Introduction
Malignant melanoma is an aggressive skin cancer that
arises from the melanocyte lineage (1). Melanoma occurs less frequently than
the majority of other malignancies, accounting for 232,000 new
cases and resulting in 55,000 cancer-related mortalities worldwide
in 2012 (2). However, it is
dangerous if diagnosed at the later stages of the disease and
causes the majority of skin cancer-related fatalities (3). Melanoma is usually caused by DNA
damage resulting from exposure to ultraviolet light from sun
exposure, or genetic predisposition (4). Early stages of melanoma are curable
by surgical removal of the tumor lesion, while later stages or
recurrent melanoma are treated with chemo- and immunotherapy, or
radiation therapy (5,6). However, to date, melanoma is one of
the most difficult types of cancer to successfully treat as it
disseminates early after the development of the initial lesion. The
majority of all melanoma patients with advanced stage disease
succumb to distant metastases 6–10 months after initial diagnosis
(7). The major obstacle in
melanoma treatment is the lack of tumor specificity. Thus, more
efficient treatment strategies specifically targeting tumor tissue
are urgently required (8).
For targeted therapy of melanoma, like the majority
of other cancer types, mesenchymal stem cells (MSCs) may have
tumor-oriented homing capacity and thus potential as an efficient
vehicle in patient-tailored cancer therapy (9,10).
Tumor-directed migration and incorporation of MSCs have been
demonstrated in a number of pre-clinical studies using in
vitro and in vivo animal tumor models (8,11,12).
For example, bone marrow mesenchymal stem cells (11), human pancreas stem cells (8) and neural stem cells (12) have the capability of tumor tracking
and this tracking capability was associated with the secretion of
cytokines and chemokines. The stem cells can migrate to and gather
around the tumor lesion with a high concentration and this feature
suggested that MSCs could be used as a carrier of enzyme/prodrug
gene in combined targeting therapy of human cancers. The homing
capacity of MSCs has previously been demonstrated in almost all
tested human cancer cell lines, including melanoma (13). Bone marrow (BM) was the first
recognized source of MSCs (14);
however, adipose tissue represents a more reliable source of MSCs
(15). Compared with BM-MSCs,
adipose-derived MSCs (ADSCs) are more suitable for tumor-gene
therapy approaches. This is because adipose tissue can be obtained
in relevant quantities by minimally invasive procedures from normal
subjects or from cancer patients (16,17).
Furthermore, tumor necrosis factor (TNF)-related
apoptosis-inducing ligand (TRAIL) is a promising anticancer death
ligand with sequence homology to TNF and FasL (18). TRAIL is one of few anticancer
proteins that can selectively induce apoptosis of transformed or
tumor cells by activation of death receptors (DR), without
affecting healthy cells (19). In
previous in vitro experiments, TRAIL was shown to induce
apoptosis of glioma, neuroblastoma, cervix uteri cancer, non-small
cell lung cancer, renal cell carcinoma, liver cancer, thyroid
cancer and melanoma cells. In addition, it was shown to exhibit a
particularly lethal effect on lung cancer cells (13), malignant glioma cells (1) and breast cancer cells (20). A previous study also showed that
TRAIL significantly inhibited the growth of hepatocellular
carcinoma cells in mice, but did not exhibit any toxic side effects
on the control mice (21). Thus,
several studies demonstrated the antitumor activity of recombinant
TRAIL (rTRAIL), but rTRAIL in vivo use is limited due to its
short half-life in the blood (22). It has been reported that ADSCs
could be used to deliver a stable source of TRAIL for cancer
therapy (23). Thus, the current
study utilized ADSCs to harbor TRAIL cDNA to facilitate TRAIL
expression and test the in vitro effects on melanoma
cells.
Materials and methods
Cell lines and culture
Human ADSCs (HUXMD-01001) were purchased from Cyagen
Biotechnology Co., Ltd. (Guangzhou, China) and cultured in
Dulbecco's modified Eagle's medium (DMEM)-F12 supplemented with 10%
Gibco fetal bovine serum (FBS; #16000044), 2 mM L-glutamine, and 1%
penicillin-streptomycin solution (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) in a humidified incubator with 5% CO2
at 37°C. The immunophenotype identification of ADSCs was tested by
flow cytometry. Adipogenic and osteogenic differentiation of ADSCs
was conducted using cell differentiation kits (HUXMD-90031 and
-90021; Cyagen Biotechnology Co., Ltd.). The constituents of the
adipogenic induction medium A were high-glucose DMEM supplemented
with 10% FBS, 2 mM L-glutamine, 100 units myllicin, 5 µg/ml
insulin, 0.5 µM 3-isobutyl-1-methylxantine, rosiglitazone
and 10 nM dexamethasone (kit HUXMD-90031). Adipogenic induction
medium B contained high-glucose DMEM supplemented with 10% FBS, 2
mM L-glutamine, 100 units myllicin and 5 µg/ml insulin (kit
HUXMD-90031). Osteogenic induction medium contained high-glucose
DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 units
myllicin, 100 µg/ml ascorbate, 10 mM β-glycerophosphate and
100 nM dexamethasone (kit HUXMD-90021). After 14–20 days induction,
the differentiated cells were fixed with 70% ethanol, then washed 3
times with PBS and stained with Alizarin red (kit HUXMD-90021), or
Oil Red O (kit HUXMD-90031) according to the manufacturer's
instructions. The A375 human melanoma cell line was obtained from
the American Type Culture Collection (Manassas, VA, USA) and was
cultured according to the manufacturer's instructions. Briefly,
A375 cells were cultured in DMEM supplemented with 10% FBS and 100
units myllicin, in a humidified incubator with 5% CO2 at
37°C.
Construction of TRAIL-carrying vector and
gene transfection
Full-length human TRAIL cDNA was cloned into the
pcDNA3.3-TOPO plasmid using a pcDNA 3.4 TOPO TA Cloning Kit
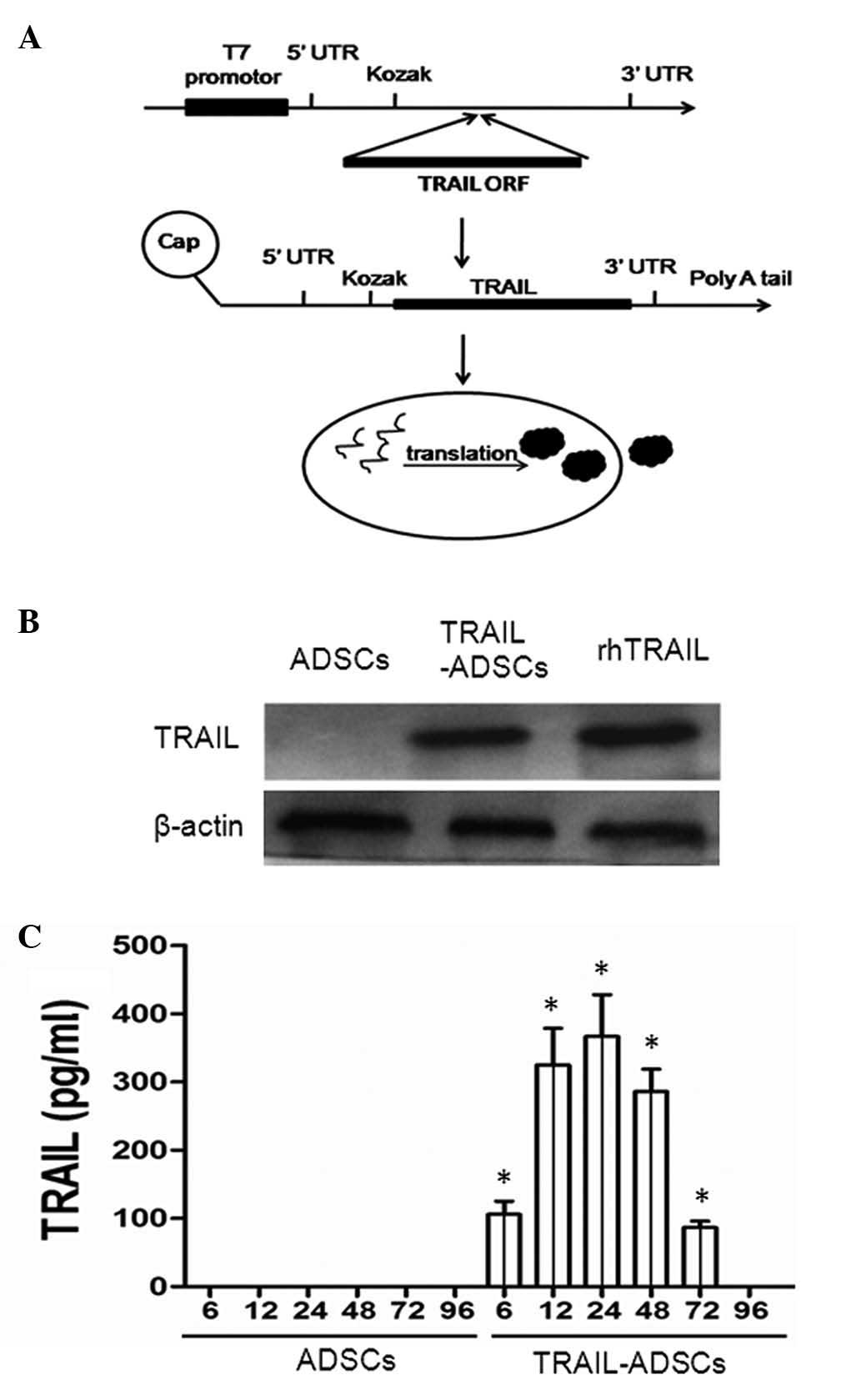
(#A14697; Thermo Fisher Scientific, Inc.) as shown in Fig. 1A. In brief, TRAIL cDNA was
amplified from MIGR1-TRAIL-GFP as described by Wiley et al
(18) and then sub-cloned into the
pcDNA3.3-TOPO plasmid. The TRAIL cDNA was connected to the pcDNA
3.3-TOPO plasmid with T4 DNA ligase (#D7006; Beyotime Institute of
Biotechnology, Beijing, China). After colony polymerase chain
reaction (PCR) amplification (the template was bacterium
suspension) and DNA sequencing confirmation (performed by Sangon
Biotech Co., Ltd., Wuhan, China), this plasmid containing TRAIL
cDNA was used for tail PCR and modified with the MEGAscript T7 kit
(Ambion; Thermo Fisher Scientific, Inc.). Then, modified TRAIL mRNA
was isolated with Ambion Anti-Reverse Cap Analog (ARCA; #AM8045)
and purified with Ambion MEGAclear spin columns (Thermo Fisher
Scientific, Inc.) and treated with Antarctic Phosphatase (New
England Biolabs, Ipswich, MA, USA) to remove residual
5′-triphosphates. The transfection of the TRAIL plasmid into ADSCs
was conducted using TransIT-mRNA (Mirus Bio LLC., Madison, WI, USA)
according to the manufacturer' instructions and ADSCs transfected
with TRAIL-cDNA were defined as TRAIL-ADSCs.
Immunofluorescence
Initially, 1×105 human foreskin
fibroblast (HFF) cells from our laboratory stocks were seeded onto
coverslips and grown overnight, and then fixed with 4%
paraformaldehyde for 30 min. For immunofluorescence, the cells were
first incubated with 3% H2O2 in
phosphate-buffered saline (PBS) solution for 30 min and then washed
with tap water and again with PBS. Next, the cells were incubated
with 5% normal goat serum (#C1771; Applygen Technologies, Inc.,
Beijing, China) in PBS for 30 min, then incubated with a polyclonal
mouse vimentin antibody (1:200; #3390S; Cell Signaling Technology,
Inc., Danvers, MA, USA) and then a secondary fluorescein
isothiocyanate (FITC)-conjugated goat anti-mouse IgG (1:500;
#A0568; Beyotime Institute of Biotechnology), and the nuclei were
stained with RedDot 1 (#40060-T; Biotium, Hayward, CA, USA). In
addition, 1×105 cells were seeded into 24-well plates
and cocultured with ADSCAs or ADSCs/TRAIL in Transwell chambers.
The dead cells were stained with prop-idium iodide (PI; #P4170;
Sigma-Aldrich, St. Louis, MO, USA) and then reviewed and scored
under a DM IRE2 fluorescent microscope (Leica Microsystems GmbH,
Wetzlar, Germany). The dead cells were shown by red fluorescence
staining and the live cells did not stain red.
Transwell cell migration assay
To assess the migration ability of ADSCs, a
Transwell migration assay using a Transwell system from Corning
(Corning, Inc., Corning, NY, USA) was performed. Specifically, A375
cells were seeded onto the plastic surface of the 24-well plates at
a density of 1×105 cells/well and grown overnight, while
HFF cells were used as a control. On the following day, parental
ADSCs and TRAIL-ADSCs were seeded on the top chambers of
Transwells, respectively and cultured in serum-free medium for 48 h
at 37°C and in 5% CO2 in an incubator. At the end of the
experiments, the up-chamber was washed with PBS and scraped gently
using a cotton swab to remove non-migrated cells. The ADSCs
migrated to the bottom side of the filter and were fixed with 70%
ethanol, stained with 0.1% crystal violet (#C0121; Beyotime
Institute of Biotechnology) and counted under a DMI3000 M inverted
manual microscope (Leica Microsystems GmbH). The average number of
migrated cells was assessed by counting five randomly selected
microscopic fields. The experiment was performed in triplicate.
Flow cytometry cell apoptosis assay
To assess the apoptosis-induction effects of
ADSC-TRAIL on A375 cells, cells were cultured under direct
co-culture conditions. In brief, A375 cells and TRAIL-ADSCs were
plated at 1:1, 1:2 or 1:5 ratios in 6-well plates and were cultured
for up to 48 h. At the end of the experiments, the cells were
stained with PI and analyzed using an EPICS XL flow cytometer
(Beckman Coulter, Danvers, MA, USA). Soluble hrTRAIL (PeproTech,
Inc., London, UK) was used as a positive control. Anti-human TRAIL
antibody diluted at different concentrations was used to neutralize
the TRAIL-induced apoptosis, which was demonstrated in co-culture
of A375 cells and TRAIL-ADSCs at a 1:1 ratio. The experiment was
performed in triplicate.
Protein extraction and western
blotting
Western blotting was performed to detect the
cellular expression of TRAIL protein in the ADSCs and
apoptosis-related proteins (caspase-3, caspase-4, caspase-8 and
caspase-9) in A375 cells. A375 cells and ADSCs were co-cultured in
a Transwell system for 48 h and then, ADSCs were collected. Equal
amounts of whole cell lysates were resolved by 12% SDS-PAGE and
electrotransferred onto a polyvinylidene difluoride membrane
(Beyotime Institute of Biotechnology). The PVDF membranes were
blotted with 5% BSA and then incubated with primary antibodies,
including monoclonal mouse anti-human sTRAIL (1:500; #500-M49;
PeproTech, Rocky Hill, NJ, USA), polyclonal rabbit anti-human
caspase-4 (1:200; #4450S; Cell Signaling Technology, Inc.),
caspase-3 (1:500; #AC030), caspase-8 (1:500; #AC056), caspase-9
(1:500; AC062), Akt (1:500; #AA326), phospho-Akt (Ser473; 1:500;
#AA329) and β-actin (1:1,000; AA128) antibodies (Beyotime Institute
of Biotechnology). After washing 3 times with Tris-buffered saline
with Tween 20, the PVDF membranes were incubated with alkaline
phosphatase-labeled goat anti-rabbit (1:1,000; #A0239) or
anti-mouse (1:1,000; #A0258) IgGs (Beyotime Institute of
Biotechnology). The immunoreactive signals were detected using a
Gel Docx2000 scanner system (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) and analyzed with Image J software, version 2.1.4.7
(imagej.nih.gov/ij).
Enzyme-linked immunosorbent assay
(ELISA)
The level of TRAIL protein in the cell-conditioned
medium was measured using a Quantikine Human TRAIL/TNFSF10 kit
(R&D Systems, Minneapolis, MN, USA) according to the
manufacturer's instructions. Briefly, the cell-conditioned medium
from the Transwell system was collected and centrifuged at 206 × g
for 3 min to remove cell debris and the supernatant was subjected
to ELISA. The experiments were performed in triplicate and repeated
three times.
Statistical analysis
Measurement data are expressed as the mean ±
standard error. Statistical differences between the means of the
different groups were evaluated using SPSS 18.0 software (Chicago,
IL, USA) using two-tailed Student's t-test. P≤0.05 was considered
to indicate a statistically significant difference.
Results
Expression of TRAIL protein in ADSCs
TRAIL-containing plasmids were transiently
transfected into ADSCs and western blot and ELISA analyses showed
expression of TRAIL protein was clearly enhanced in transfected
ADSCs (Fig. 1B) and in the
conditioned medium (Fig. 1C) in a
time-dependent manner. Specifically, soluble TRAIL could be
detected after gene transfection at 6 h (126.8±18.4 pg/ml), the
levels increased at 12 h (325.1±53.6 pg/ml) and peaked at 24 h
(366.4±57.5 pg/ml) and reduced at 96 h (Fig. 1C), suggesting that the TRAIL mRNA
was synthesized effectively.
Characteristics of ADSC differentiation
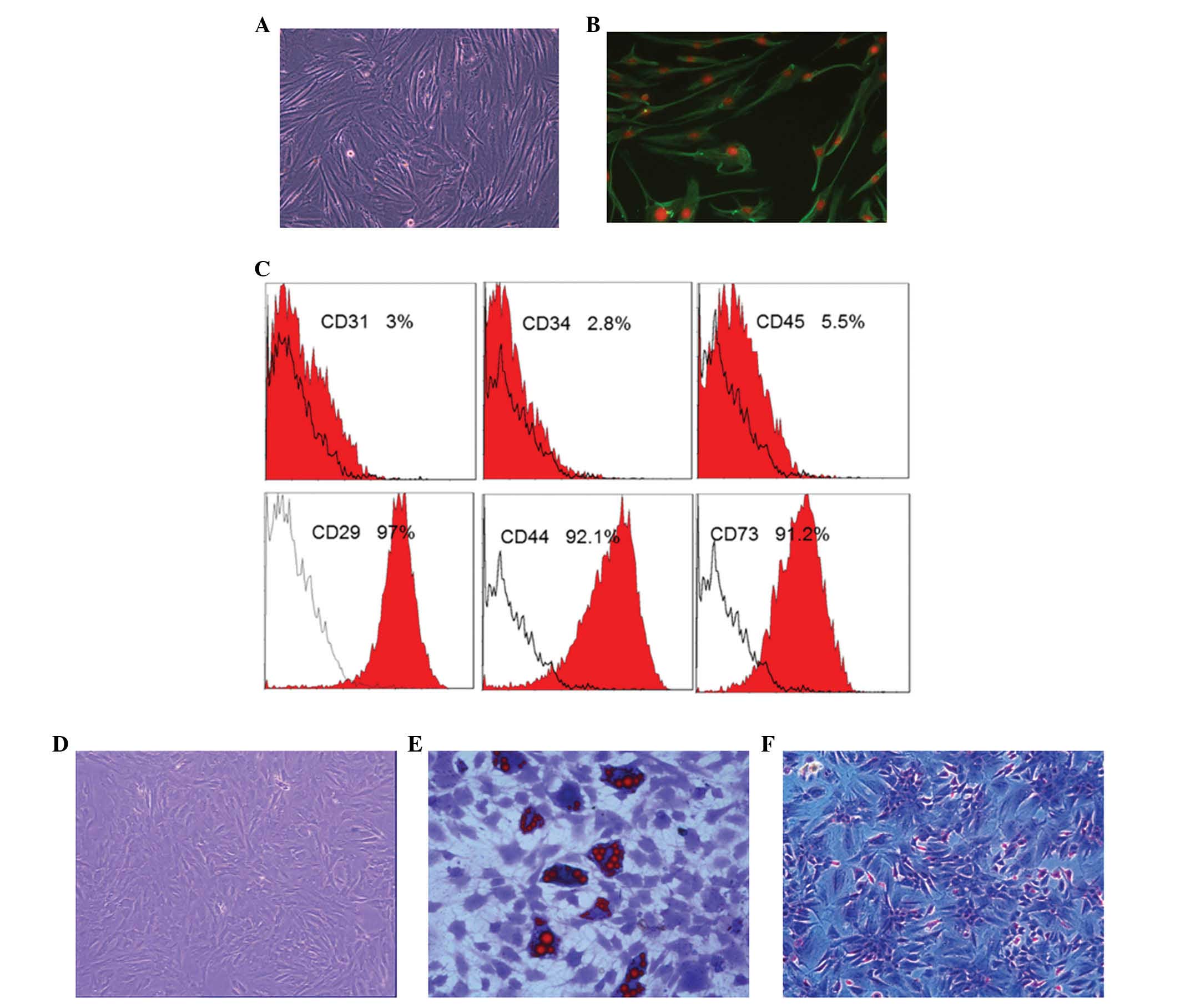
in vitro
Firstly, the properties of ADSCs and A375 cells
in vitro were characterized, while HFF cells, which have
long spindle morphology and are vimentin-positive, were used as a
control (Fig. 2A and B). The ADSCs
used in the present study exhibited positive expression of
CD44+, CD73+, CD29+ and were
negative for CD31−, CD34− and
CD45− (Fig. 2C). ADSC
differentiation was induced using the different kit that could
induce ADSCs differentiated into adipogenic and osteogenic cells
under specific differentiation conditions (Fig. 2D–F). Specifically, when cultured in
adipogenic differentiation medium for 10–14 days, there were
numerous lipid cells with big fat droplets positive for oil red
staining (Fig. 2E). By contrast,
following culture in osteogenic differentiation medium for 14–21
days, there were numerous osteoblast cells positive for Alizarin
red staining (Fig. 2F).
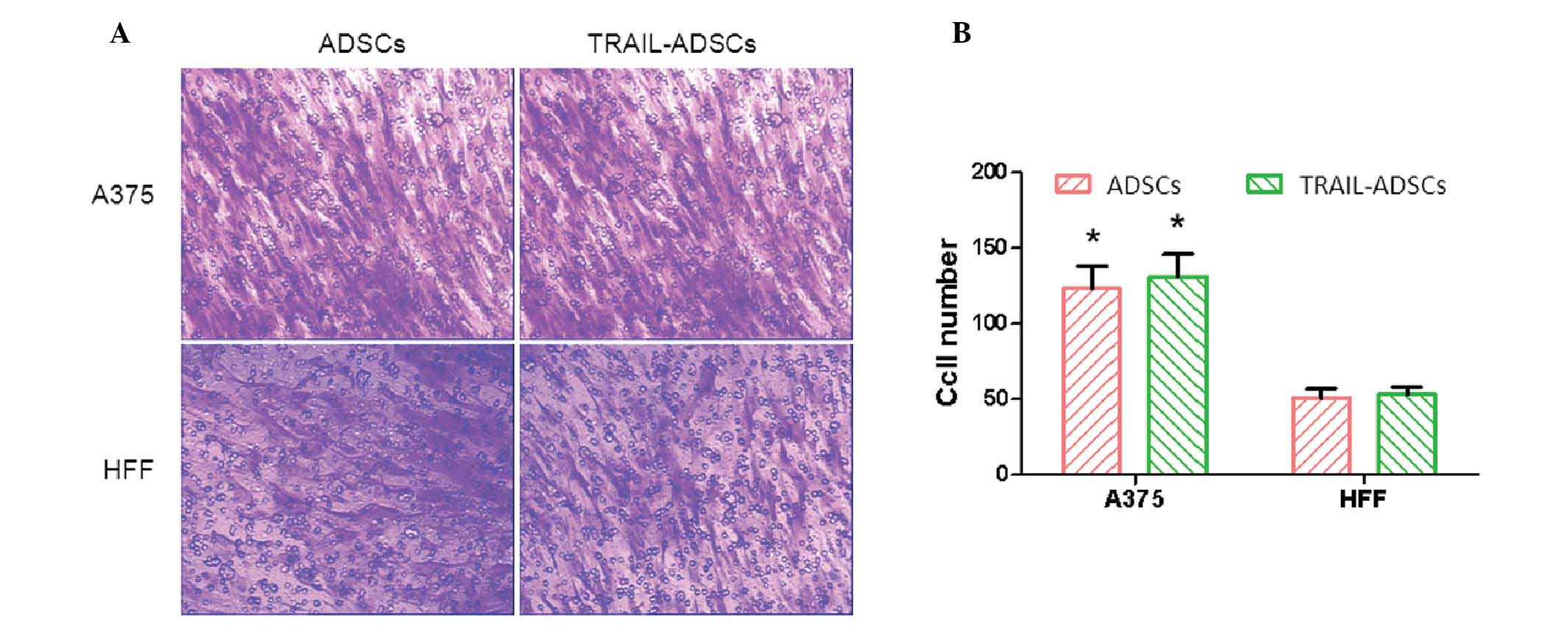
Effects of melanoma cells on the
regulation of ADSC migration
ADSC migration was determined by a Transwell assay
and HFF cells were used as a control. After co-culture for 48 h, a
considerable number of ADSCs migrated across the Transwell membrane
(Fig. 3A). ADSCs and TRAIL-ADSCs
exhibited significantly stronger migration ability towards A375
melanoma cells (123±15 and 131±16, respectively) compared with HFF
cells (51±6 and 53±5, respectively) (P<0.05). Compared with the
control ADSCs, TRAIL cDNA transfection did not obstruct ADSC
migration towards A375 cells (Fig. 3A
and B). These results suggest that ADSCs and TRAIL-ADSCs have a
specific migration ability toward tumor cells, and the transfection
of TRAIL mRNA did not affect this ability.
Effects of TRAIL expression facilitated
by ADSCs on the regulation of melanoma cell viability
Indirect co-culture was used to determine the
effects of TRAIL-ADSCs on the viability of A375 cells using PI
staining. As shown in Fig. 4A,
TRAIL-ADSCs induced A375 cell death, which was represented by a
reduction in the number of adherent A375 cells and the presence of
cellular debris. These features were even more prominent after 48 h
of co-culture. However, the parental ADSCs did not exhibit this
effect. The PI-positive dead A375 cells were observed in the
TRAIL-ADSCs group (Fig. 4A). As
shown in Fig. 4B, apoptosis of
A375 cells was only detectable in co-culture with TRAIL-ADSCs
starting from the 1:1 ratio (35.2±3.3, P<0.01). This effect
increased significantly as the ratio of ADSCs increased: Tumor
cells increased (42.4±5.3 and 56.8±6.5 for 1:2 and 1:5,
respectively, P<0.05) and the mortality was more prominent than
that of rTRAIL (32.1±2.7, P<0.05). For all the considered
ratios, parental ADSCs did not induce A375 cell death (9.4±1.1,
10.2±1.3 and 12.3±1.5 for 1:1, 1:2 and 1:5, respectively),
indicating specific action of TRAIL. To further confirm that A375
cell apoptosis is specifically due to the antitumor effect of
TRAIL, anti-TRAIL was added to the coculture medium (Fig. 4C) and starting from 1 µg/ml
(41.1±3.7), a significant (P<0.05) reduction of A375 cell death
was identified. This effect was more prominent at the highest
concentration (35.3±4.2, 28.2±3.3 and 15.3±1.8 for 2.5, 5 and 10
µg/ml, respectively) where A375 death was comparable to that
of the control group (8.4±1.1, P<0.05).
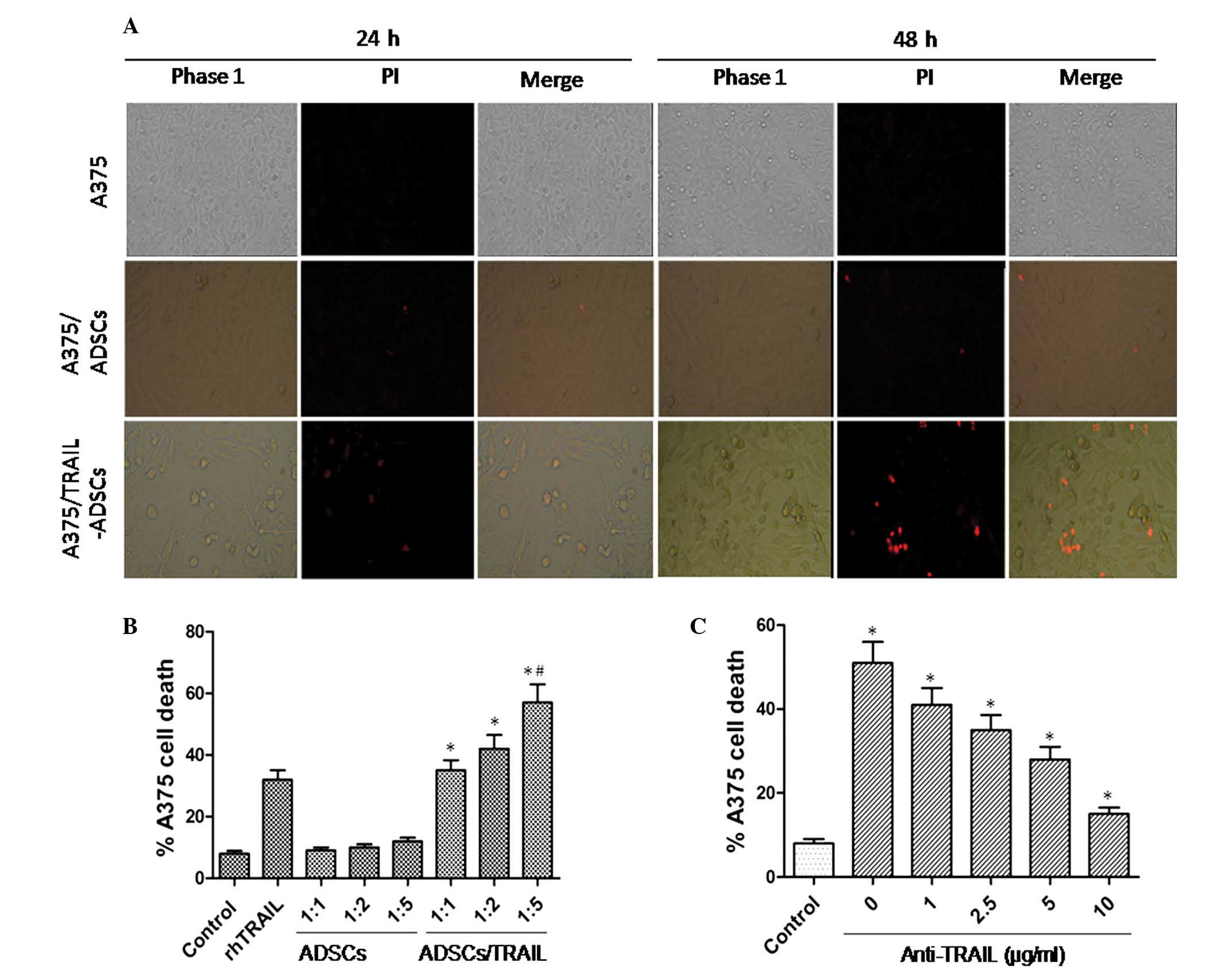 | Figure 4Induction of A375 cell apoptosis after
indirect co-culture with TRAIL-ADSCs. (A) A375 cells were
cocultured with ADSCs or TRAIL-ADSCs in a Transwell system, with
A375 cells as control. After 24 or 48 h culture, the cells were
stained with PI. Red staining indicates dead cells. Phase 1, whole
population of cells that still attached to the culture surface
assessed under the phase contrast microscope; PI, apoptotic cells
stained with PI show red; merge, merged images of Phase 1 and PI.
Original magnification, ×100. (B) Fluorescence-activated flow
cytometry was used to detect A375 cell apoptosis after staining
with PI in co-culture with TRAIL-ADSCs or ADSCs at a different
ratio in the Transwell system. rTRAIL served as a positive control
and A375 cells alone served as a negative control. Ratio indicates
the cell number of A375 cells to ADSCs or TRAIL-ADSCs. (C)
Anti-human TRAIL antibody diluted at different concentrations was
used to neutralize the TRAIL-induced apoptosis, which was shown in
co-culture of A375 cells and TRAIL-ADSCs at a 1:1 ratio.
*P<0.05 vs. control, #P<0.05 vs.
rhTRAIL. TRAIL, tumor necrosis factor-related apoptosis-inducing
ligand; ADSCs, adipose-derived stem cells. |
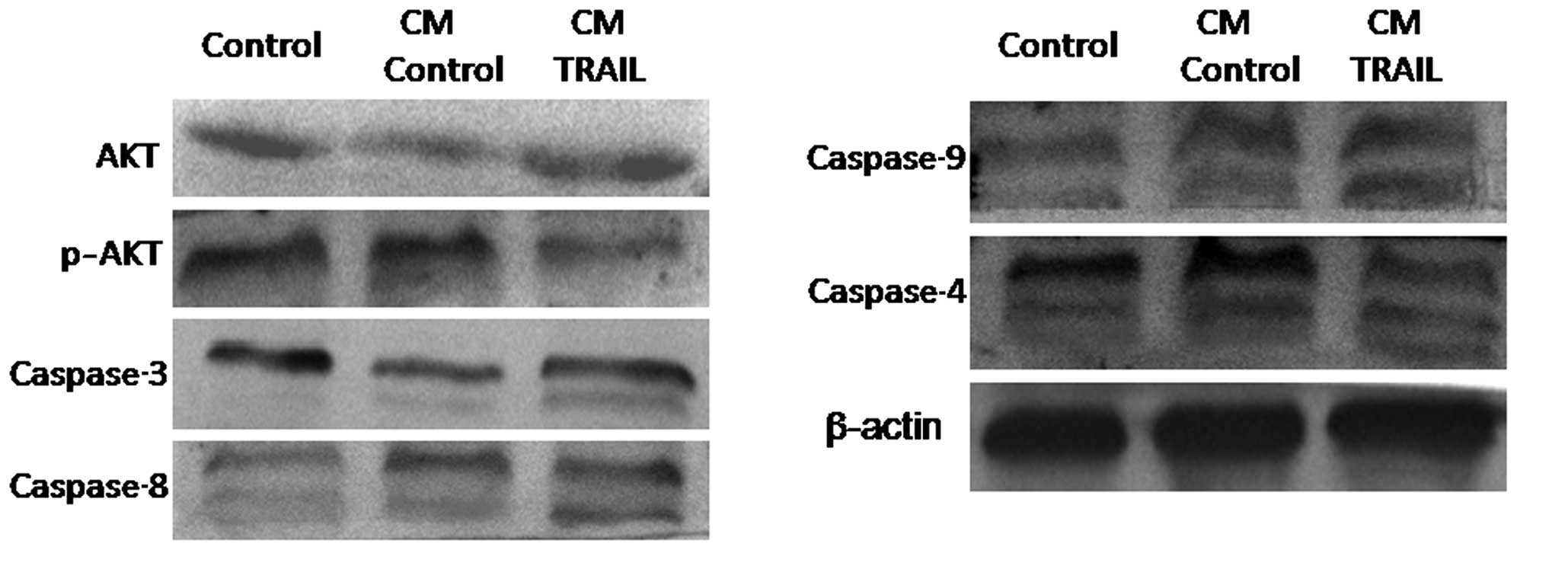
TRAIL-induces caspase activation
To further analyze the molecular events of
TRAIL-induced cell apoptosis, A375 cells were indirectly
co-cultured with TRAIL-ADSCs or parental ADSCs in a Transwell
system for 48 h, and then lysed to be subjected to western blot
analysis. Fig. 5 shows that
compared with parental ADSCs, the activity of caspase-4, -8, -9,
and -3 in A375 cells was significantly induced in TRAIL-ADSC
co-culture.
Discussion
Gene and targeted therapy is advancing rapidly and a
large number of targeted therapies have been developed in the
recent decade. However, current targeting agents exhibit frequent
and severe toxicities as traditional cytotoxic agents (24). In the current study, ADSCs were
used to induce TRAIL expression to mediate melanoma therapy. Such a
modification has advantages compared with other strategies. For
example, unlike DNA-based plasmid (pDNA) gene therapy (25,26),
TRAIL was first transfected into ADSCs. Our previous experiments
modified 5′-cDNA ARCAs and 3′-poly(A) tails, which led to better
transfection efficiency (27,28)
and translation into a functional protein in the cytoplasm
directly. Furthermore, such a modification would not lead to any
effect on the host genome and could be degraded by the host cell
quickly (often within 2–3 days). Therefore, the risk of insertion
mutagenesis is omitted.
In the current study, ADSCs were used as a carrier
to deliver TRAIL protein, as the ADSCs express TRAIL protein which
is then secreted into the extracellular space. Data showed that
TRAIL protein was expressed in ADSCs and the active expression was
retained for up to 72 h with the peak activity 48 h after
transfection. These results indicated that the TRAIL protein
synthesized in ADSCs was active and that the expression level is
consistent with DNA-based vectors (3). The current study also demonstrated
that TRAIL cDNA transfection did not alter the migration ability of
ADSCs. Data further demonstrated that ADSCs migrated toward A375
cells compared with the HFFs, which indicated that ADSCs have
tumor-oriented homing capacity. Thus, the current data provide a
basis for the use of ADSCs in TRAIL-mediated anticancer
therapy.
Furthermore, the TRAIL-ADSC antitumor activity in
A375 cells was assessed and it was demonstrated that under
co-culture conditions, TRAIL induced A375 cell apoptosis. This
finding is consistent with previously published data on TRAIL
activity (8,21). The underlying molecular mechanism
of TRAIL-ADSCs-mediated A375 cell death was also investigated and
it was demonstrated that TRAIL-ADSCs altered the expression of
members of the PI3K-AKT signaling pathway, including AKT,
caspase-3, caspase-4, caspase-8 and caspase-9. The phosphorylated
form of AKT (pAKT) was markedly downregulated following co-culture
with TRAIL-ADSCs. Caspase-3, caspase-4, caspase-8 and caspase-9
were activated after co-culture with TRAIL-ADSCs. Consistent with a
study by Mao et al (13)
TRAIL-induced apoptosis of human melanoma cells was observed to
involve the activation of caspase-4. This appeared to be mediated
by caspase-3, in that caspase-4 was activated later than caspase-8,
-9 and -3, and that inhibition of caspase-3 blocked TRAIL-induced
caspase-4 activation (13). These
results indicated that the TRAIL-ADSCs-mediated melanoma cell death
was associated the PI3K-AKT signaling pathway.
However, the current study was a proof-of-principle
study and further investigation is required before this TRAIL-ADSCs
system can be considered as a clinical strategy. Future studies
will investigate the effects of TRAIL-ADSCs antitumor activity
in vivo. In addition, investigation of the half-life or
differentiation potential of TRAIL-ADSCs is also required. In
conclusion, the strategy of this ADSC-mediated gene therapy may
offer a double-targeted killing effect on tumor cells, consisting
the tropism property of ADSCs and engineered anticancer agents. It
is able to exert killing effects locally and consistently. This
strategy also holds the potential for the use of the patient's own
ADSCs and to switch tumor attackers corresponding to the individual
situation. The present in vitro study provides an essential
base for the use of synthesized mRNA in clinical trials. Further
in vivo studies particularly combined with the use of
clinical tumor samples may lead to development of a clinically
meaningful patient-tailored anticancer therapy.
References
|
1
|
Vandamme N and Berx G: Melanoma cells
revive an embryonic transcriptional network to dictate phenotypic
heterogeneity. Front Oncol. 4:3522014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stewart BW and Wild CP: World Health
Organization World Cancer Report 2014. Chapter5.14; pp. 495–502.
2014
|
|
3
|
Jerant AF, Johnson JT, Sheridan CD and
Caffrey TJ: Early detection and treatment of skin cancer. Am Fam
Physician. 62:357–368. 375–376. 381–382. 2000.PubMed/NCBI
|
|
4
|
Halachmi S and Gilchrest BA: Update on
genetic events in the pathogenesis of melanoma. Cur Opin Oncol.
13:129–136. 2001. View Article : Google Scholar
|
|
5
|
Kanavy HE and Gerstenblith MR: Ultraviolet
radiation and melanoma. Semin Cutan Med Surg. 30:222–228. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jost LM; ESMO Guidelines Task Force: ESMO
minimum clinical recommendations for diagnosis, treatment and
follow-up of cutaneous malignant melanoma. Ann Oncol. 14:1012–1013.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Balch CM, Soong SJ, Gershenwald JE,
Thompson JF, Reintgen DS, Cascinelli N, Urist M, McMasters KM, Ross
MI, Kirkwood JM, et al: Prognostic factors analysis of 17,600
melanoma patients: Validation of the American joint committee on
cancer melanoma staging system. J Clin Oncol. 19:3622–3634.
2001.PubMed/NCBI
|
|
8
|
Sun XY, Nong J, Qin K, Lu H, Moniri MR,
Dai LJ and Warnock GL: MSC(TRAIL)-mediated HepG2 cell death in
direct and indirect co-cultures. Anticancer Res. 31:3705–3712.
2011.PubMed/NCBI
|
|
9
|
Loebinger MR and Janes SM: Stem cells as
vectors for anti-tumour therapy. Thorax. 65:362–369. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dai LJ, Moniri MR, Zeng ZR, Zhou JX, Rayat
J and Warnock GL: Potential implications of mesenchymal stem cells
in cancer therapy. Cancer Lett. 305:8–20. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang B, Wu X, Mao Y, Bao W, Gao L, Zhou P,
Xie R, Zhou L and Zhu J: Dual-targeted antitumor effects against
brainstem glioma by intravenous delivery of tumor necrosis
factor-related, apoptosis-inducing, ligand-engineered human
mesenchymal stem cells. Neurosurgery. 65:610–624. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ivanov DP, Parker TL, Walker DA, Alexander
C, Ashford MB, Gellert PR and Garnett MC: In vitro co-culture model
of medulloblastoma and human neural stem cells for drug delivery
assessment. J Biotechnol. 205:3–13. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mao ZG, Jiang CC, Yang F, Thorne RF,
Hersey P and Zhang XD: TRAIL-induced apoptosis of human melanoma
cells involves activation of caspase-4. Apoptosis. 15:1211–1222.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Friedenstein AJ, Deriglasova UF, Kulagina
NN, Panasuk AF, Rudakowa SF, Luriá EA and Ruadkow IA: Precursors
for fibroblasts in different populations of hematopoietic cells as
detected by the in vitro colony assay method. Exp Hematol. 2:83–92.
1974.PubMed/NCBI
|
|
15
|
Zuk PA, Zhu M, Mizuno H, Huang J, Futrell
JW, Katz AJ, Benhaim P, Lorenz HP and Hedrick MH: Multilineage
cells from human adipose tissue: Implications for cell-based
therapies. Tissue Eng. 7:211–228. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zuk PA, Zhu M, Ashjian P, De Ugarte DA,
Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P and Hedrick
MH: Human adipose tissue is a source of multipotent stem cells. Mol
Biol Cell. 13:4279–4295. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kern S, Eichler H, Stoeve J, Klüter H and
Bieback K: Comparative analysis of mesenchymal stem cells from bone
marrow, umbilical cord blood, or adipose tissue. Stem Cells.
24:1294–1301. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wiley SR, Schooley K, Smolak PJ, Din WS,
Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA,
et al: Identification and characterization of a new member of the
TNF family that induces apoptosis. Immunity. 3:673–682. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu GS: TRAIL as a target in anti-cancer
therapy. Cancer Lett. 285:1–5. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rosenecker J, Huth S and Rudolph C: Gene
therapy for cystic fibrosis lung disease: Current status and future
perspectives. Curr Opin Mol Ther. 8:439–445. 2006.PubMed/NCBI
|
|
21
|
Yamashita Y, Shimada M, Tanaka S,
Okamamoto M, Miyazaki J and Sugimachi K: Electroporation-mediated
tumor necrosis factor-related apoptosis-inducing ligand
(TRAIL)/Apo2L gene therapy for hepatocellular carcinoma. Hum Gene
Ther. 13:275–286. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashkenazi A, Pai RC, Fong S, Leung S,
Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert
A, et al: Safety and antitumor activity of recombinant soluble Apo2
ligand. J Clin Invest. 104:155–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grisendi G, Bussolari R, Cafarelli L,
Petak I, Rasini V, Veronesi E, De Santis G, Spano C, Tagliazzucchi
M, Barti-Juhasz H, et al: Adipose-derived mesenchymal stem cells as
stable source of tumor necrosis factor-related apoptosis-inducing
ligand delivery for cancer therapy. Cancer Res. 70:3718–3729. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dy GK and Adjei AA: Understanding,
recognizing, and managing toxicities of targeted anticancer
therapies. CA Cancer J Clin. 63:249–279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zangi L, Lui KO, von Gise A, Ma Q, Ebina
W, Ptaszek LM, Später D, Xu H, Tabebordbar M, Gorbatov R, et al:
Modified mRNA directs the fate of heart progenitor cells and
induces vascular regeneration after myocardial infarction. Nat
Biotechnol. 31:898–907. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshioka N, Gros E, Li HR, Kumar S, Deacon
DC, Maron C, Muotri AR, Chi NC, Fu XD, Yu BD and Dowdy SF:
Efficient generation of human iPSCs by a synthetic self-replicative
RNA. Cell Stem Cell. 13:246–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leonhardt C, Schwake G, Stögbauer TR,
Rappl S, Kuhr JT, Ligon TS and Rädler JO: Single-cell mRNA
transfection studies: Delivery, kinetics and statistics by numbers.
Nanomedicine. 10:679–688. 2014.
|
|
28
|
Stepinski J, Waddell C, Stolarski R,
Darzynkiewicz E and Rhoads RE: Synthesis and properties of mRNAs
containing the novel 'anti-reverse' cap analogs 7-methyl
(3′-O-methyl) GpppG and 7-methyl (3′-deoxy) GpppG. RNA.
7:1486–1495. 2001.PubMed/NCBI
|