Introduction
Leukemia is the most common malignancy in children
(0–14 years) and young adults (15–39 years). According to GLOBOCAN
2012 data, 351,965 new cases of leukemia were diagnosed in 2012
(2.5% of total cancer cases), and leukemia accounts for ~265,471
cases of cancer-associated mortality worldwide every year. Among
these newly diagnosed cases, 30% (105,436/351,965 cases) occurred
in the USA and China (1). In
addition, leukemia has the highest incidence and mortality rate of
any cancer in children in China. The majority of cases of leukemia
are acute myeloid leukemia (AML). AML represents a heterogeneous
hematological malignancy characterized by uncontrollable
proliferation and survival, and impaired differentiation of
neoplastic cells or blasts (2). In
the USA, 19,950 cases of AML and 10,430 cases of AML-associated
mortality are projected in 2016 (3). AML is often fatal, due to the
following reasons: Firstly, the majority of cases of AML initially
respond to traditional chemotherapy (4); however, relapse is common indicating
resistance of malignant cells to chemotherapy (4–7); and
secondly, the acuity and severity of AML is often poor at
diagnosis. Recently, studies have focused on identifying novel
genes, which target tumor cell growth and survival signaling
pathways (8–11). Identification of these genes may
provide a novel strategy for the optimization of AML therapy or
diagnosis (12).
The ribosome is comprised of two ribonucleoprotein
subunits: 40S and 60S, which are known as the 'small' and 'large'
subunits, respectively. It has previously been reported that
ribosomal proteins exhibit extra-ribosomal functions, which
include, but are not limited to, DNA repair, cell death,
inflammation, tumorigenesis and transcriptional regulation
(13). Ribosomal protein S15a
(RPS15A), which is a component of the 40S ribosomal subunit, is
able to promote mRNA/ribosome interaction during the early stage of
translation. RPS15A has been shown to suppress the cdc33 mutation
growth arrest phenotype by binding and stabilizing eukaryotic
initiation factor 4E (14), thus
indicating that RPS15A has a function in cell growth regulation. In
addition, RPS15A has been identified as a novel internal control
gene for use in quantitative polymerase chain reaction (qPCR)
analysis of prepubertal bovine mammary tissue (15). Akiyama et al (16) demonstrated that RPS15A is
upregulated in response to transforming growth factor-β in the A549
human lung carcinoma cell line, which has a central role in the
regulation of proliferation, differentiation, apoptosis and
carcinogenesis. Furthermore, Zeller et al (17) identified RPS15A as a responsive
gene of the Myc oncogenic transcription factor. Krüppel-like factor
4 is a tumor suppressor (18),
which is able to inhibit the GC content of the RPS15A promoter to
downregulate its expression (19).
In addition, RPS15A was shown to be overexpressed in hepatitis B
virus-encoded X antigen-positive cells, and overexpression of
RPS15A stimulated cell growth, colony formation and tumor formation
in SCID mice in vivo (20),
thus indicating that RPS15A may have a key role in hepatocellular
carcinogenesis. A meta-analysis of cancer gene expression
signatures revealed that RPS15A is highly expressed in astrocytoma,
colorectal cancer and prostate cancer (21).
Despite reports indicating that RPS15A may stimulate
growth in yeast, plants and human cancer (22), its functional role in AML remains
unknown. The present study demonstrated that knockdown of RPS15A by
lentivirus-mediated short hairpin RNA (shRNA) was able to inhibit
AML cell proliferation and induce apoptosis in vitro.
Furthermore, RPS15A knockdown suppressed cell cycle progression via
G0/G1 phase arrest. Collectively, RPS15A may
modulate AML cell growth and have a prominent role in AML;
therefore, RPS15A may be considered a potential therapeutic target
for the treatment of AML.
Materials and methods
Cell lines and culture conditions
Human embryonic kidney 293T (HEK 293T) cells were
cultured in Dulbecco's modified Eagle's medium (Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal bovine serum (FBS; Biowest SAS, Nuaillé, France). Human acute
myeloid leukemia (AML) U937 cells were cultured in RPMI-1640
(Hyclone; GE Healthcare Life Sciences) supplemented with 10% FBS.
The cell lines were purchased from the Cell Bank of Chinese Academy
of Science (Shanghai, China), and were maintained at 37°C in a
humidified incubator containing 5% CO2.
Preparation of shRNA-expressing
lentiviruses and cell infection
Two targeted sequences (S1, S2) designed to be
homologous to RPS15A were cloned into the lentiviral expression
vector pFH-L (Shanghai Hollybio, Shanghai, China), which was
digested by NheI and PacI restriction enzymes (Takara
Bio, Inc., Otsu, Japan). The sequences of the two shRNA targeting
RPS15A were as follows: S1,
5′-GTGCAACTCAAAGACCTGGAACTCGAGTTCCAGGTCTTTGAGTTGCACTTTTT-3′ and S2,
5′-GCATGGTTACATTGGCGAATTCTCGAGAATTCGCCAATGTAACCATGCTTTTT-3′
(Shanghai Hollybio). The lentiviruses containing S1 and S2
sequences were classified as Lv-shRPS15A (S1) and Lv-shRPS15A (S2),
respectively. As a negative control, a scrambled shRNA was used
(Shanghai Hollybio), which had the following sequence:
5′-GCGGAGGGTTGAAAGAATATCTCGAGATATTCTTTCAAACCCTCCGCTTTTTT-3′. This
sequence was classified as Lv-shCon. The uninfected control cells
were classified as Con. Each single-stranded oligonucleotide became
a double-stranded oligonucleotide using an annealing system.
Briefly, the annealing reaction mixture, containing 5 µl
each of forward and reverse primers (Shanghai Hollybio), 10
µl 5X annealing buffer (Beyotime Institute of Biotechnology,
Shanghai, China) and 30 µ1 double-distilled H2O
(ddH2O), was run on a Bioer TC-96/G/H(b)A (BIOER,
Hangzhou, China) thermal cycler with the following program: Initial
denaturation at 94°C for 1 min, followed by 50 cycles of
denaturation at 80°C for 30 sec and extension at 30°C for 30 sec,
followed by cooling to 4°C at the end of the PCR. Subsequently, the
double-stranded oligonucleotide was cloned into the linearized
pFH-L vector.
The ligation product was used to transform the
Escherichia coli DH5α strain, and was extracted using a
plasmid purification kit [Tiangen Biotech (Beijing) Co., Ltd.,
Beijing, China]. The plasmid was confirmed by PCR and sequencing.
The shRNA-expressing lentiviruses were produced by co-transfection
of 10 µg recombinant expression shRNA vectors and packaging
pHelper plasmids (7.5 µg pVSVG-I and 5 µg pCMVΔR8.92;
Shanghai Hollybio) into the HEK 293T cells using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
according to the manufacturer's protocol. Supernatants containing
either the lentivirus expressing the RPS15A shRNA or the control
shRNA were harvested 24, 48 and 72 h post-transfection. The
lentiviruses were purified using ultracentrifugation, and the titer
of the lentiviruses was determined as previously described
(23). U937 cells were infected
with the concentrated virus at a multiplicity of infection of 80
and mock-infected cells were used as negative controls. After 96 h
of infection, the expression of green fluorescent protein was
observed using a fluorescent microscope (CKX41; Olympus
Corporation, Tokyo, Japan) in order to assess the infection
efficiency. The efficiency of RPS15A knockdown was evaluated by
reverse transcription (RT)-qPCR and western blot analysis.
RNA isolation and RT-qPCR
U937 cells were harvested 5 days post-lentiviral
infection. Total RNA was extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The purity and integrity of the RNA was
assessed by spectrophotometry (Epoch Microplate Spectrophotometer;
Biotek Instruments, Inc., Winooski, VT, USA) and 3% agarose gel
electrophoresis, respectively. First-strand cDNA was synthesized
from 2 µg total RNA using RT reagents containing 1 µl
Oligo dT (0.5 µg/µl), 4 µl M-MLV Buffer, 1.25
µl dNTPs, 0.5 µl RNasin, 0.75 µl M-MLV-RTase
and Nuclease-free water, to a final volume of 20 µl (Promega
Corporation, Madison, WI, USA). PCR primers (Shanghai Hollybio)
were designed to amplify fragments that span intron/exon boundaries
and the sequences were as follow: RPS15A, forward
5′-TGACGTGCAACTCAAAGACC-3′, reverse 5′-CCAGAGTCCATGAGGCATT-3′;
β-actin, forward 5′-GTGGACATCCGCAAAGAC-3′, and reverse
5′-AAAGGGTGTAACGCAACTA-3′. RT-qPCR was performed in the linear
range using the SYBR Green Core Reagents kit (Takara Bio, Inc.) on
a Bio-Rad CFX96 Touch™ Real-Time PCR system (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The PCR reaction mixture consisted of 5
µl cDNA, 10 µl 2X SYBR Premix Ex Taq (Takara Bio,
Inc.), 0.8 µl PCR primers (2.5 µM) and 4.2 µl
ddH2O. The PCR cycling conditions were as follows:
Initial denaturation at 95°C for 60 sec, 45 cycles of denaturation
at 95°C for 5 sec, annealing and extension at 60°C for 20 sec,
followed by cooling to 4°C. The absorbance values were read at the
extension stage. β-actin was used as the internal control for all
normalizations and the relative expression levels were calculated
using the 2−ΔΔCq method (24).
Western blot analysis
U937 cells were harvested 5 days post-lentiviral
infection, were washed twice with ice-cold phosphate-buffered
saline (PBS), and were lysed in ice-cold 2× sodium dodecyl sulfate
(SDS) Lysis Buffer [100 mM Tris-HCl (pH 6.8), 10 mM EDTA, 4% SDS,
10% Glycine]. The protein concentration of the cell lysates was
determined using the bicinchoninic acid protein assay kit (Thermo
Scientific Pierce, Rockford, IL, USA). Total protein samples (30
µg) were separated by 10% SDS-polyacrylamide gel
electrophoresis and were transferred onto nitrocellulose membranes.
The membranes were blocked with Tris-buffered saline and Tween 20
(Sigma-Aldrich, St. Louis, MO, USA) containing 5% non-fat milk at
room temperature for 2 h, after which they were incubated with the
following antibodies: Anti-RPS15A (1:1,000; cat. no. AP4804a;
Abgent, San Diego, CA, USA) and anti-glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) (1:40,000; cat. no. 10494-1-AP; Proteintech
Group, Inc., Chicago, IL, USA) at 4°C overnight. Subsequently, the
membranes were incubated with horseradish peroxidase-conjugated
goat anti-rabbit secondary antibodies (1:5,000; cat. no. sc-2054;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Signals were
detected using an enhanced chemiluminescence test kit (Amersham; GE
Healthcare Life Sciences, Chalfont, UK). GAPDH served as the
internal standard. Density analysis was performed using Quantity
One software, version 4.62 (Bio-Rad Laboratories, Inc.).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay-growth curve
In vitro cell viability was analyzed using
the MTT assay. U937 cells were seeded at a density of 3,500
cells/well in 96-well plates 96 h post-lentiviral infection. After
24 h, 20 µl 5 mg/ml MTT solution (Sigma-Aldrich) was added
to each well daily between days 1 and 5, and the plates were
incubated for 4 h at 37°C. Subsequently, 100 µl stop buffer
(0.012 M HCl, 10% SDS, 5% isopropanol) was added to each well and
gently agitated for 10 min. Absorbance values were measured at a
wavelength of 595 nm using the Epoch Microplate
Spectrophotometer.
Cell cycle analysis
To determine cell cycle distribution,
8×104 U937 cells were seeded in 6 cm dishes a total of 6
days after lentiviral infection. Following a 40 h culture, the
cells were washed twice with ice-cold PBS and were resuspended in
PBS containing 50 µg/ml RNase A (Sigma-Aldrich) and 50
µg/ml propidium iodide (Sigma-Aldrich). Cells were incubated
at 37°C in the dark for 1 h. The percentage of cells in each phase
of the cell cycle was measured using FACScan (BD Biosciences, San
Diego, CA, USA) and results were analyzed using ModFit software,
version 3.2 (Verity Software House, Topsham, ME, USA).
Apoptosis analysis
To assess the apoptotic rate, 1×105 U937
cells were seeded in 6 cm dishes. Apoptosis was detected 6 days
following lentiviral infection. Cells were harvested and the
experiment was conducted according to the Annexin V-allophycocyanin
(APC)/7-aminoactinomycin D (7-AAD) Apoptosis Assay kit (Nanjing
KeyGen Biotech Co., Ltd., Nanjing, China). A total of
1×106 cells were resuspended in 100 µl 1X Annexin
V binding buffer with 5 µl Annexin V-APC and 5 µl
7-AAD and were incubated for 15 min at room temperature in the
dark. The cells were analyzed on a FACSCalibur (BD Biosciences)
using CellQuest Pro software (BD Biosciences). The percentage of
each quadrant was calculated.
Statistical analysis
Results are presented as the mean ± standard
deviation. Differences between the groups were assessed using the
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference. Statistical analyses were
performed using SPSS 13.0 statistical software (SPSS, Inc.,
Chicago, IL, USA).
Results
Lentivirus-mediated shRNA inhibits the
expression of RPS15A in U937 cells
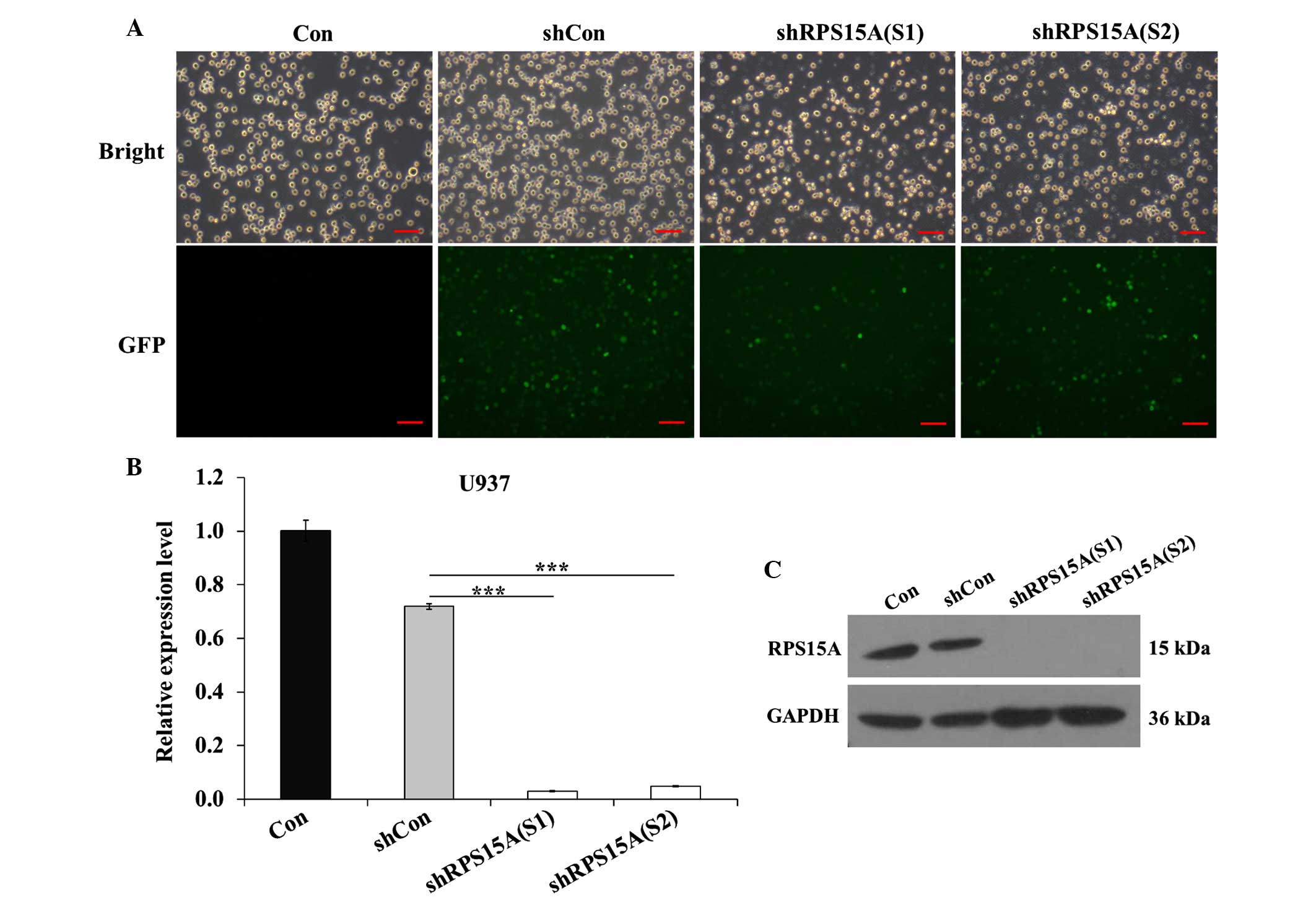
The U937 cell line was infected with shCon, shRPS15A
(S1) and shRPS15A (S2) lentiviral particles. Infection efficiency
was >70%, as determined by detecting the expression of green
fluorescent protein 96 h post-infection (Fig. 1A). qPCR analysis demonstrated that
the RPS15A mRNA expression levels were significantly reduced in the
shRPS15A (S1; P<0.001) and shRPS15A (S2; P<0.001) groups
compared with the shCon and control groups (Fig. 1B). The protein expression levels of
RPS15A were also markedly decreased in the shRPS15A (S1) and
shRPS15A (S2) groups compared with the shCon and control groups
(Fig. 1C). The RPS15A knockdown
efficacy of shRPS15A (S1) and shRPS15A (S2) was 96.0 and 93.2%,
respectively. These results suggest that lentivirus-mediated shRNA
knockdown of RPS15A expression was specific, and the off-target
effects were eliminated. These results indicate that
lentivirus-mediated RPS15A shRNA was able to significantly
downregulate RPS15A expression in U937 cells.
Knockdown of RPS15A inhibits
proliferation of U937 cells
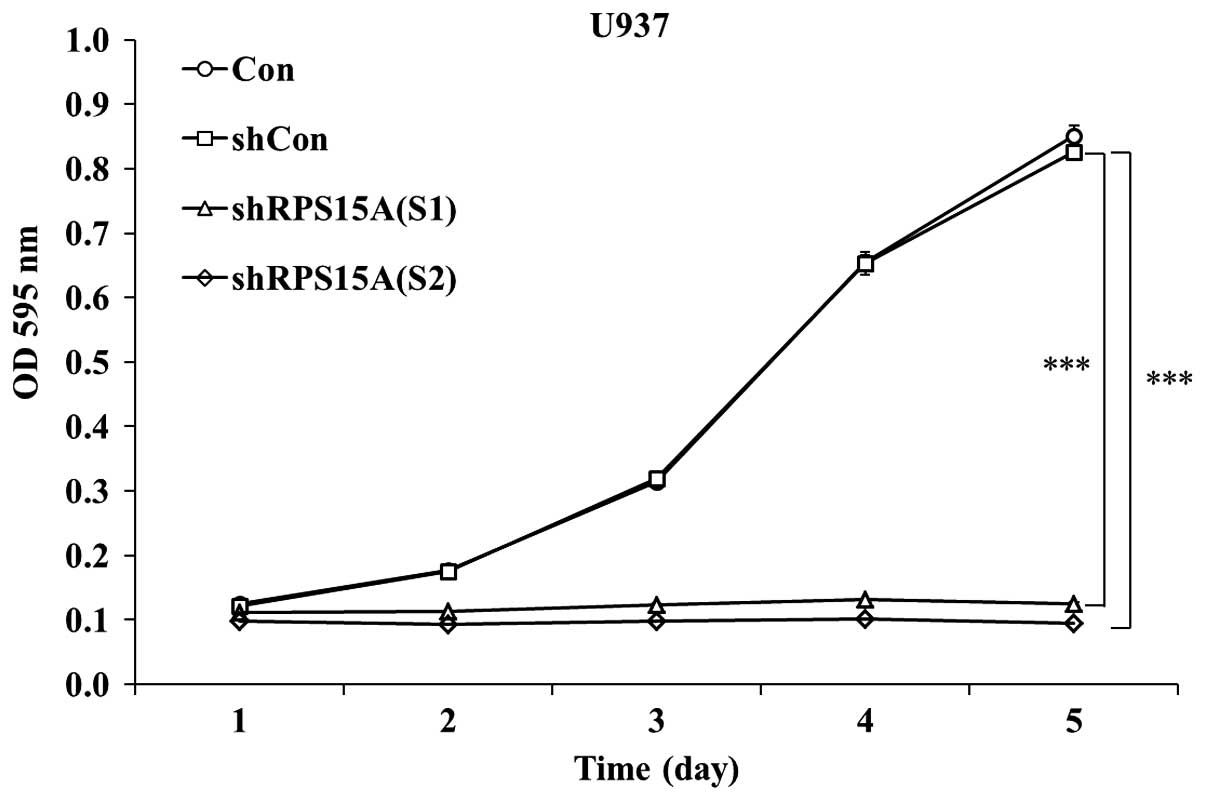
To examine the effects of RPS15A knockdown on U937
cell growth, shRPS15A (S1)-, shRPS15A (S2)- and shCon-infected U937
cells were subjected to the MTT assay. As shown in Fig. 2, cell proliferation in the shRPS15A
(S1) and shRPS15A (S2) groups was significantly reduced;
proliferation decreased by >80% in the shRPS15A (S1) group
(P<0.001) and by >84.5% in the shRPS15A (S2) group
(P<0.001; Fig. 2) at days 4 or
5. Since proliferation inhibition occurred to a greater extent in
the shRPS15A (S2) group, these cells were selected for further
experimentation. Notably, the proliferative index [proliferative
index = (S + G2/M)/(G0/G1 + S +
G2/M)] of shRPS15A (S2)-transduced U937 cells was
slightly lower than that of the shCon and control cells (35.45±3.2
vs. 50±3.8 and 51.3±4.2%; data from Fig. 3). These results indicate that the
proliferation of U937 cells was significantly inhibited following
RPS15A knockdown compared with in the shCon and control groups.
Knockdown of RPS15A arrests cell cycle
progression of U937 cells
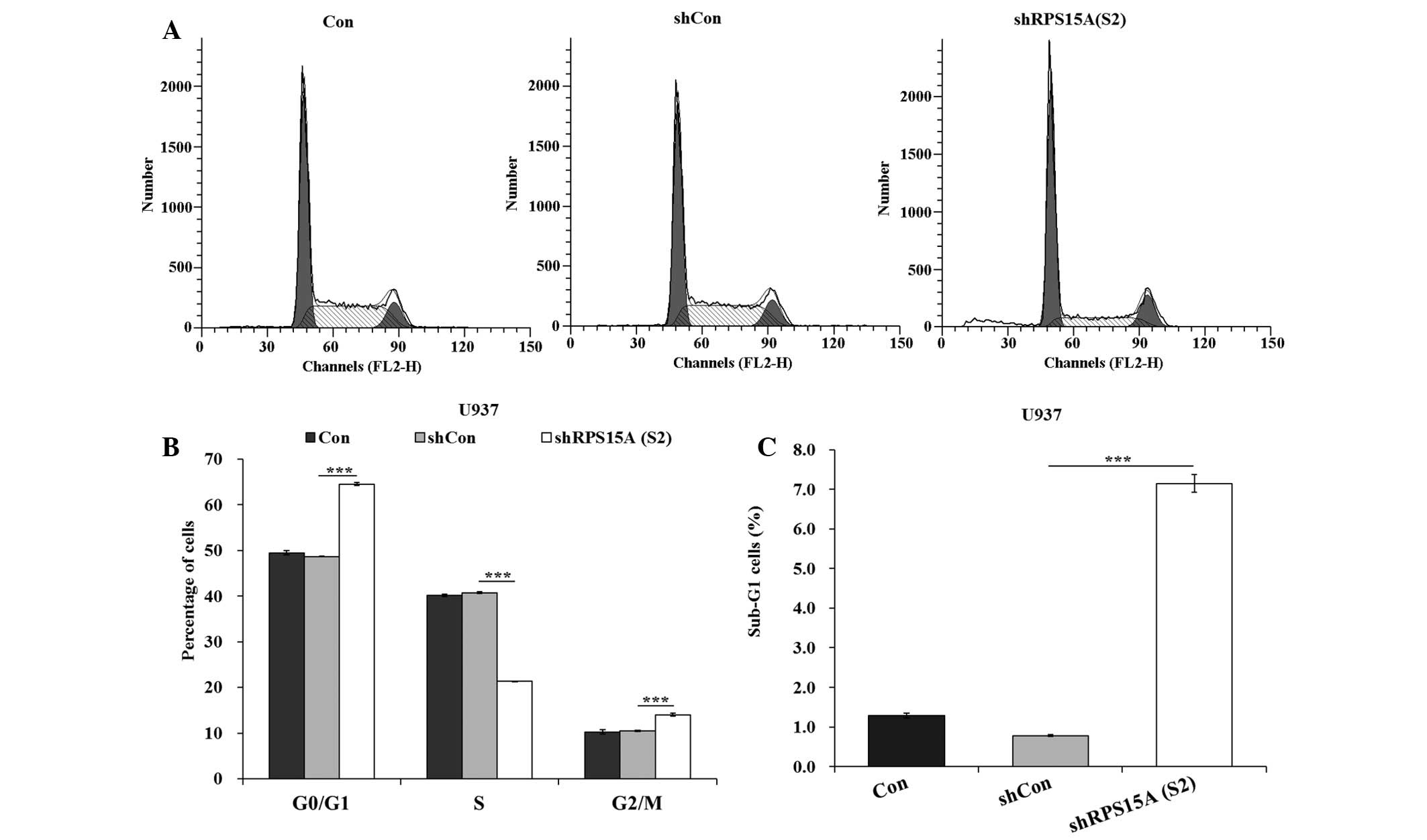
To investigate whether cell cycle arrest contributed
to growth inhibition, flow cytometric analysis was conducted. The
proportion of cells in G0/G1 phase was
significantly increased (P<0.001), and the percentage of cells
in S phase were significantly decreased (P<0.001), in the
shRPS15A (S2) group compared with the shCon and control groups
(Fig. 3A and B). These results
indicate that shRPS15A (S2)-induced growth suppression may be
partly mediated by cell cycle arrest at G0/G1
phase. In addition, the proportion of cells in sub-G1
was significantly increased in the shRPS15A (S2) group compared
with the shCon and control groups (P<0.001; Fig. 3C).
Knockdown of RPS15A enhances apoptosis of
U937 cells
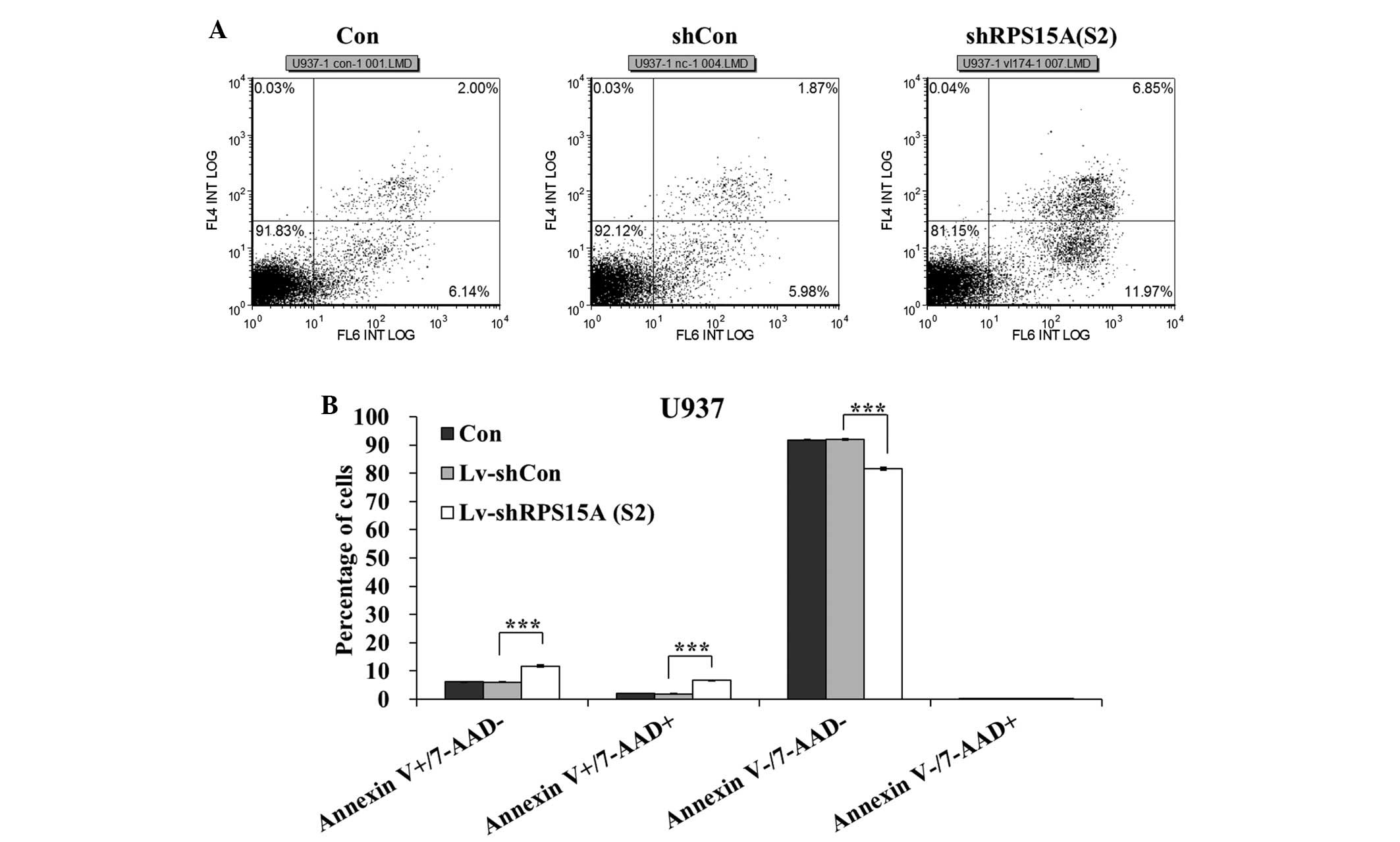
Results of the flow cytometric analysis indicated
that the percentage of early apoptotic (Annexin
V-positive/7-AAD-negative) and late apoptotic (Annexin
V-positive/7-AAD-positive) cells was significantly higher in the
shRPS15A (S2)-transduced cells compared with in the shCon and
control cells (P<0.001; Fig. 4A and
B). Following transduction, the apoptotic rate (early and late
apoptotic cells) of U937 cells in the shRPS15A (S2) group was
18.35±0.28%, which was significantly higher than in the shCon and
control groups (7.97±0.12 and 8.2±0.06%). The apoptotic rate of
U937 cells was significantly increased following RPS15A knockdown,
as compared with that in the shCon and control groups.
Discussion
AML has the lowest survival rate among all types of
leukemia (25), and knowledge
regarding its basic biology remains to be completely elucidated.
Numerous molecules have been identified as potential targets;
however, only a few have pivotal roles in AML cell proliferation
and survival (12,26–33).
Therefore, identification of novel therapeutic targets, and the
development of novel therapeutic regimens that more effectively
regulate cellular function are of central importance. RPS15A has
been reported to be overexpressed and have an important role in
regulating carcinogenesis in several types of human cancer.
Elevation of RPS15A expression in tumor cells leads to phenotype
changes that are characteristic of more aggressive malignancy
(20,22).
It has been reported that RPS15A gene expression was
upregulated in leukemia tissues at the mRNA level (34–36).
Furthermore, in order to ensure the specificity of RPS15A
silencing, two RPS15A shRNA expression vectors were used, which
resulted in a marked decrease in RPS15A expression in U937 cells.
Knockdown of RPS15A inhibited proliferation of U937 cells, and led
to cell cycle arrest at G0/G1 phase, as
determined by flow cytometry. Notably, downregulation of RPS15A in
hepatocellular carcinoma cells has previously been shown to
potently suppress cell growth via cell cycle arrest at
G0/G1 phase (22). The results of the present study
also indicated that RPS15A may have a crucial role in regulating
AML cell growth. In addition, the present study demonstrated that
the majority of shRPS15A-transduced cells underwent apoptosis.
These results strongly suggested that RPS15A may have a central
role in AML carcinogenesis and the maintenance of malignant
phenotypes. It may be hypothesized that RPS15A dysregulation may
affect the translation of proteins that specifically govern the
cell cycle. Therefore, the identification of RPS15A downstream
target proteins through high-throughput proteomics is the focal
point of our future research, which will facilitate the elucidation
of the mechanisms underlying the effects of RPS15A on AML
development.
In conclusion, to the best of our knowledge, this is
the first study to examine the function of RPS15A in AML cells. The
results demonstrated that inhibition of RPS15A significantly
reduced U937 cell proliferation, and induced
G0/G1 phase arrest and apoptosis, thus
providing a future target for AML therapy. Further investigations
regarding the regulatory mechanisms underlying the effects of
RPS15A on AML may help to better understand AML carcinogenesis.
Acknowledgments
The authors of the present study are grateful for
the financial support received from the National Natural Science
Foundation of China (grant no. 81573772) and the Shandong
Provincial Science and Technology Development Projects (grant no.
2014GSF118141).
References
|
1
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality
Worldwide: IARC CancerBase No. 11 (Internet). International Agency
for Research on Cancer; Lyon, France: 2013, http://globocan.iarc.fr.
Accessed December 12, 2013.
|
|
2
|
Smith M, Barnett M, Bassan R, Gatta G,
Tondini C and Kern W: Adult acute myeloid leukaemia. Crit Rev Oncol
Hematol. 50:197–222. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Showel MM and Levis M: Advances in
treating acute myeloid leukemia. F1000Prime Rep. 6:962014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Swords R, Freeman C and Giles F: Targeting
the FMS-like tyrosine kinase 3 in acute myeloid leukemia. Leukemia.
26:2176–2185. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Döhner H, Estey EH, Amadori S, Appelbaum
FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson
RA, et al European LeukemiaNet: Diagnosis and management of acute
myeloid leukemia in adults: Recommendations from an international
expert panel, on behalf of the European LeukemiaNet. Blood.
115:453–474. 2010. View Article : Google Scholar
|
|
7
|
Cornelissen JJ, van Putten WL, Verdonck
LF, Theobald M, Jacky E, Daenen SM, van Marwijk Kooy M, Wijermans
P, Schouten H, Huijgens PC, et al: Results of a HOVON/SAKK donor
versus no-donor analysis of myeloablative HLA-identical sibling
stem cell transplantation in first remission acute myeloid leukemia
in young and middle-aged adults: Benefits for whom? Blood.
109:3658–3666. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu Q, Ding W, Mirza A, Van Arsdale T, Wei
I, Bishop WR, Basso A, McClanahan T, Luo L, Kirschmeier P, et al:
Integrative genomics revealed RAI3 is a cell growth-promoting gene
and a novel P53 transcriptional target. J Biol Chem.
280:12935–12943. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tan M, Wang Y, Guan K and Sun Y:
PTGF-beta, a type beta transforming growth factor (TGF-beta)
superfamily member, is a p53 target gene that inhibits tumor cell
growth via TGF-beta signaling pathway. Proc Natl Acad Sci USA.
97:109–114. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hiss D: Optimizing molecular-targeted
therapies in ovarian cancer: The renewed surge of interest in
ovarian cancer biomarkers and cell signaling pathways. J Onco.
2012:7379812012.
|
|
11
|
Lee SG, Su ZZ, Emdad L, Sarkar D, Franke
TF and Fisher PB: Astrocyte elevated gene-1 activates cell survival
pathways through PI3K-Akt signaling. Oncogene. 27:1114–1121. 2008.
View Article : Google Scholar
|
|
12
|
Bouchet S, Tang R, Fava F, Legrand O and
Bauvois B: Targeting CD13 (aminopeptidase-N) in turn downregulates
ADAM17 by internalization in acute myeloid leukaemia cells.
Oncotarget. 5:8211–8222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Warner JR and McIntosh KB: How common are
extraribosomal functions of ribosomal proteins? Mol Cell. 34:3–11.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lavoie C, Tam R, Clark M, Lee H, Sonenberg
N and Lasko P: Suppression of a temperature-sensitive cdc33
mutation of yeast by a multicopy plasmid expressing a Drosophila
ribosomal protein. J Biol Chem. 269:14625–14630. 1994.PubMed/NCBI
|
|
15
|
Piantoni P, Bionaz M, Graugnard DE,
Daniels KM, Akers RM and Loor JJ: Gene expression ratio stability
evaluation in prepubertal bovine mammary tissue from calves fed
different milk replacers reveals novel internal controls for
quantitative polymerase chain reaction. J Nutr. 138:1158–1164.
2008.PubMed/NCBI
|
|
16
|
Akiyama N, Matsuo Y, Sai H, Noda M and
Kizaka-Kondoh S: Identification of a series of transforming growth
factor beta-responsive genes by retrovirus-mediated gene trap
screening. Mol Cell Biol. 20:3266–3273. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeller KI, Jegga AG, Aronow BJ, O'Donnell
KA and Dang CV: An integrated database of genes responsive to the
Myc oncogenic transcription factor: Identification of direct
genomic targets. Genome Biol. 4:R692003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Katz JP, Perreault N, Goldstein BG, Actman
L, McNally SR, Silberg DG, Furth EE and Kaestner KH: Loss of Klf4
in mice causes altered proliferation and differentiation and
precancerous changes in the adult stomach. Gastroenterology.
128:935–945. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Whitney EM, Ghaleb AM, Chen X and Yang VW:
Transcriptional profiling of the cell cycle checkpoint gene
krüppel-like factor 4 reveals a global inhibitory function in
macromolecular biosynthesis. Gene Expr. 13:85–96. 2006. View Article : Google Scholar :
|
|
20
|
Lian Z, Liu J, Li L, Li X, Tufan NL, Wu
MC, Wang HY, Arbuthnot P, Kew M and Feitelson MA: Human S15a
expression is upregulated by hepatitis B virus X protein. Mol
Carcinog. 40:34–46. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kavak E, Unlü M, Nistér M and Koman A:
Meta-analysis of cancer gene expression signatures reveals new
cancer genes, SAGE tags and tumor associated regions of
co-regulation. Nucleic Acids Res. 38:7008–7021. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu M, Wang Y, Chen L, Pan B, Chen F, Fang
Y, Yu Z and Chen G: Down-regulation of ribosomal protein S15A mRNA
with a short hairpin RNA inhibits human hepatic cancer cell growth
in vitro. Gene. 536:84–89. 2014. View Article : Google Scholar
|
|
23
|
Tiscornia G, Singer O and Verma IM:
Production and purification of lentiviral vectors. Nat Protoc.
1:241–245. 2006. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Deschler B and Lübbert M: Acute myeloid
leukemia: Epidemiology and etiology. Cancer. 107:2099–2107. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun K, Li Y, Lu Z, Zhang L, Gao Z and Jin
Q: Suppression of titanium particle-induced TNF-alpha expression
and apoptosis in human U937 macrophages by siRNA silencing. Int J
Artif Organs. 36:522–527. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yao K, Xing H, Yang W, Liao A, Wu B, Li Y,
Zhang R and Liu Z: Knockdown of RLIP76 expression by RNA
interference inhibits proliferation, enhances apoptosis, and
increases chemosensitivity to daunorubicin in U937 leukemia cells.
Tumour Biol. 35:8023–8031. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Long M, Hao M, Dong K, Shen J, Wang X, Lin
F, Liu L, Wei J, Liang Y, Yang J, et al: AEG-1 overexpression is
essential for maintenance of malignant state in human AML cells via
up-regulation of Akt1 mediated by AURKA activation. Cell Signal.
25:1438–1446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bai Y, Qiu GR, Zhou F, Gong LY, Gao F and
Sun KL: Overexpression of DICER1 induced by the upregulation of
GATA1 contributes to the proliferation and apoptosis of leukemia
cells. Int J Oncol. 42:1317–1324. 2013.PubMed/NCBI
|
|
30
|
Gao H, Jiang Q, Han Y, Peng J and Wang C:
shRNA-mediated EMMPRIN silencing inhibits human leukemic monocyte
lymphoma U937 cell proliferation and increases chemosensitivity to
adriamycin. Cell Biochem Biophys. 71:827–835. 2015. View Article : Google Scholar
|
|
31
|
Ye P, Zhao L, McGirr C and Gonda TJ: MYB
down-regulation enhances sensitivity of U937 myeloid leukemia cells
to the histone deacetylase inhibitor LBH589 in vitro and in vivo.
Cancer Lett. 343:98–106. 2014. View Article : Google Scholar
|
|
32
|
Watanabe N, Narita M, Yamahira A,
Taniguchi T, Furukawa T, Yoshida T, Miyazawa T, Nashimoto M and
Takahashi M: Induction of apoptosis of leukemic cells by TRUE gene
silencing using small guide RNAs targeting the WT1 mRNA. Leuk Res.
37:580–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu S, Chen R, Man X, Feng X, Cen J, Gu W,
He H, Li J, Chai Y and Chen Z: Function and expression of
insulin-like growth factor-binding protein 7 (IGFBP7) gene in
childhood acute myeloid leukemia. Pediatr Hematol Oncol.
28:279–287. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Haferlach T, Kohlmann A, Wieczorek L,
Basso G, Kronnie GT, Béné MC, De Vos J, Hernández JM, Hofmann WK,
Mills KI, et al: Clinical utility of microarray-based gene
expression profiling in the diagnosis and subclassification of
leukemia: Report from the International Microarray Innovations in
Leukemia Study Group. J Clin Oncol. 28:2529–2537. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Valk PJ, Verhaak RG, Beijen MA, Erpelinck
CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM,
Beverloo HB, Moorhouse MJ, van der Spek PJ, Löwenberg B and Delwel
R: Prognostically useful gene-expression profiles in acute myeloid
leukemia. N Engl J Med. 350:1617–1628. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Andersson A, Ritz C, Lindgren D, Edén P,
Lassen C, Heldrup J, Olofsson T, Råde J, Fontes M, Porwit-Macdonald
A, et al: Microarray-based classification of a consecutive series
of 121 childhood acute leukemias: Prediction of leukemic and
genetic subtype as well as of minimal residual disease status.
Leukemia. 21:1198–1203. 2007. View Article : Google Scholar : PubMed/NCBI
|