Introduction
Acinetobacter baumannii (A. baumannii)
has emerged as a major pathogen of nosocomial infections and is
associated with high rates of morbidity and mortality in recent
years (1,2). A nationwide surveillance program,
including hospitals from 14 geographically different regions in
China revealed that the ratio of A. baumannii is on the
increase annually (3).
Carbapenem has good antibacterial activity against
A. baumannii and was the first choice in treatment of
infection caused by A. baumannii in the past years (4). However, the emergence of resistance
to carbapenem was reported in 1991 (5), followed by similar reports from
different parts of the world (6,7). In
China, 57 and 61% of Acinetobacter spp. (A. baumannii
accounted for 89.6%) showed resistance to imipenem and meropenem,
respectively (3). International
studies in China as well as in other parts of the world focused
only on evaluating the resistance of A. baumannii to various
antimicrobials (8–10). However, to the best of our
knowledge, few studies have investigated the molecular mechanism
underlying drug resistance. Additionally, no data are available on
the epidemiological characteristics of imipenem-resistant A.
baumannii in Shanghai.
Thus, A. baumannii clinical isolates were
collected from three tertiary hospitals in Shanghai and their drug
resistance pattern to a spectrum of antimicrobials, molecular
mechanisms (including carbapenemase, efflux pumps and membrane
proteins) behind their resistance and multilocus sequence analysis
(MLST) were analyzed to assess their molecular epidemiology.
Materials and methods
Bacterial strains
During the period July, 2011 to June, 2012, 46
non-duplicate imipenem-resistant A. baumannii strains were
collected from three tertiary hospitals located in Shanghai,
China.
Reconfirmation of strains
The collected strains were subjected to gram
staining, biochemical tests, and recA gene and 16S-23S rRNA
gene intergenic spacer region to reconfirm them as A.
baumannii (7).
Antimicrobial susceptibility and efflux
phenotype tests
The collected A. baumannii isolates were
subjected to an antimicrobial susceptibility test against imipenem,
meropenem, amikacin, piperacillin, ceftazidime, cefotaxime,
mino-cycline, ciprofloxacin, ampicillin/sulbactam, sulbactam,
cefoperazone/sulbactam, piperacillin/tazobactam, colistin,
tigecycline and trimethoprim/sulfamethoxazole using agar dilution
method. Escherichia coli strain ATCC25922 and Pseudomonas
aeruginosa (P. aeruginosa) strain ATCC27853 were used as
reference strains.
Strains in which efflux pump operation was detected
by agar dilution method where imipenem- and meropenem-resistant
isolates were cultured in Mueller-Hinton agar contained the efflux
pump inhibitor phenylalanine-arginine β-naphthylamide (PAβN) at a
final concentration of 20 mg/l (11,12).
A ≥4-fold reduction of imipenem or meropenem minimal inhibitory
concentrations (MICs) in the presence of PAβN possessed an
operating drug efflux pump.
Analysis of genes responsible for drug
resistance, drug efflux and outer membrane protein
Polymerase chain reaction (PCR) was performed for
the genes, blaKPC, blaIMP,
blaNDM, blaOXA-51,
blaOXA-23, blaOXA-24,
blaOXA-58, blaSHV,
blaGIM and blaVIM, CarO,
adeA, adeB, adeC, adeS and adeR.
Thus, obtained amplicons were subjected to sequencing analysis.
A fresh and pure bacterial colony was suspended in
distilled water and boiled at 100°C for 15 min. After
centrifu-gation at 8,000 × g for 15 min, 1 µl of the
supernatant was used for PCR analysis with the primers (Table I). PCR was performed in a total
volume of 50 µl containing 0.25 µl Taq DNA polymerase
(Takara Bio, Inc., Tokyo, Japan), 5 µl 10X PCR buffer
(Mg2+ Plus), 4 µl dNTP mixture (2.5 mM each), 2.5
µl DNA template, 1 µl of each primer (20 µM),
and 36.25 µl ddH2O. The PCR thermal cycle
consisted of initial denaturation at 94°C for 5 min, followed by 30
cycles of 94°C for 30 sec, annealing 55°C for 1 min and 72°C for 1
min and a final extension at 72°C for 7 min. The PCR products were
electrophoresed in 1% agarose gel and visualized under ultraviolet
light, and subsequently sequenced (Sangon Biotech Co., Ltd.,
Shanghai, China).
 | Table IGene-specific primers used in this
study. |
Table I
Gene-specific primers used in this
study.
| Genes | Primer
sequences |
|---|
| recA | F:
CCTGAATCTTCTGGTAAAAC
R: GTTTCTGGGCTGCCAAACATTAC |
| ITS | F:
CATTATCACGGTAATTAGTG
R: AGAGCACTGTGCACTTAAG |
|
blaKPC | F:
TTACTGCCCGTTGACGCCCAATCC
R: TCGCTAAACTCGAACAGG |
|
blaIMP | F:
AACCAGTTTTGCCTTACCAT
R: CTACCGCAGCAGAGTCTTTG |
|
blaNDM | F:
CCGCCCAGATCCTCAACT
R: ATCAGGCAGCCACCAAAA |
|
blaOXA-51 | F:
TAATGCTTTGATCGGCCTTG
R: TGGATTGCACTTCATCTTGG |
|
blaOXA-23 | F:
GATCGGATTGGAGAACCAGA
R: ATTTCTGACCCATTTCCAT |
|
blaOXA-24 | F:
GGTTAGTTGGCCCCCTTAAA
R: AGTTGAGCGAAAAGGGGATT |
|
blaOXA-58 | F:
AAGTATTGGGGCTTGTGCTG
R: CCCCTCTGCGCTCTACATAC |
|
blaSHV | F:
GGTTATGCGTTATATTCGCC
R: TTAGCGTTGCCAGTGCTC |
|
blaGIM | F:
AGAACCTTGACCGAACGCAG
R: ACTCATGACTCCTCACGAGG |
|
blaVIM | F:
TCCACGCACTTTCATGACGA
R: AGACGTGCGTGACAACTCAT |
| adeA | F:
GAAATCCGTCCGCAAGTC
R: ACACGCACATACATACCC |
| adeB | F:
AAAGACTTCAAAGAGCGG
R: TCACGCATTGCTTCACCC |
| adeC | F:
ATTTCAGGTCGTAGCATT
R: CTTGATAAGTAGAGTAGGGATT |
| adeS | F:
ACTGTTATCTTCTGTGGCTGTA
R: GTGGACGTTAGGTCAAGTTCTG |
| adeR | F:
AAACGGTTGGGAAGTATTA
R: ATGGCTATCTACGGTTCG |
| CarO | F:
AAGGAGAAAACGATGA
R: TTATTACGTGGTTATGG |
| gltA | F:
AATTTACAGTGGCACATTAGGTCCC
R: GCAGAGATACCAGCAGAGATACACG |
| gyrB | F:
TGAAGGCGGCTTATCTGAGT
R: GCTGGGTCTTTTTCCTGACA |
| gdhB | F:
ACCACATGCTTTGTTATG
R: GTTGGCGTATGTTGTGC |
| recA | F:
CCTGAATCTTCYGGTAAAAC
R: GTTTCTGGGCTGCCAAACATTAC |
| cpn60 | F:
GGTGCTCAACTTGTTCGTGA
R: CACCGAAACCAGGAGCTTTA |
| Gpi | F:
GAAATTTCCGGAGCTCACAA
R: TCAGGAGCAATACCCCACTC |
| rpoD | F:
ACCCGTGAAGGTGAAATCAG
R: TTCAGCTGGAGCTTTAGCAAT |
MLST
Seven housekeeping genes including homologous
recombination factor (recA), citrate synthase (gltA),
DNA gyrase subunit (gyrB), glucose-6-phosphate isomerase
isomerase (gpi), glucose dehydrogenase B (gdhB),
60-kDa chaperonin (cpn60), and RNA polymerase 70 factor
(rpoD) were amplified in PCR using relevant primers
(Table I) and appropriate thermal
conditions. The amplicons were sequenced and the sequences were
submitted to the MLST database (http://pubmlst.org.net) to compare them with sequences
submitted from other parts of the world. Each strain was then
characterized by a pattern of numbers defining its allelic
profile.
Results
Antimicrobial susceptibility
A. baumannii resistant to imipenem
simultaneously showed resistance to several other common
antimicrobials. The resistance rate was >80% for all the
antimicrobials except minocycline and colistin. Antibiotic
susceptibility of the 46 clinical isolates is shown in Table II. Thirteen imipenem-resistant
A. baumannii isolates were positive for efflux pump.
 | Table IIThe drug-resistant rates of
imipenem-resistant Acinetobacter baumannii. |
Table II
The drug-resistant rates of
imipenem-resistant Acinetobacter baumannii.
| Drug | Resistance
rate
No. of resistant strains (%) |
|---|
| Meropenem | 41 (89) |
| Amikacin | 38 (83) |
| Piperacillin | 46 (100) |
| Ceftazidime | 46 (100) |
| Minocycline | 34 (74) |
| Ciprofloxacin | 45 (98) |
|
Ampicillin/sulbactam | 43 (93) |
|
Piperacillin/tazobactam | 46 (100) |
| Colistin | 1 (2) |
|
Trimethoprim/sulfamethoxazole | 43 (93) |
| Cefotaxime | 45 (98) |
Detection of genes involved in drug
resistance, drug efflux and outer membrane protein
Of the various drug resistance genes tested,
blaOXA-51 was present in 46 isolates,
blaOXA-23 gene was present in 44 isolates and
blaNDM gene was found in only one strain. Other
drug-resistant genes including blaKPC,
blaIMP, blaOXA-24,
blaOXA-58, blaSHV,
blaGIM and blaVIM were not
detected in the isolates.
Of the five genes associated with the drug efflux
pump tested, all five were found to be present in the isolates.
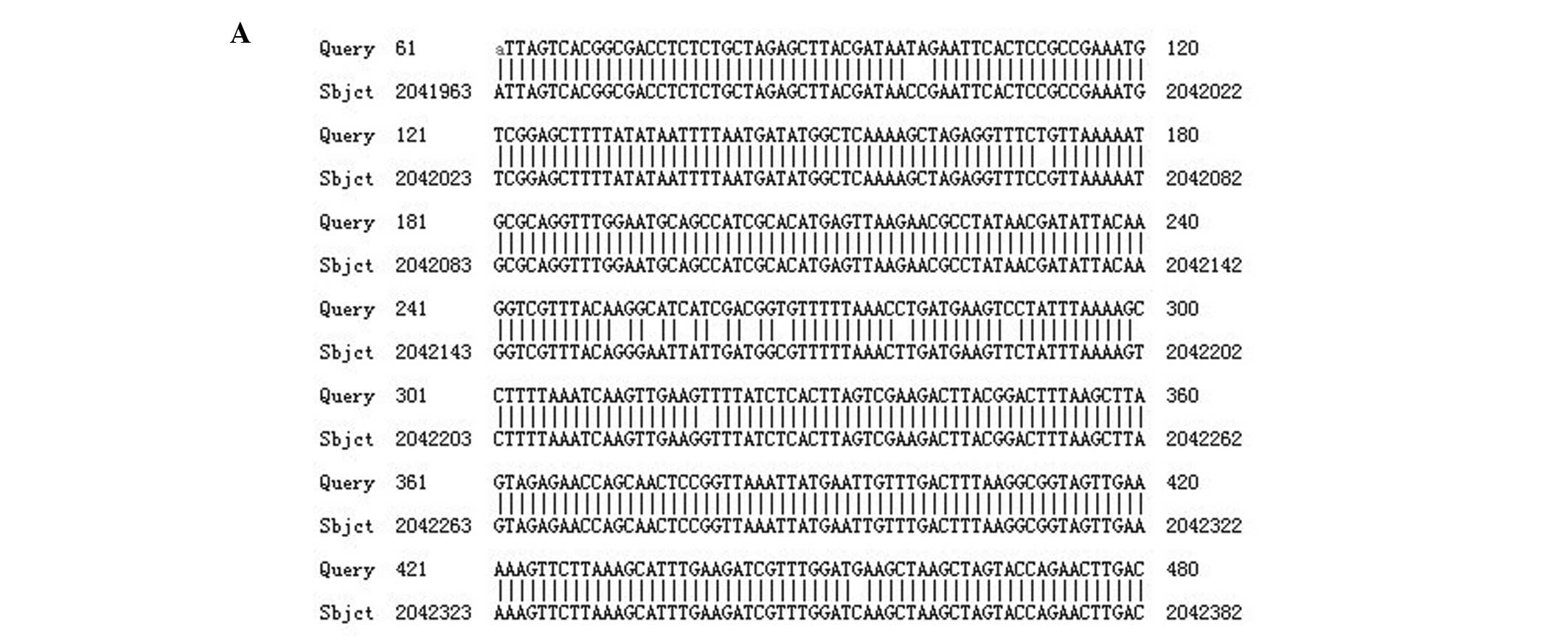
Several mutations were found in the sequences of adeS gene
in isolates with efflux phenotype. Differences were observed at
three places when nucleotide sequences were translated into an
amino acid sequence. This amino acid sequence was then compared to
the amino acid sequence of the reference strain ATCC17978 (Fig. 1).
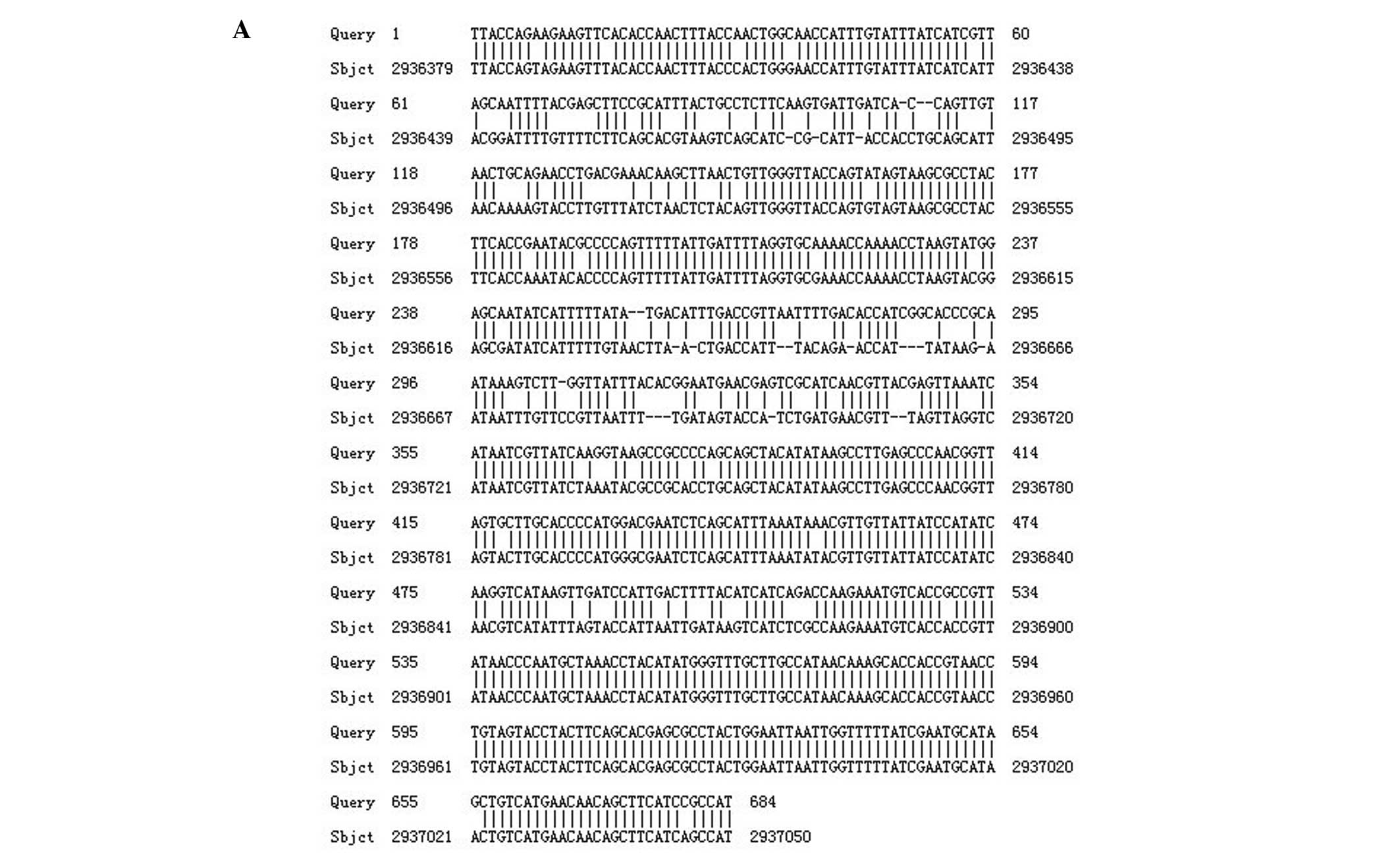
Similarly, the nucleotide sequence of the outer
membrane protein encoding gene CarO, when compared with the
nucleotide sequence of reference strain ATCC17978, harbored
mutations that were reflected in the amino acid sequence (Fig. 2).
Genotyping of isolates by MLST
The MLST analysis revealed that the isolates were
clustered in 11 different genotypes or STs. The ST208 genotype was
shared by the majority of isolates (58.7%, 27/46), followed by
ST191 (10.9%, 5/46) and ST451 (6.5%, 3/46). We also detected some
other STs shared by certain isolates such as ST75 (2.1%, 1/46),
ST90 (4.2%, 2/46), ST92 (2.1%, 1/46), ST108 (2.1%, 31/46), ST109
(2.1%, 1/46), ST172 (2.1%, 1/46), ST368 (4.2%, 2/46) and ST69
(4.2%, 2/46). These STs were grouped into the three clonal
complexes, CC92, CC109 and CC28.
Discussion
A. baumannii develops resistance to imipenem
through a variety of mechanisms. Carbapenemase is an important
factor responsible for imipenem resistance. In the present study,
common carbapenemases were detected in the isolates, including
blaOXA-51, blaOXA-23,
blaOXA-24, blaOXA-58,
blaKPC, blaIMP,
blaSHV, blaGIM,
blaNDM and blaVIM. OXA-type
enzymes are naturally present in Acinetobacter spp. and are
usually expressed in small amounts (13). The expression of such genes is
markedly higher under the effect of a strong promoter (insertion
sequence ISAba1 is the most shared) and induce drug resistance only
when combined with a reduction in outer membrane permeability
and/or activation of the efflux pump (14). In the present study,
blaOXA-51 and blaOXA-23 genes
were prevalent among the isolates, results that are consistent with
other reports (15–17). Carbapenemases that are different
from OXA, such as KPC, IMP, SHV, GIM, NDM and VIM have strong
carbapenem-hydrolysing activity (14). However, such types of
carbapenemases were rarely detected in A. baumannii. The
blaNDM gene was identified in only one strain
while the remaining resistant genes were not detected. NDM was
first identified in Escherichia coli and Klebsiella
pneumoniae in 2008 in India (18). This finding was followed by reports
on NDM-producing P. aeruginosa, Enterobacter cloacae,
Citrobacter freundii and Enterococcus faecium
(19–23). In China, NDM-producing A.
baumannii was first reported in 2011. Of the anti microbials
tested one NDM-positive isolate in the present study was identified
that was multidrug-resistant, and only susceptible to amikacin,
colistin and minocycline.
Drug efflux systems including AdeABC, AdeIJK, AdeDE
and AdeXYZ (RND family) have been found in A. baumannii. Of
these, the AdeABC efflux system is common in A. baumannii
(24). This efflux pump, together
with other resistant mechanisms, can lead to high-level imipenem
resistance. Although mediated by the substrate, its expression may
increase when a single point mutation occurs in the adeR or
adeS gene (25). PAβN was
proven to be an effective inhibitor of drug efflux. In the present
study, adeA, adeB and adeC were present in all
of the isolates because when PAβN was added the MICs inherent to
imipenem in 13 isolates were decreased. The adeS gene
differed from the adeS of standard strain and this is the
possible reason for increased drug efflux associated with drug
resistance.
Few studies concerning the impact of changes on
membrane proteins in A. baumannii are available. In 2002, a
laboratory in Argentina advocated for the first time that inducible
resistance by imipenem can trigger loss of a 29-kDa membrane
protein. In 2005, the same laboratory furthering their study,
demonstrated that the outer membrane protein is encoded by the
CarO gene and when there is an insertion mutation or any
other mutation in the CarO gene makes it off and thus the
strain become resistant to certain drugs (26). In the present study, the sequence
of CarO gene had nucleotide insertions, deletions and point
mutations in comparison with the standard strains and there were
also differences in their nucleotide and amino acid sequences.
In summary, for a global epidemiologic analysis, a
comparison of the results between different laboratories is
required. MLST is a powerful tool used to transfer typing data and
compare results via relevant databases. The MLST analysis revealed
that the major epidemic clone of A. baumannii in Shanghai
was ST208 (CC92 clone complex), which differed from the results
obtained in other regions in China (27).
Acknowledgments
The project was supported by a grant from the
Natural Science Foundation of Shanghai Science and Technology
Committee (no. 12ZR1426200), the Medical Guide Program of Shanghai
Science and Technology Committee (no. 14411962900), Key project of
Shanghai Municipal Health and Family Planning Commission (no.
201540367) and Central Universities Basic Research Program (no.
1511219024).
References
|
1
|
Perez F, Hujer AM, Hujer KM, Decker BK,
Rather PN and Bonomo RA: Global challenge of multidrug-resistant
Acinetobacter baumannii. Antimicrob Agents Chemother. 51:3471–3484.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Giamarellou H, Antoniadou A and
Kanellakopoulou K: Acinetobacter baumannii: A universal threat to
public health? Int J Antimicrob Agents. 32:106–119. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang F, Zhu D, Hu F, Jiang X, Hu Z, Li Q,
Sun Z, Chen Z, Xu Y, Zhang X, et al: CHINET 2012 surveillance of
bacterial resistance in China. China J Infect Chemother.
13:321–330. 2013.Chinese.
|
|
4
|
Kim YJ, Kim SI, Hong KW, Kim YR, Park YJ
and Kang MW: Risk factors for mortality in patients with
carbapenem-resistant Acinetobacter baumannii bacteremia: Impact of
appropriate antimicrobial therapy. J Korean Med Sci. 27:471–475.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Go ES, Urban C, Burns J, Kreiswirth B,
Eisner W, Mariano N, Mosinka-Snipas K and Rahal JJ: Clinical and
molecular epidemiology of acinetobacter infections sensitive only
to polymyxin B and sulbactam. Lancet. 344:1329–1332. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pournaras S, Markogiannakis A, Ikonomidis
A, Kondyli L, Bethimouti K, Maniatis AN, Legakis NJ and Tsakris A:
Outbreak of multiple clones of imipenem-resistant Acinetobacter
baumannii isolates expressing OXA-58 carbapenemase in an intensive
care unit. J Antimicrob Chemother. 57:557–561. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kuo LC, Teng LJ, Yu CJ, Ho SW and Hsueh
PR: Dissemination of a clone of unusual phenotype of
pandrug-resistant Acinetobacter baumannii at a university hospital
in Taiwan. J Clin Microbiol. 42:1759–1763. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu J, Sun Z, Li Y and Zhou Q: Surveillance
and correlation of antibiotic consumption and resistance of
Acinetobacter baumannii complex in a tertiary care hospital in
northeast China, 2003–2011. Int J Environ Res Public Health.
10:1462–1473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kwon NY, Kim JD and Pai HJ: The resistance
mechanisms of b-lactam antimicrobials in clinical isolates of
Acinetobacter baumannii. Korean J Intern Med. 17:94–99. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
García-Quintanilla M, Pulido MR,
Moreno-Martínez P, Martín-Peña R, López-Rojas R, Pachón J and
McConnell MJ: Activity of host antimicrobials against
multidrug-resistant Acinetobacter baumannii acquiring colistin
resistance through loss of lipopolysaccharide. Antimicrob Agents
Chemother. 58:2972–2975. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen TL, Siu LK, Wu RC, Shaio MF, Huang
LY, Fung CP, Lee CM and Cho WL: Comparison of one-tube multiplex
PCR, automated ribotyping and intergenic spacer (ITS) sequencing
for rapid identification of Acinetobacter baumannii. Clin Microbiol
Infect. 13:801–806. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clinical Laboratory Standards Institute
(CLSI): Performance standards for antimicrobial susceptibility
testing; Twenty-second informational supplement. CLSI document
M100-S22. CLSI; Wayne, PA: 2012
|
|
13
|
Zhang JP, Zhu W, Tian SF, Chu YZ and Chen
BY: Molecular characteristics and resistant mechanisms of
imipenem-resistant Acinetobacter baumannii isolates in Shenyang.
China J Microbiol. 48:689–694. 2010. View Article : Google Scholar
|
|
14
|
Merkier AK and Centrón D: bla(OXA-51)-type
beta-lactamase genes are ubiquitous and vary within a strain in
Acinetobacter baumannii. Int J Antimicrob Agents. 28:110–113. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He C, Xie Y, Zhang L, Kang M, Tao C, Chen
Z, Lu X, Guo L, Xiao Y, Duo L, et al: Increasing imipenem
resistance and dissemination of the ISAba1-associated blaOXA-23
gene among Acinetobacter baumannii isolates in an intensive care
unit. J Med Microbiol. 60:337–341. 2011. View Article : Google Scholar
|
|
16
|
Martínez P and Mattar S:
Imipenem-resistant Acinetobacter baumannii carrying the ISAba1-bla
OXA-23,51 and ISAba1-bla ADC-7 genes in Monteria, Colombia. Braz J
Microbiol. 43:1274–1280. 2012. View Article : Google Scholar
|
|
17
|
Corvec S, Poirel L, Naas T, Drugeon H and
Nordmann P: Genetics and expression of the carbapenem-hydrolyzing
oxacil-linase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob
Agents Chemother. 51:1530–1533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yong D, Toleman MA, Giske CG, Cho HS,
Sundman K, Lee K and Walsh TR: Characterization of a new
metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin
esterase gene carried on a unique genetic structure in Klebsiella
pneumoniae sequence type 14 from India. Antimicrob Agents
Chemother. 53:5046–5054. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cabanes F, Lemant J, Picot S, Simac C,
Cousty J, Jalin L, Naze F, Boisson V, Cresta MP, André H, et al:
Emergence of Klebsiella pneumoniae and Salmonella
metallo-beta-lactamase (NDM-1) producers on reunion island. J Clin
Microbiol. 50:38122012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Savard P, Gopinath R, Zhu W, Kitchel B,
Rasheed JK, Tekle T, Roberts A, Ross T, Razeq J, Landrum BM, et al:
First NDM-positive Salmonella sp. strain identified in the United
States. Antimicrob Agents Chemother. 55:5957–5958. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jovcic B, Lepsanovic Z, Suljagic V, Rackov
G, Begovic J, Topisirovic L and Kojic M: Emergence of NDM-1
metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates
from Serbia. Antimicrob Agents Chemother. 55:3929–3931. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Poirel L, Dortet L, Bernabeu S and
Nordmann P: Genetic features of blaNDM-1-positive
Enterobacteriaceae. Antimicrob Agents Chemother. 55:5403–5407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH,
Ang I, Tong AH, Bao JY, Lok S and Lo JY: Complete sequencing of
pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant
Escherichia coli strain isolated in Hong Kong. PLoS One.
6:e179892011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chu YW, Chau SL and Houang ET: Presence of
active efflux systems AdeABC, AdeDE and AdeXYZ in dierent AbeM
(MATE) Ade ABC Acinetobacter genomic DNA groups. J Med Microbiol.
55:477–478. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marchand I, Damier-Piolle L, Courvalin P
and Lambert T: Expression of the RND-type efflux pump AdeABC in
Acineto-bacter baumannii is regulated by the AdeRS two-compo nent
system. Antimicrob Agents Chemother. 48:3298–3304. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mussi MA, Limansky AS and Viale AM:
Acquisition of resistance to carbapenems in multidrug-resistant
clinical strains of Acinetobacter baumannii: Natural insertional
inactivation of a gene encoding a member of a novel family of
β-barrel outer membrane proteins. Antimicrob Agents Chemother.
49:1432–1440. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou Z, Du X, Wang L, Yang Q, Fu Y and Yu
Y: Clinical carbapenem-resistant Acinetobacter baylyi strain
coharboring blaSIM-1 and blaOXA-23 from China. Antimicrob Agents
Chemother. 55:5347–5349. 2011. View Article : Google Scholar : PubMed/NCBI
|