mitochondrial oxidative phosphorylation (23). Therefore, dysregulated anaerobic
glycolysis has been associated with drug resistance and
radioresistance in cancer (23).
It has been reported that LDHA contributes to paclitaxel (17) and trastuzumab (24) resistance. Furthermore, radiation
induces aerobic glycolysis through reactive oxygen species
(25), suggesting that cancer
cells with active glycolysis may acquire radioresistance.
Currently, as a clinically relevant area in cancer research, the
dysregulation of glycolysis and radiosensitivity has not been
specifically addressed. The results of the present study
demonstrated that radioresistant cancer cells exhibit upregulated
levels of glycolysis and downregulated expression levels of
miR-34a. To the best of our knowledge, the present study is the
first to describe a direct association between glycolysis and
radioresistance in human liver cancer. A TargetScan database search
in the present study identified LDHA as a target of miR-34a, and
the results demonstrated that overexpression of miR-34a inhibited
glycolysis by targeting LDHA, leading to sensitization of
radioresistant cancer cells to radiation.
Radiation therapy generates free radicals, which
damage DNA in order to induce cancer cell apoptosis. Generally,
ionizing radiation induces reactive oxygen species in targeted
cells, leading to genomic instability and lipid peroxidation
(26). Of note, excessive lactate
produced by cancer cells has been reported to be an antioxidant,
which may counteract the reactive oxygen species generated by
irradiation (27). Therefore, it
is possible that the accumulation of lactate surrounding a tumor
may contribute to its radioresistance. The results of the present
study demonstrated a significant correlation between the expression
of LDHA and radioresistance. The inhibition of LDHA by miR-34a
resensitized the radio-resistant cells to irradiation, and the
restoration of LDHA in the miR-34a-overexpressing cells reversed
the resensitization, suggesting that targeting LDHA may be a
therapeutic strategy for the treatment of radiation resistance.
Future investigation aim to identify potential glycolysis
inhibitors, in order to investigate whether the administration of
these inhibitors in combination with radiation treatments exhibits
synergistic effects on hepatocellular carcinoma cells. In addition,
the use of a mouse model offers potential in verifying the
phenotypes.
In conclusion, the present study demonstrated a
marked correlation between miR-34a-mediated glycolysis and
radiation resistance, and provides novel insights into the role of
LDHA in anti-radioresistance therapy.
Acknowledgments
The authors of the present study would like to thank
the staff and the faculty members working in the Department of
General Surgery and the Department of Vascular Surgery, The Second
People's Hospital of Yunnan Province and Fourth Affiliated Hospital
of Kunming Medical University (Kunming, China). The present study
was supported by funding from the Departments of General Surgery
and Vascular Surgery, The Second People's Hospital of Yunnan
Province and Fourth Affiliated Hospital of Kunming Medical
University.
References
|
1
|
Karaman B, Battal B, Sari S and Verim S:
Hepatocellular carcinoma review: Current treatment, and
evidence-based medicine. World J Gastroenterol. 20:18059–18060.
2014.PubMed/NCBI
|
|
2
|
Olsen SK, Brown RS and Siegel AB:
Hepatocellular carcinoma: Review of current treatment with a focus
on targeted molecular therapies. Therap Adv Gastroenterol. 3:55–66.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hong TS: Radiotherapy for hepatocellular
carcinoma with tumor vascular thrombus: Ready for prime time? J
Clin Oncol. 31:1619–1620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Singh S, Singh PP, Roberts LR and Sanchez
W: Chemopreventive strategies in hepatocellular carcinoma. Nat Rev
Gastroenterol Hepatol. 11:45–54. 2014. View Article : Google Scholar
|
|
5
|
Klein J and Dawson LA: Hepatocellular
carcinoma radiation therapy: Review of evidence and future
opportunities. Int J Radiat Oncol Biol Phys. 87:22–32. 2013.
View Article : Google Scholar
|
|
6
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nelson KM and Weiss GJ: MicroRNAs and
cancer: Past, present and potential future. Mol Cancer Ther.
7:3655–3660. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bouyssou JM, Manier S, Huynh D, Issa S,
Roccaro AM and Ghobrial IM: Regulation of microRNAs in cancer
metastasis. Biochim Biophys Acta. 1845:255–265. 2014.PubMed/NCBI
|
|
10
|
Dumortier O, Hinault C and Van Obberghen
E: MicroRNAs and metabolism crosstalk in energy homeostasis. Cell
Metab. 18:312–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Misso G, Di Martino MT, De Rosa G, Farooqi
AA, Lombardi A, Campani V, Zarone MR, Gullà A, Tagliaferri P,
Tassone P and Caraglia M: Mir-34: A new weapon against cancer? Mol
Ther Nucleic Acids. 3:e1942014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang TC, Wentzel EA, Kent OA,
Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M,
Ferlito M, Lowenstein CJ, et al: Transactivation of miR-34a by p53
broadly influences gene expression and promotes apoptosis. Mol
Cell. 26:745–752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cole KA, Attiyeh EF, Mosse YP, Laquaglia
MJ, Diskin SJ, Brodeur GM and Maris JM: A functional screen
identifies miR-34a as a candidate neuroblastoma tumor suppressor
gene. Mol Cancer Res. 6:735–742. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li XJ, Ren ZJ and Tang JH: MicroRNA-34a: A
potential therapeutic target in human cancer. Cell Death Dis.
5:e13272014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Miao P, Sheng S, Sun X, Liu J and Huang G:
Lactate dehydrogenase A in cancer: A promising target for diagnosis
and therapy. IUBMB Life. 65:904–910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hennessey D, Martin LM, Atzberger A, Lynch
TH, Hollywood D and Marignol L: Exposure to hypoxia following
irradiation increases radioresistance in prostate cancer cells.
Urol Oncol. 31:1106–1116. 2013. View Article : Google Scholar
|
|
17
|
Zhou M, Zhao Y, Ding Y, Liu H, Liu Z,
Fodstad O, Riker AI, Kamarajugadda S, Lu J, Owen LB, et al: Warburg
effect in chemo-sensitivity: Targeting lactate dehydrogenase-A
re-sensitizes taxol-resistant cancer cells to taxol. Mol Cancer.
9:332010. View Article : Google Scholar
|
|
18
|
Shimura T, Noma N, Sano Y, Ochiai Y,
Oikawa T, Fukumoto M and Kunugita N: AKT-mediated enhanced aerobic
glycolysis causes acquired radioresistance by human tumor cells.
Radiother Onco. 112:302–307. 2014. View Article : Google Scholar
|
|
19
|
Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ,
Zhao JH and Tang JH: MicroRNA-34a modulates chemosensitivity of
breast cancer cells to adriamycin by targeting Notch1. Arch Med
Res. 43:514–521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Akao Y, Noguchi S, Iio A, Kojima K, Takagi
T and Naoe T: Dysregulation of microRNA-34a expression causes
drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer
Lett. 300:197–204. 2011. View Article : Google Scholar
|
|
21
|
Zhao J, Kelnar K and Bader AG: In-depth
analysis shows synergy between erlotinib and miR-34a. PLoS One.
9:e891052014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kojima K, Fujita Y, Nozawa Y, Deguchi T
and Ito M: MiR-34a attenuates paclitaxel-resistance of
hormone-refractory prostate cancer PC3 cells through direct and
indirect mechanisms. Prostate. 70:1501–1512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martinez-Outschoorn UE, Lin Z, Ko YH,
Goldberg AF, Flomenberg N, Wang C, Pavlides S, Pestell RG, Howell
A, Sotgia F and Lisanti MP: Understanding the metabolic basis of
drug resistance: Therapeutic induction of the warburg effect kills
cancer cells. Cell Cycle. 10:2521–2528. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao Y, Liu H, Liu Z, Ding Y, Ledoux SP,
Wilson GL, Voellmy R, Lin Y, Lin W, Nahta R, et al: Overcoming
trastuzumab resistance in breast cancer by targeting dysregulated
glucose metabolism. Cancer Res. 71:4585–4597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhong J, Rajaram N, Brizel DM, Frees AE,
Ramanujam N, Batinic-Haberle I and Dewhirst MW: Radiation induces
aerobic glycolysis through reactive oxygen species. Radiother
Oncol. 106:390–396. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Baskar R, Lee KA, Yeo R and Yeoh KW:
Cancer and radiation therapy: Current advances and future
directions. Int J Med Sci. 9:193–199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Le A, Cooper CR, Gouw AM, Dinavahi R,
Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL and Dang
CV: Inhibition of lactate dehydrogenase A induces oxidative stress
and inhibits tumor progression. Proc Natl Acad Sci USA.
107:2037–2042. 2010. View Article : Google Scholar : PubMed/NCBI
|