Introduction
Polycystic ovary syndrome (PCOS), characterized by
irregular menses, hyperandrogenism and polycystic ovaries, is the
most common type of endocrine disorder affecting women of
reproductive age (1). It has been
reported that 5–11% of women of reproductive age have PCOS
worldwide, and 15–20% of Reproductive Medicine criteria are used
(2). There are several proposed
diagnostic criteria for PCOS: National Institutes of Health (NIH)
consensus criteria 1990, Rotterdam criteria 2003, and the AES
definition 2008 (3). The clinical
manifestations of PCOS include oligomenorrhea or amenorrhea,
hirsutism and frequently infertility and type 1 diabetes, type 2
diabetes and gestational diabetes are the predominant risk factors
for PCOS (4). Insulin resistance
is a major cause of comorbidity, including metabolic syndrome,
hypertension, dyslipidemia, glucose intolerance and diabetes, among
women with PCOS (5). Studies have
also shown that women with PCOS are at a high risk for developing
cardiovascular diseases and exhibit endothelial dysfunction
(6–9). In addition, mental health disorders
are common in women with PCOS (10). Due to the extensive detrimental
consequences of PCOS, it is necessary to investigate the
pathogenetic mechanism of this disease.
Although several studies have focussed on the
pathogenesis of PCOS, the underlying etiology of PCOS remains to be
elucidated (11–13). Previous evidence has demonstrated
that important regulators, including transcription factors (TFs)
and microRNAs, are important in the development of PCOS. For
example, AR, a nuclear transcription factor and member of the
steroid receptor superfamily, is important in hyperandrogenism,
which is a common syndrome of PCOS, and hyperactive AR is also
considered an important marker for PCOS diagnosis (14). Insulin resistance is a key
pathophysiological marker of PCOS, and it is reported that insulin
resistance occurs in 50–70% of women with PCOS (15). The transcription factor, cAMP
response element-binding protein (CREB) and its coactivator,
CREB-regulated transcriptional coactivator 2 are reported to be
involved in the control of hepatic gluconeogenesis in insulin
resistance (16). Therefore, the
investigation of these TFs is necessary for the diagnosis and
therapy of PCOS.
MicroRNAs have gained increased attention in
research. MicroRNAs, which are 21–25 nucleotides long, non-coding
RNA molecules, function in the transcriptional and
post-transcriptional regulation of gene expression, and they are
involved in various diseases (17). Certain microRNAs have been reported
in metabolic disorders of PCOS, including miRNA-21, miRNA-27b,
miRNA-103 and miRNA-155; and these four microRNAs have been
reported to be involved in the metabolic and immune processes of
PCOS (18). Roth et al
reported that rno-miR-221, rno-miR-222, rno-miR-25 and rno-miR-26b
are differentially expressed between rats with PCOS and control
rats (19). Hossain et al
found that 24% of 349 investigated microRNAs were differentially
expressed between a PCOS rat model and a control group (20). Sang et al also found that
miR-132 and miR-320 are expressed at significantly lower levels in
the follicular fluid of patients with PCOS compared with healthy
controls (21). Another study
found that miRNA-93 is overexpressed and inhibits GLUT4, which is
implicated in the insulin resistance of PCOS (22).
The above studies indicated the importance of TFs
and microRNAs in PCOS, however, the majority of the underlying
mechanism remains to be fully elucidated. Furthermore, the
synergistic regulatory action of TFs and microRNAs in PCOS have not
been clearly demonstrated. The present study aimed to construct a
microRNA-differentially expressed gene (DEG) network, from which a
TF-DEG network was constructed, based on the DEGs identified from
the PCOS samples and normal control samples. The regulatory
associations between the TFs and their targets, and the
associations between microRNAs and their targets, were obtained
from the CHIPBase and miRTarBase databases (23,24).
Integrating the above two networks was then used to establish a
TF-microRNA synergistic regulatory network, from which key
microRNAs and TFs in PCOS can be identified. The results may
provide insights into revealing the potential mechanism of PCOS on
transcriptional regulations levels in the context of the
TF-microRNA synergistic regulatory network.
Materials and methods
Microarray data and DEG analysis
The publicly available microarray dataset, GSE34526,
was obtained from the Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo/) of the
National Center of Biotechnology Information. This profile
contained a total of 10 samples, including seven samples from
patients with PCOS and three normal samples, based on human
granulosa cells isolated from ovarian aspirates from women with and
without PCOS (25). These samples
were profiled using the Affymetrix Human Genome U133 Plus 2.0 Array
platform (HG-U133_Plus_2; Affymetrix, Santa Clara, CA, USA).
The raw microarray data and the probe annotation
files were downloaded (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34526)
for further analysis. The probes were converted into gene Entrez
Gene IDs using the Database for Annotation, Visualization and
Integrated Discovery (DAVID) tool (26), and the fold-change method was used
to identify the DEGs. DAVID is a high-throughput and integrated
data-mining environment, and can be used to analyze a given gene
list derived from genomic experiments. As several probes may be
mapped to a single gene, the expression value of a given gene was
computed by calculating the average expression value of all probes
of the corresponding gene. The genes with a fold-change >2 or
<0.5 were defined as DEGs.
Functional enrichment analysis
To implement functional annotation with different
regulatory networks, the present study used the DAVID tool
(26). The DAVID tool was used to
implement Kyoto Encyclopedia of Genes and Genomes and Gene Ontology
(GO) enrichment analysis, based on hypergeometric distribution.
P<0.01 was selected as the cutoff criterion for statistically
significant pathways or GO terms associated with PCOS.
TF-DEG network and microRNA-DEG network
construction
To construct the TF regulatory network, the TF-mRNA
associations for 329 TFs were downloaded from the CHIPBase
database, which is an integrated resource and platform for decoding
TF binding maps, expression profiles and transcriptional regulation
of several types of RNA from ChIP-Seq data (27). A total of 4,845 associations were
obtained, which consisted of 329 human TFs and 1,658 targets, and
an original network of TFs and their targets was constructed. The
DEGs of PCOS were mapped to this original network and the largest
connected component was extracted, which produced a TF-DEG network,
which included 164 TFs and 274 DEGs as targets. If one node acted
as a TF and a DEG, it was marked as a DEG. For the microRNA-DEGs
network, the experimental verified associations between human
microRNAs and their targets were downloaded from miRTarBase, which
has accumulated >50,000 miRNA-target interactions collected by
manually surveying pertinent literature following systematic data
mining of the text (23). Similar
to the TF-DEG network, an original network was constructed and the
DEGs were mapped to this network. Finally, the microRNA-DEG network
was obtained, including 432 microRNAs and 1,524 DEGs as
targets.
TF-microRNA synergistic regulatory
network construction
The final TF-microRNA synergistic regulatory network
was constructed based on the TF-DEG network and microRNA-DEG
network. The DEGs, which were targeted by microRNAs and TFs were
selected, and the microRNAs and TFs which regulated them were
extracted. Finally, a synergistic regulatory network was
established in which the nodes were DEGs, microRNAs and TFs, and
the DEGs were regulated by the synergistic regulatory action of the
other two types of nodes. These three networks were visualized
using Cytoscape software (National Institute of General Medical
Sciences, Bethesda, MD, USA; version 3.0.1).
Results
DEG analysis between patients with PCOS
and healthy controls
In order to identify the DEGs of PCOS, the present
study obtained the microarray dataset (GSE34526) of PCOS samples
and normal samples from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The fold-change
method was then used to identify DEGs the between the patients with
PCOS and the controls. A total of 7,027 genes were considered
differentially expressed.
TF-microRNA synergistic regulatory
network
To construct the TF-microRNA network, TF-DEG and
microRNA-DEG networks were first constructed. Based on the
miRTarBase and CHIPbase databases, the TFs and microRNAs which
regulated the DEGs of PCOS were identified. A total of 1,008 pairs
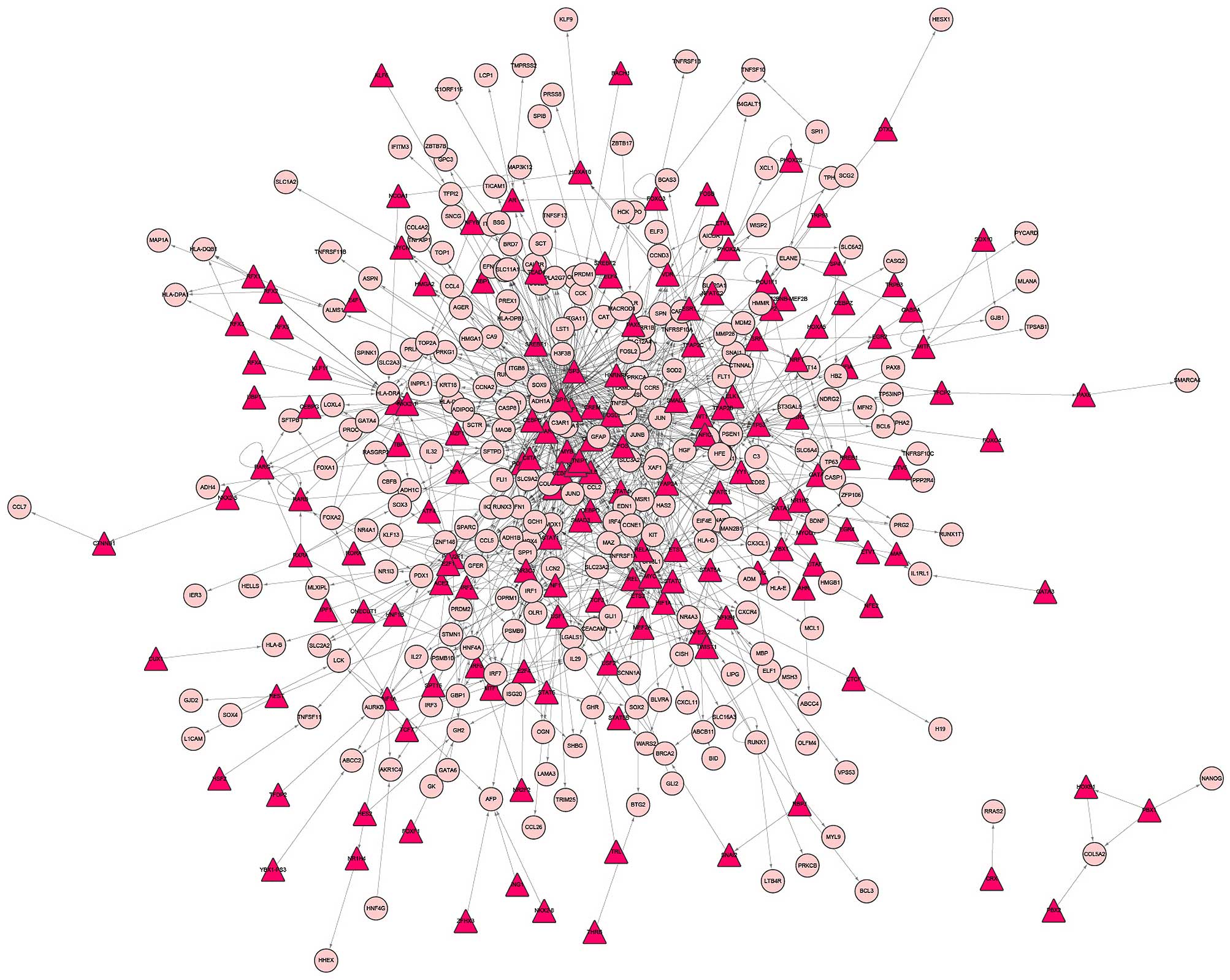
were obtained, including 164 TFs and 274 DEGs (Fig. 1). In addition, a total of 5,632
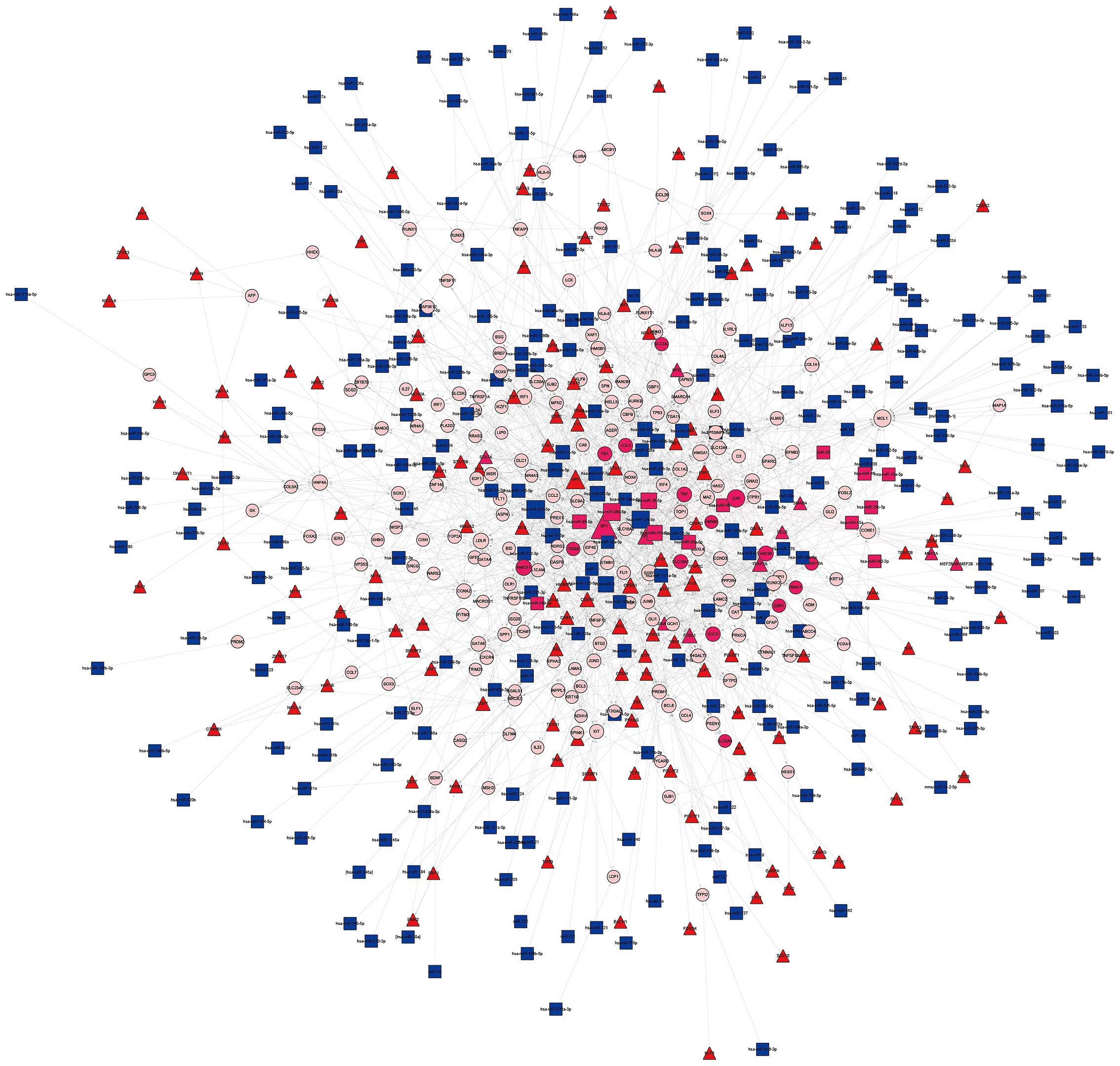
associations between 432 microRNAs and 1,524 DEGs were found
(Fig. 2). Finally, the
co-regulated DEGs, which were regulated by microRNA and TF, and
their corresponding regulators (microRNA and TF) were extracted.
This network contained 195 DEGs, 136 TFs and 283 microRNAs, with
730 associations between the TFs and DEGs and 1,032 associations
between the microRNAs and DEGs (Fig.
3).
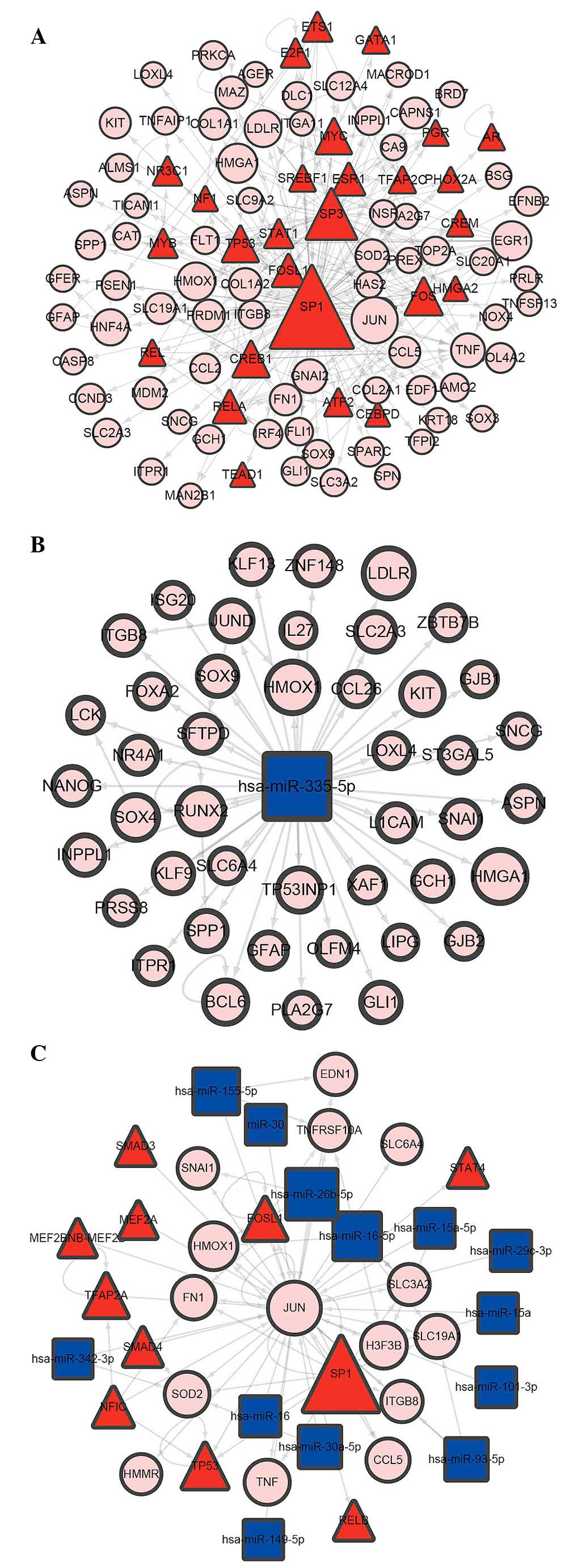
The top 10 DEG nodes, TF nodes and microRNA nodes
with the highest degrees in this synergistic regulatory network are
listed in Table I. The connected
nodes for the DEG, TF and microRNAs with the highest degree were
JUN, SP1 and mir-355-5p, which were considered to be potentially
important regulators in the development of PCOS. In addition, the
sub-networks of JUN, SP1 and mir-355-5p, respectively, were
constructed (Fig. 4).
 | Table ITop 10 TF, microRNA and DEG nodes with
highest degree in the TF-microRNA synergistic regulatory
network. |
Table I
Top 10 TF, microRNA and DEG nodes with
highest degree in the TF-microRNA synergistic regulatory
network.
| Rank | TF | microRNA | DEG |
|---|
| 1 | SP1 | hsa-miR-335-5p | JUN |
| 2 | SP3 | hsa-miR-124-3p | MCL1 |
| 3 | FOS | hsa-miR-26b-5p | EGR1 |
| 4 | CREB1 | hsa-miR-16-5p | HMGA1 |
| 5 | TP53 | hsa-miR-1 | CCNE1 |
| 6 | MYC | hsa-miR-155-5p | HMOX1 |
| 7 | TFAP2A | hsa-miR-98-5p | IRF1 |
| 8 | FOSL1 | hsa-let-7b-5p | LDLR |
| 9 | RELA | hsa-miR-92a-3p | HNF4A |
| 10 | ESR1 | hsa-miR-193b-3p | H3F3B |
Functional enrichment results
To examine the biological functions of different
sets of DEGs, the present study performed KEGG and GO functional
enrichment analyses of the co-regulated DEGs. The results of the
KEGG analysis (Table II) revealed
seven significant KEGG pathways, including Extracellular matrix
(ECM)-receptor interaction, Focal adhesion and Pathways in cancer.
The results of the GO analysis are shown in Table III, and included the positive
regulation of macromolecule metabolic process. The results of the
GO analysis of the mir-355-5p and SP1 sub-network are shown in
Tables IV and V, respectively. To further examine the
biological roles of JUN, the nodes in the JUN sub-network were
annotated to the KEGG pathway, a crucial pathway associated with
WNT signaling, transforming growth factor (TGF) signaling and cell
cycle, were identified, which may be involved in PCOS (Fig. 5).
 | Table IIResults of Kyoto Encyclopedia of Genes
and Genomes pathway enrichment analysis of differentially expressed
genes in the transcription factor-microRNA synergistic regulatory
network. |
Table II
Results of Kyoto Encyclopedia of Genes
and Genomes pathway enrichment analysis of differentially expressed
genes in the transcription factor-microRNA synergistic regulatory
network.
| Term | P-value |
|---|
|
hsa04512:Extracellular matrix-receptor
interaction | 4.03E-07 |
| hsa04510:Focal
adhesion | 4.70E-06 |
| hsa05200:Pathways in
cancer | 3.61E-05 |
|
hsa04060:Cytokine-cytokine receptor
interaction | 1.09E-04 |
| hsa04650:Natural
killer cell mediated cytotoxicity | 8.90E-04 |
| hsa04620:Toll-like
receptor signaling pathway | 0.002948018 |
|
hsa04621:Nucleotide-binding and
oligomerization domain-like receptor signaling pathway | 0.006244647 |
 | Table IIIThe results of GO enrichment analysis
of differentially expressed genes in the transcription
factor-microRNA synergistic regulatory network. |
Table III
The results of GO enrichment analysis
of differentially expressed genes in the transcription
factor-microRNA synergistic regulatory network.
| Category | Term | P-value |
|---|
| GOTERM_BP_FAT | GO:0010604:positive
regulation of macromolecule metabolic process | 1.44E-17 |
| GOTERM_BP_FAT |
GO:0042127:regulation of cell
proliferation | 5.70E-16 |
| GOTERM_BP_FAT | GO:0031328:positive
regulation of cellular biosynthetic process | 6.54E-16 |
| GOTERM_BP_FAT | GO:0009891:positive
regulation of biosynthetic process | 1.12E-15 |
| GOTERM_BP_FAT | GO:0051173:positive
regulation of nitrogen compound metabolic process | 2.60E-15 |
| GOTERM_CC_FAT |
GO:0044421:extracellular region part | 1.02E-12 |
| GOTERM_CC_FAT |
GO:0005615:extracellular space | 7.63E-12 |
| GOTERM_CC_FAT |
GO:0005576:extracellular region | 2.72E-07 |
| GOTERM_CC_FAT | GO:0045121:membrane
raft | 3.16E-07 |
| GOTERM_CC_FAT |
GO:0005578:proteinaceous extracellular
matrix | 1.68E-05 |
| GOTERM_MF_FAT |
GO:0003700:transcription factor
activity | 4.55E-12 |
| GOTERM_MF_FAT |
GO:0043565:sequence-specific DNA
binding | 9.55E-10 |
| GOTERM_MF_FAT |
GO:0030528:transcription regulator
activity | 5.69E-09 |
| GOTERM_MF_FAT |
GO:0016563:transcription activator
activity | 1.03E-08 |
| GOTERM_MF_FAT |
GO:0042802:identical protein binding | 1.93E-07 |
 | Table IVResults of GO enrichment analysis of
targets of SP1 in the transcription factor-microRNA synergistic
regulatory network. |
Table IV
Results of GO enrichment analysis of
targets of SP1 in the transcription factor-microRNA synergistic
regulatory network.
| Category | Term | P-value |
|---|
| GOTERM_BP_FAT | GO:0010604:positive
regulation of macromolecule metabolic process | 2.45E-16 |
| GOTERM_BP_FAT |
GO:0043067:regulation of programmed cell
death | 2.79E-14 |
| GOTERM_BP_FAT |
GO:0010941:regulation of cell death | 3.08E-14 |
| GOTERM_BP_FAT |
GO:0042127:regulation of cell
proliferation | 9.07E-14 |
| GOTERM_BP_FAT |
GO:0042981:regulation of apoptosis | 1.56E-13 |
| GOTERM_CC_FAT | GO:0045121:membrane
raft | 5.76E-07 |
| GOTERM_CC_FAT |
GO:0044421:extracellular region part | 1.54E-06 |
| GOTERM_CC_FAT |
GO:0005667:transcription factor
complex | 1.71E-06 |
| GOTERM_CC_FAT |
GO:0005615:extracellular space | 1.51E-05 |
| GOTERM_CC_FAT |
GO:0031974:membrane-enclosed lumen | 2.90E-05 |
| GOTERM_MF_FAT |
GO:0043565:sequence-specific DNA
binding | 1.33E-14 |
| GOTERM_MF_FAT |
GO:0003700:transcription factor
activity | 1.28E-13 |
| GOTERM_MF_FAT |
GO:0030528:transcription regulator
activity | 1.53E-10 |
| GOTERM_MF_FAT |
GO:0016563:transcription activator
activity | 2.63E-07 |
| GOTERM_MF_FAT | GO:0003677:DNA
binding | 3.64E-07 |
 | Table VResults of GO enrichment analysis of
targets of mir-355-5p in the transcription factor-microRNA
synergistic regulatory network. |
Table V
Results of GO enrichment analysis of
targets of mir-355-5p in the transcription factor-microRNA
synergistic regulatory network.
| Category | Term | P-value |
|---|
| GOTERM_BP_FAT | GO:0010557:positive
regulation of macromolecule biosynthetic process | 2.59E-06 |
| GOTERM_BP_FAT | GO:0031328:positive
regulation of cellular biosynthetic process | 4.06E-06 |
| GOTERM_BP_FAT | GO:0009891:positive
regulation of biosynthetic process | 4.67E-06 |
| GOTERM_BP_FAT | GO:0010604:positive
regulation of macromolecule metabolic process | 5.52E-06 |
| GOTERM_BP_FAT | GO:0010628:positive
regulation of gene expression | 6.63E-06 |
| GOTERM_CC_FAT |
GO:0005615:extracellular space | 1.70E-06 |
| GOTERM_CC_FAT |
GO:0044421:extracellular region part | 6.94E-06 |
| GOTERM_CC_FAT |
GO:0005576:extracellular region | 0.006937 |
| GOTERM_CC_FAT | GO:0045121:membrane
raft | 0.007474 |
| GOTERM_CC_FAT | GO:0031981:nuclear
lumen | 0.016719 |
| GOTERM_MF_FAT | GO:0003702:RNA
polymerase II transcription factor activity | 5.83E-06 |
| GOTERM_MF_FAT |
GO:0003700:transcription factor
activity | 1.30E-05 |
| GOTERM_MF_FAT |
GO:0030528:transcription regulator
activity | 2.24E-04 |
| GOTERM_MF_FAT |
GO:0043565:sequence-specific DNA
binding | 3.07E-04 |
| GOTERM_MF_FAT |
GO:0016563:transcription activator
activity | 0.00112 |
Discussion
In the present study, based on the GSE34526 dataset
from the GEO database, TF-target associations from the CHIPBase
database and microRNA-target associations from the miRTarBase
database, a TF-microRNA synergistic regulatory network was
constructed, which included 136 TFs, 283 microRNAs and 195 DEGs.
The interactions among the TFs, microRNAs and DEGs were then
collected, and their potential roles in the development of PCOS
were investigated in terms of their level of transcription. The
DEGs in this network were examined through KEGG pathway and GO
enrichment analyses. From this, seven pathways were identified,
including ECM-receptor interaction, Focal adhesion and Pathways in
cancer. For example, the ECM-receptor regulates TGF-β signaling,
which is reported to contribute to the pathogenesis of PCOS
(28). In addition, Toll-like
receptor (TLR) 4 is involved in insulin resistance, a major cause
of several co-morbidities in PCOS (29). Several other signaling associated
with inflammation are also involved in PCOS, with the exception of
ECM- and TLRs (18). The results
of the GO analysis suggested that certain GO terms, including the
Positive regulation of the macromolecule metabolic process and
Extracellular region, may be involved in the development of PCOS.
Table III shows the top five
significant terms for the biological process, cellular component
and molecular function, respectively.
The present study also demonstrated that, in this
TF-microRNA synergistic regulatory network, certain nodes were
connected to several other types of nodes. For the TF nodes, SP1
was connected to up to 104 target genes, suggesting its importance
in PCOS. Studies have indicated that SP1 is a key mediator of gene
expression induced by insulin, which suggested that SP1 may
interact with genes of insulin resistance in PCOS (30,31).
To further examine this, the present study extracted the SP1
sub-network, in which the nodes were SP1 and its targets from the
TF-microRNA synergistic regulatory network, and GO analysis of the
nodes in this sub-network were implemented. Certain terms
associated with cell death (GO:0043067: regulation of programmed
cell death and GO:0010941: regulation of cell death) were
identified. The annotated genes in these terms predominantly
included DLC1, E2F1, TNF, TNFSF13, KIT, SOX9, GLI1 and FOS. Studies
have shown that the overexpression of anti-apoptotic factors and
the downregulation of apoptosis in PCOS may contribute to the
ovarian polycystic appearance (32,33).
The most connected microRNA nodes identified in the
present study were mir-355-5p, which targeted 44 DEGs. Similar to
SP1, an mir-355-5p network was also constructed, and GO analysis
was implemented. Although no direct evidence of the role of
mir-355-5p in PCOS have been reported until now, its targets,
including RUNX2, were considered as candidate genetic markers in
the monitoring of embryo quality for patients with PCOS (34). Furthermore, the present study
identified certain GO terms, including GO:0010557: Positive
regulation of macromolecule biosynthetic process, in which the
targeted DEGs of mir-355-5p were enriched (Table V). The biological functions of the
mir-355-5p targets also indicated the mechanism of PCOS
development.
Notably, certain DEGs in the TF-microRNA synergistic
regulatory network also acted as TFs. These nodes were not only
differentially expressed between these PCOS and normal samples, but
also regulated several targets and were regulated by microRNAs. For
example, JUN, the DEG node with the highest degree, was found to
interact directly with specific target DNA sequences to regulate
gene expression (35). Another
study showed that the expression level of JUN was decreased in PCOS
(24). The present study annotated
JUN to the KEGG pathways. The results revealed that it was involved
in the WNT signaling pathway, which is a potent regulator of
adipogenesis and has also been found to be differentially expressed
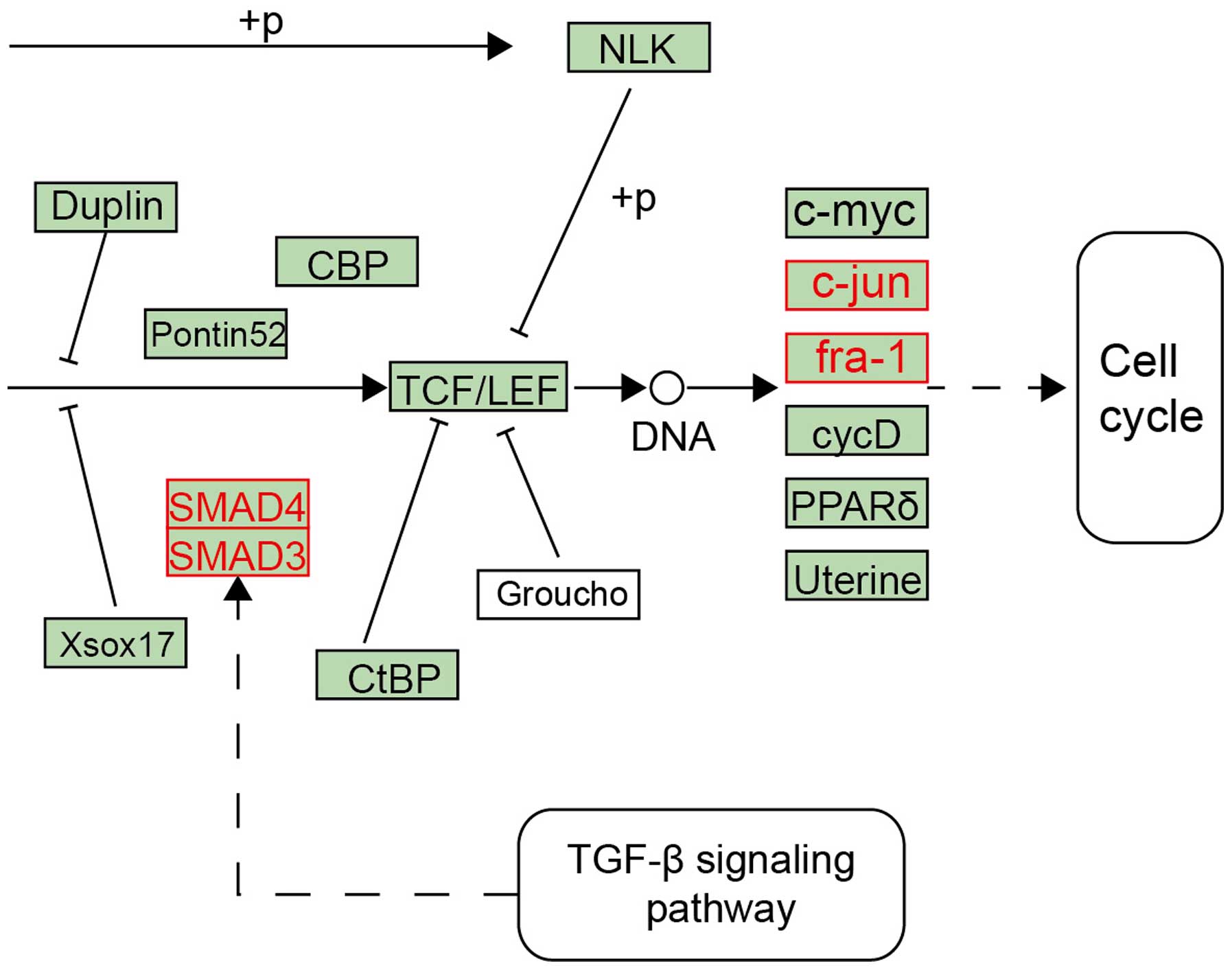
in adipose tissue from non-obese patients with PCOS (36). As shown in Fig. 5, a local region in the WNT
signaling pathway was observed, in which red nodes represented DEGs
of PCOS, including small mothers against decapentaplegic (SMAD)3,
SMAD4, fra-1 and JUN. These genes were located in the crosstalk of
the WNT signaling pathway, TGF-β signaling pathway and cell cycle.
Of these genes, the JUN and fra-1 mediated cell cycle pathway;
SMAD3 and SMAD4 were downstream of TGF-β, which was found to be
increased by 3–4 fold in PCOS. This its dysregulation may
contribute to (1) the fetal
origins of PCOS, (2) reproductive
abnormalities in PCOS and (4)
cardiovascular and metabolic abnormalities in PCOS (28,37),
suggesting JUN-associated pathways may be important in PCOS.
In conclusion, the present study constructed a
PCOS-associated TF-microRNA synergistic regulatory network and
revealed the potential mechanism of PCOS on transcriptional
regulation levels. Furthermore, certain crucial regulators in PCOS
were identified. These findings provided novel insights for
understanding the mechanism of PCOS and provide a potential
reference for therapeutic strategies in the treatment of PCOS.
Acknowledgments
This study was supported by the Project on Science
and Technology Department of Guangdong Province (grant no.
2013B022000022) and by the Medical Scientific Research Foundation
of Guangdong Province (grant no. A2014316).
References
|
1
|
Sirmans SM and Pate KA: Epidemiology,
diagnosis and management of polycystic ovary syndrome. Clin
Epidemiol. 6:1–13. 2013. View Article : Google Scholar
|
|
2
|
Raja-Khan N, Stener-Victorin E, Wu X and
Legro RS: The physiological basis of complementary and alternative
medicines for polycystic ovary syndrome. American journal of
physiology. Am J Physiol Endocrinol Metab. 301:E1–E10. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Trivax B and Azziz R: Diagnosis of
polycystic ovary syndrome. Clin Obstet Gynecol. 50:168–177. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Joham AE, Ranasinha S, Zoungas S, Moran L
and Teede HJ: Gestational diabetes and type 2 diabetes in
reproductive-aged women with polycystic ovary syndrome. J Clin
Endocrinol Metab. 99:447–452. 2014.
|
|
5
|
Nasrat H, Patra SK, Goswami B, Jain A and
Raghunandan C: Study of association of leptin and insulin
resistance markers in patients of PCOS. Indian J Clin Biochem.
31:104–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kao YH, Chiu WC, Hsu MI and Chen YJ:
Endothelial progenitor cell dysfunction in polycystic ovary
syndrome: Implications for the genesis of cardiovascular diseases.
Int J Fertil Steril. 6:208–213. 2013.
|
|
7
|
Chang AY, Oshiro J, Ayers C and Auchus RJ:
Influence of race/ethnicity on cardiovascular risk factors in
polycystic ovary syndrome, the Dallas Heart Study. Clin Endocrinol
(Oxf). Nov 26–2015.Epub ahead of print. View Article : Google Scholar
|
|
8
|
Cheng C, Zhang H, Zhao Y, Li R and Qiao J:
Paternal history of diabetes mellitus and hypertension affects the
prevalence and phenotype of PCOS. J Assist Reprod Genet.
32:1731–1739. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee YJ, Jeong JE, Joo JK and Lee KS: A
case of idiopathic intracranial hypertension associated with PCOS.
Clin Exp Obstet Gynecol. 42:547–549. 2015.PubMed/NCBI
|
|
10
|
Bazarganipour F, Ziaei S, Montazeri A,
Foroozanfard F, Kazemnejad A and Faghihzadeh S: Psychological
investigation in patients with polycystic ovary syndrome. Health
Qual Life Outcomes. 11:1412013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qi X, Pang Y and Qiao J: Role of
Anti-Müllerian Hormone in the pathogenesis and pathophysiological
characteristics of polycystic ovary syndrome. Eur J Obstet Gynecol
Reprod Biol. 199:82–87. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding L, Gao F, Zhang M, Yan W, Tang R,
Zhang C and Chen ZJ: Higher PDCD4 expression is associated with
obesity, insulin resistance, lipid metabolism disorders, and
granulosa cell apoptosis in polycystic ovary syndrome. Fertil
Steril. Feb 8–2016.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Palioura E and Diamanti-Kandarakis E:
Polycystic ovary syndrome (PCOS) and endocrine disrupting chemicals
(EDCs). Rev Endocr Metab Disord. Jan 29–2016.Epub ahead of print.
PubMed/NCBI
|
|
14
|
Rajender S, Carlus SJ, Bansal SK, Negi MP,
Sadasivam N, Sadasivam MN and Thangaraj K: Androgen receptor CAG
repeats length polymorphism and the risk of polycystic ovarian
syndrome (PCOS). PLoS One. 8:e757092013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gul OO, Cander S, Gul B, Acikgoz E,
Sarandol E and Ersoy C: Evaluation of insulin resistance and plasma
levels for visfatin and resistin in obese and non-obese patients
with polycystic ovary syndrome. Eur Cytokine Netw. 26:73–78.
2015.
|
|
16
|
Rice S, Elia A, Jawad Z, Pellatt L and
Mason HD: Metformin inhibits follicle-stimulating hormone (FSH)
action in human granulosa cells: Relevance to polycystic ovary
syndrome. J Clin Endocrinol Metab. 98:E1491–E1500. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li D, Li C, Xu Y, Xu D, Li H, Gao L, Chen
S, Fu L, Xu X, Liu Y, et al: Differential expression of microRNAs
in the ovaries from letrozole-induced rat model of polycystic ovary
syndrome. DNA Cell Biol. Jan 8–2016.Epub ahead of print. View Article : Google Scholar
|
|
18
|
Ojeda-Ojeda M, Murri M, Insenser M and
Escobar-Morreale HF: Mediators of low-grade chronic inflammation in
polycystic ovary syndrome (PCOS). Curr Pharm Des. 19:5775–5791.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roth LW, McCallie B, Alvero R, Schoolcraft
WB, Minjarez D and Katz-Jaffe MG: Altered microRNA and gene
expression in the follicular fluid of women with polycystic ovary
syndrome. J Assist Reprod Genet. 31:355–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hossain MM, Cao M, Wang Q, Kim JY,
Schellander K, Tesfaye D and Tsang BK: Altered expression of miRNAs
in a dihydrotestosterone-induced rat PCOS model. J Ovarian Res.
6:362013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sang Q, Yao Z, Wang H, Feng R, Wang H,
Zhao X, Xing Q, Jin L, He L and Wu L: Identification of microRNAs
in human follicular fluid: Characterization of microRNAs that
govern steroidogenesis in vitro and are associated with poly-cystic
ovary syndrome in vivo. J Clin Endocrinol Metab. 98:3068–3079.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen YH, Heneidi S, Lee JM, Layman LC,
Stepp DW, Gamboa GM, Chen BS, Chazenbalk G and Azziz R: miRNA-93
inhibits GLUT4 and is overexpressed in adipose tissue of polycystic
ovary syndrome patients and women with insulin resistance.
Diabetes. 62:2278–2286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic acids research.
42:D78–D85. 2014. View Article : Google Scholar :
|
|
24
|
Yang JH, Li JH, Jiang S, Zhou H and Qu LH:
ChIPBase: A database for decoding the transcriptional regulation of
long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic
Acids Res. 41:D177–D187. 2013. View Article : Google Scholar :
|
|
25
|
Kaur S, Archer KJ, Devi MG, Kriplani A,
Strauss JF III and Singh R: Differential gene expression in
granulosa cells from polycystic ovary syndrome patients with and
without insulin resistance: Identification of susceptibility gene
sets through network analysis. J Clin Endocrinol Metab.
97:E2016–E2021. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic acids research.
37:1–13. 2009. View Article : Google Scholar
|
|
27
|
Yang JH, Li JH, Jiang S, Zhou H and Qu LH:
ChIPBase: A database for decoding the transcriptional regulation of
long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic
Acids Res. 41:D177–D187. 2013. View Article : Google Scholar :
|
|
28
|
Raja-Khan N, Urbanek M, Rodgers RJ and
Legro RS: The role of TGF-β in polycystic ovary syndrome. Reprod
Sci. 21:20–31. 2014. View Article : Google Scholar
|
|
29
|
Uchimura K, Hayata M, Mizumoto T, Miyasato
Y, Kakizoe Y, Morinaga J, Onoue T, Yamazoe R, Ueda M, Adachi M, et
al: The serine protease prostasin regulates hepatic insulin
sensitivity by modulating TLR4 signalling. Nat Commun. 5:34282014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Solomon SS, Majumdar G, Martinez-Hernandez
A and Raghow R: A critical role of Sp1 transcription factor in
regulating gene expression in response to insulin and other
hormones. Life Sci. 83:305–312. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Anjali G, Kaur S, Lakra R, Taneja J,
Kalsey GS, Nagendra A, Shrivastav TG, Gouri Devi M, Malhotra N,
Kriplani A and Singh R: FSH stimulates IRS-2 expression in human
granulosa cells through cAMP/SP1, an inoperative FSH action in PCOS
patients. Cell Signal. 27:2452–2466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jansen E, Laven JS, Dommerholt HB, Polman
J, van Rijt C, van den Hurk C, Westland J, Mosselman S and Fauser
BC: Abnormal gene expression profiles in human ovaries from
polycystic ovary syndrome patients. Mol Endocrinol. 18:3050–3063.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee BS, Oh J, Kang SK, Park S, Lee SH,
Choi D, Chung JH, Chung YW and Kang SM: Insulin protects cardiac
myocytes from doxorubicin toxicity by Sp1-mediated transactivation
of survivin. PLoS One. 10:e01354382015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang X, Hao C, Shen X, Zhang Y and Liu X:
RUNX2, GPX3 and PTX3 gene expression profiling in cumulus cells are
reflective oocyte/embryo competence and potentially reliable
predictors of embryo developmental competence in PCOS patients.
Reprod Biol Endocrinol. 11:1092013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang A, Al-Kuhlani M, Johnston SC, Ojcius
DM, Chou J and Dean D: Transcription factor complex AP-1 mediates
inflammation initiated by Chlamydia pneumoniae infection. Cell
Microbiol. 15:779–794. 2013. View Article : Google Scholar :
|
|
36
|
Chazenbalk G, Chen YH, Heneidi S, Lee JM,
Pall M, Chen YD and Azziz R: Abnormal expression of genes involved
in inflammation, lipid metabolism and Wnt signaling in the adipose
tissue of polycystic ovary syndrome. J Clin Endocrinol Metab.
97:E765–E770. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Saikumar P, Selvi VK, Prabhu K, Venkatesh
P and Krishna P: Anti mullerian hormone: A potential marker for
recruited non growing follicle of ovarian pool in women with
polycystic ovarian syndrome. J Clin Diagn Res. 7:1866–1869.
2013.PubMed/NCBI
|