Introduction
Glaucoma is a complex, heterogeneous ocular disorder
with multifactorial etiology characterized by structural damage to
the optic nerve, visual field defects (1) and commonly relatively high
intraocular pressure (IOP) (2). It
is a leading cause of irreversible blindness worldwide (3,4) with
~20% of cases occurring secondary to other ocular or systemic
diseases (1). Based on anatomical
changes in the anterior chamber angle, primary glaucoma may be
classified as primary angle closure glaucoma (PACG) or primary
open-angle glaucoma (POAG), which may be further subdivided into
juvenile open-angle glaucoma (JOAG) and adult onset POAG (1,5).
Glaucoma is a treatable disease if detected early; however, many
patients are diagnosed during routine examinations or only
following advanced field loss, as glaucoma is typically
asymptomatic in the early stages. Therefore, the development of an
accurate test for the detection of presymptomatic carriers at risk
is important for the management of glaucoma.
A family history of glaucoma is a well-known risk
factor. Therefore genetic background is considered important for
the development of the disease (6–8). The
following genes have been reported to be associated with primary
glaucoma: myocilin (MYOC) (9), optineurin (OPTN)
(10), WD repeat domain 36
(11,12), neurotrophin 4 (13), optic atrophy 1 (14,15),
cytochrome P450 family 1 subfamily B member 1 (16) and latent transforming growth
factor β (17). To date,
mutations in these genes account for only ~5% of patients with POAG
(18), and the influence of
mutations in these genes on patients with PACG remain controversial
(19). The MYOC gene,
located at the GLC1A locus on chromosome 1q24.3–q25.2, has been
confirmed to be associated with JOAG and POAG, although mutation
frequencies vary between ethnic groups (20). The altered protein product secreted
into the extracellular matrix of the trabecular meshwork causes a
severe form of autosomal dominant JOAG associated with very high
IOP (9). Although up to 20% of
JOAG may be associated with MYOC mutations, mutations in
this gene have been identified in only 3 to 5% of POAG patients
(7,21). Mutations in the OPTN gene,
located at the GLC1E locus on chromosome 10p14–p15, result in
normal tension glaucoma (NTG) without elevated IOP, a subtype of
POAG (10).
To investigate the underlying genetic influences of
primary glaucoma, molecular analysis of the MYOC and
OPTN genes was performed in 112 Korean sporadic primary
glaucoma patients, with the results compared to healthy
controls.
Patients and methods
Patients
All 112 patients with primary glaucoma from
unrelated Korean patients were recruited from the ophthalmology
clinic of Seoul St. Mary's Hospital (Seoul, Korea), including 17
POAG, 19 JOAG and 76 NTG patients. Healthy, unrelated Korean
individuals (n=100) served as the non-selected population control.
All patients and healthy controls in the study provided written
informed consent for clinical and molecular analyses, and the study
protocol was approved by the institutional review board of The
Catholic University of Korea (Seoul, Korea). The diagnostic
criteria of POAG were an IOP >21 mmHg by Goldmann applanation
tonometry (GAT), presence of glaucomatous visual field defect or
glaucomatous optic neuropathy and open angle. Where onset occurred
at 5–35 years of age, patients were diagnosed with JOAG (22). The diagnostic criteria of NTG were
an IOP <22 mmHg by GAT, presence of glaucomatous visual field
defect or glaucomatous optic neuropathy and open angle. Patients
with secondary glaucoma resulting from trauma, uveitis, neovascular
or steroid-induced glaucoma (SIG), and other associated ocular or
systemic anomalies were excluded from the present study. Healthy
controls had IOP <20 mmHg, a normal optic disc appearance
without suspicious glaucomatous changes, and no family or personal
history of primary glaucoma.
Molecular analysis of the MYOC and OPTN
genes
Genomic DNA samples were extracted from the
peripheral blood using the QIAamp DNA Mini kit (Qiagen GmbH,
Hilden, Germany). Entire coding exons and flanking intronic
sequences of MYOC and OPTN were amplified by
polymerase chain reaction (PCR) using different combinations of
forward and reverse primers designed by the authors (Table I). Briefly, the PCR reaction had a
total volume of 15 μl, comprising 10 μl Taq
PCR Master Mix kit (Qiagen GmbH) containing Taq DNA
Polymerase, 2X Qiagen PCR Buffer, 3 mM MgCl2, and 400
μM of each deoxynucleotide, 2 μl 5X Q-Solution, 1
μl primer mix, and 2 μl genomic DNA (50
ng/μl). The PCR was amplified with a C1000 thermal cycler
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) under the following
conditions: 5 min at 95°C, followed by 35 cycles of 95°C for 30
sec, 60°C for 30 sec and 72°C for 1 min, and a final step of 72°C
for 5 min. Direct sequencing of PCR products was performed with the
BigDye Terminator version 3.1 Cycle Sequencing kit (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
the products were resolved on ABI 3130XL Genetic Analyzer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The resulting sequence
electropherogram was analyzed using Sequencher software version 4.9
(Gene Codes Corporation, Ann Arbor, MI, USA). Sequence data were
analyzed using reference sequences in GenBank (www.ncbi.nlm.nih.gov/genbank/). The
RefSeq IDs NM_000261.1 for MYOC and NM_001008211.1 for
OPTN were used for cDNA nucleotide numbering. All identified
variants were confirmed by bidirectional resequencing.
 | Table IOligonucleotide primer sequences used
for mutation analysis of MYOC and OPTN genes. |
Table I
Oligonucleotide primer sequences used
for mutation analysis of MYOC and OPTN genes.
| Gene | Exon | Primer sequence (5′
to 3′)
| Size (bp) |
|---|
| Forward | Reverse |
|---|
| MYOC | 1a |
TCTTGCTGGCAGCGTG |
CTGGTCCAAGGTCAATTGGT | 626 |
| MYOC | 1b |
AGCACCCAACGCTTAGACCT |
TCTGTCTTGTGCTAGCTGTGC | 497 |
| MYOC | 2 |
TGCCACCACATCCAGCTAAT |
CTCTGCTCCCAGGGAAGTTA | 495 |
| MYOC | 3a |
ACCCAGACGATTTGTCTCCA |
GCCTCATCGGTGCTGTAAAT | 586 |
| MYOC | 3b |
CGCTGAGTCCAGAACTGTCA |
CGCCCTCAGACTACAATTCC | 685 |
| OPTN | 1 |
CGGACAGCGAGGGTGGGTA |
GCGGGTACCGTTTTCAGG | 445 |
| OPTN | 2 |
TCCACATGGATGCCTCTACA |
TTCCCATGCAAATCTTCAAA | 459 |
| OPTN | 3 |
TGTTAGCCAGGATGGTCTCA |
AGAGGTTGATGGGACATTGC | 391 |
| OPTN | 4 |
CACACACACACTTTTCTGAAGC |
CCCCACCAGCTACCACCTAT | 497 |
| OPTN | 5 |
CTTCGTCTTTTTGCTGCTGA |
CTTCCAAGACCAGGCAAAAC | 492 |
| OPTN | 6 |
TGTAAAGATGGGGGTCTTGC |
GAAAATGAGAGCCAATTTATCTTTG | 491 |
| OPTN | 7 |
CTTGGGTTGCATGTCACAAA |
CAGTGTGAGCCAAACAGGAA | 398 |
| OPTN | 8 |
GACCAGCTGTGCTTGTTCAC |
CAGACAGTGAGTGCTGTTTGG | 495 |
| OPTN | 9 |
TTTCACTTGCCTTTTACCTCTG |
GACACAGAGCAGGACAAGGA | 498 |
| OPTN | 10 |
TTGGGGTATTGTCAAAGTTGG |
ATGCCCCTAAATGGCAGAAT | 486 |
| OPTN | 11 |
TCATAAACCCTACAGCCCTAAAA |
TGCTAGGACTCCTTCAGATAAGTG | 398 |
| OPTN | 12 |
GCTAGTAGGTCGTGGGGTGA |
GGAAAACAACCTTTGAAACCA | 346 |
| OPTN | 13 |
CCGGCCAGAGCTGATAAT |
TTTTAATACACTCACGGGTGAAA | 394 |
| OPTN | 14 |
AGCAGGATTGTGCATCTGTG |
GCGCGAACACAGCTATTCTT | 369 |
| OPTN | 15 |
GGTTTTTATGAACCTTGGCAGT |
GATTCGGTGGGTAATGGATG | 381 |
| OPTN | 16 |
TGCATCGTGATGACTTCAGTT |
CTCAAACCCTGACCCCAAGT | 500 |
Genetic variations were contrasted with Human Gene
Mutation Database Public (www.hgmd.cf.ac.uk), 1,000 genomes project browser
(browser.1000genomes.org) and the single
nucleotide polymorphism database (www.ncbi.nlm.nih.gov/projects/SNP). Mutation
pathogenicity was predicted through PolyPhen-2 version 2.2.2
(genetics.bwh.harvard.edu/pph2)
(23) and PROVEAN version 1.1
(provean.jcvi.org) (24), the first predictor that includes
in-frame insertions/deletions. To assess the extent of conservation
of a novel variant of MYOC and OPTN hypothesized to
be associated with disease, the amino acid sequence was aligned
with protein sequences of various mammalian species using Clustal
Omega software (www.ebi.ac.uk/Tools/msa/clustalo/) (25).
Haplotype analysis of the MYOC and OPTN
genes
Haplotypes of each gene were constructed based on
the genotype data of the MYOC and OPTN variants
obtained from the 112 patients and 100 healthy controls. Haplotype
reconstruction and frequency estimation were performed using
haplo.em, an implementation of an Expectation-Maximization
algorithm included in the R package haplo.stats, which computes
maximum likelihood estimates of haplotype probabilities from
unphased genotypes measured on unrelated individuals (R Foundation
for Statistical Computing, Vienna, Austria; www.r-project.org). The frequencies of variant status
between the patients and healthy controls were compared by
χ2 or Fisher's exact test using MedCalc software version
12.7.2 (MedCalc Software bvba, Ostend, Belgium). P<0.05 was
considered to indicate a statistically significant difference.
Results
A total of three MYOC and four OPTN
variants potentially associated with primary glaucoma were
identified in 4 and 18 patients, respectively (Table II).
 | Table IIFrequencies of MYOC and
OPTN variants identified in primary glaucoma. |
Table II
Frequencies of MYOC and
OPTN variants identified in primary glaucoma.
| Gene | Variant | Zygosity | Primary glaucoma,
n=112
| Healthy control,
n=100 |
|---|
| POAG, n=17 | JOAG, n=19 | NTG, n=76 |
|---|
| MYOC | c.-83G>A and
c.764T>C;p.Leu255Pro | Combined
heterozygous | 0 | 1 | 0 | 0 |
| MYOC |
c.1058C>T;p.Thr353Ile | Heterozygous | 0 | 1 | 0 | 0 |
| MYOC |
c.369C>T;p.Thr123= and
c.864C>T;p.Ile288= | Combined
heterozygous | 0 | 1 | 1 | 0 |
| OPTN | c.102G>A;
p.Thr34= | Homozygous | 1 | 1 | 2 | 0 |
| OPTN |
c.293T>A;p.Met98Lys | Homozygous | 1 | 1 | 2 | 0 |
| OPTN |
c.811C>T;p.Arg271Cys | Heterozygous | 0 | 0 | 1 | 0 |
| OPTN |
c.102G>A;p.Thr34= and
c.1634G>A;p.Arg545Gln | Combined
heterozygous | 2 | 2 | 5 | 2 |
Molecular analysis of MYOC
Missense variants of MYOC were identified in
two JOAG patients (P107 and P044) with a family history of primary
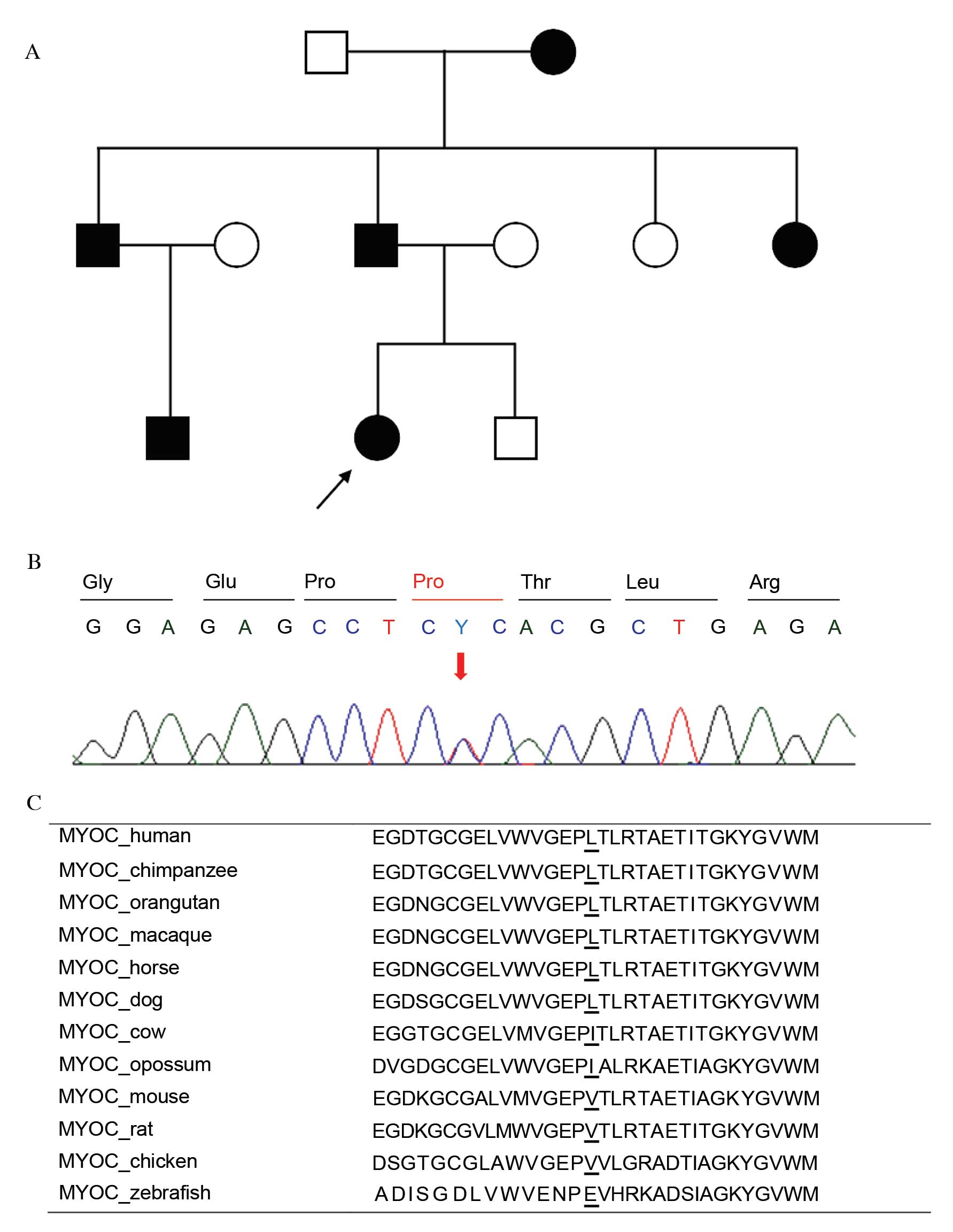
glaucoma (pedigree analysis of P107 is presented in Fig. 1A). One was determined to be a novel
missense variant, p.Leu255Pro, (P107; Fig. 1B) which was predicted to be
'probably damaging' with a score of 0.970 by PolyPhen-2 and was
expected to be 'deleterious' with a score of −2.887 by PROVEAN. The
position of changed amino acid was conserved among other mammalian
species (Fig. 1C). No healthy
control exhibited the same variant. The other variant identified,
p.Thr353Ile, has previously been reported in Korean POAG
(26). In addition, combined
heterozygous variants p.[Thr123=;Ile288=] were identified in
2 of 112 (2%) patients but not in healthy controls;
p.Thr123=[rs75682756; minor allele frequency (MAF)/minor
allele count (MAC) by 1000 Genomes=0.0014/7],
p.Ile288=(rs181923440; MAF/MAC=0.0010/5). The MYOC
promoter variant c.-83G>A (rs2075648; MAF/MAC=0.1380/691) was
detected only as a heterozygous state in a JOAG patient with
p.Leu255Pro and in five NTG patients, but not in the 100
healthy controls (Table III).
Clinical manifestations of primary glaucoma with MYOC are
summarized in Table IV.
 | Table IIIGenotype frequencies of MYOC
gene identified in korean patients with primary glaucoma. |
Table III
Genotype frequencies of MYOC
gene identified in korean patients with primary glaucoma.
| Location | cDNA change | Protein change | Genotype
frequencies (%)
| rs IDs |
|---|
| Primary glaucoma,
n=112 | Normal control,
n=100 |
|---|
| 5′-UTR | c.-83G>A | – | GG 106/112
(94.6) | GA 6/112 (5.4) | AA 0/112 (0) | GG 100/100
(100) | GA 0/100 (0) | AA 0/100 (0) | rs2075648 |
| Exon1 | c.57G>T |
p.Gln19His | GG 110/112
(98.2) | GT 2/112 (1.8) | TT 0/112 (0) | GG 100/100
(100) | GT 0/100 (0) | TT 0/100 (0) | rs2234925 |
| Exon1 | c.227G>A |
p.Arg76Lys | GG 102/112
(91.1) | GA 10/112
(8.9) | AA 0/112 (0) | GG 96/100 (96) | GA 4/100 (4) | AA 0/100 (0) | rs2234926 |
| Exon1 | c.369C>T |
p.Thr123= | CC 110/112
(98.2) | CT 2/112 (1.8) | TT 0/112 (0) | CC 100/100
(100) | CT 0/100 (0) | TT 0/100 (0) | rs75682756 |
| Exon2 | c.624C>G |
p.Asp208Glu | CC 109/112
(97.3) | CG 3/112 (2.7) | GG 0/112 (0) | CC 98/100 (98) | CG 2/100 (2) | GG 0/100 (0) | rs2234927 |
| Exon3 | c.764T>C |
p.Leu255Pro | TT 111/112
(99.1) | TC 1/112 (0.9) | CC 0/112 (0) | TT 100/100
(100) | TC 0/100 (0) | CC 0/100 (0) | – |
| Exon3 | c.864C>T |
p.Ile288= | CC 110/112
(98.2) | CT 2/112 (1.8) | TT 0/112 (0) | CC 100/100
(100) | CT 0/100 (0) | TT 0/100 (0) | rs181923440 |
| Exon3 | c.1058C>T |
p.Thr353Ile | CC 111/112
(99.1) | CT 1/112 (0.9) | TT 0/112 (0) | CC 100/100
(100) | CT 0/100 (0) | TT 0/100 (0) | rs137853277 |
| Exon3 | c.1464C>T |
p.Ala488= | CC 111/112
(99.1) | CT 1/112 (0.9) | TT 0/112 (0) | CC 100/100
(100) | CT 0/100 (0) | TT 0/100 (0) | rs2234929 |
| 3′-UTR |
c.*73G>C | – | GG 103/112
(92) | GC 8/112 (7.1) | CC 1/112 (0.9) | GG 98/100 (98) | GC 1/100 (1) | CC 1/100 (1) | rs74403899 |
 | Table IVClinical manifestations in four
primary glaucomas with MYOC variants. |
Table IV
Clinical manifestations in four
primary glaucomas with MYOC variants.
| Patient | Diagnosis | Variant 1 | Variant 2 | Sex | Age at Dx
(year) | BCVA
| Max IOP (mmHg)
| VCDR
| VFMD
|
|---|
| OD | OS | OD | OS | OD | OS | OD | OS |
|---|
| P107 | JOAG | c.-83G>A |
c.764T>C;p.Leu255Pro | F | 14 | 1 | 0.04 | 39 | 40 | 0.6 | 0.8 | −5.17 | −5.32 |
| P044 | JOAG |
c.1058C>T;p.Thr353Ile | None | F | 12 | 1 | 0.8 | 27 | 26 | 0.6 | 0.6 | −3.33 | −4.24 |
| P006 | JOAG |
c.369C>T;p.Thr123= |
c.864C>T;p.Ile288= | M | 28 | 1 | 1 | 23 | 23 | 0.5 | 0.6 | −0.26 | −1.11 |
| P039 | NTG |
c.369C>T;p.Thr123= |
c.864C>T;p.Ile288= | F | 22 | 1 | 1 | 16 | 15 | 0.6 | 0.5 | −1.07 | −1.02 |
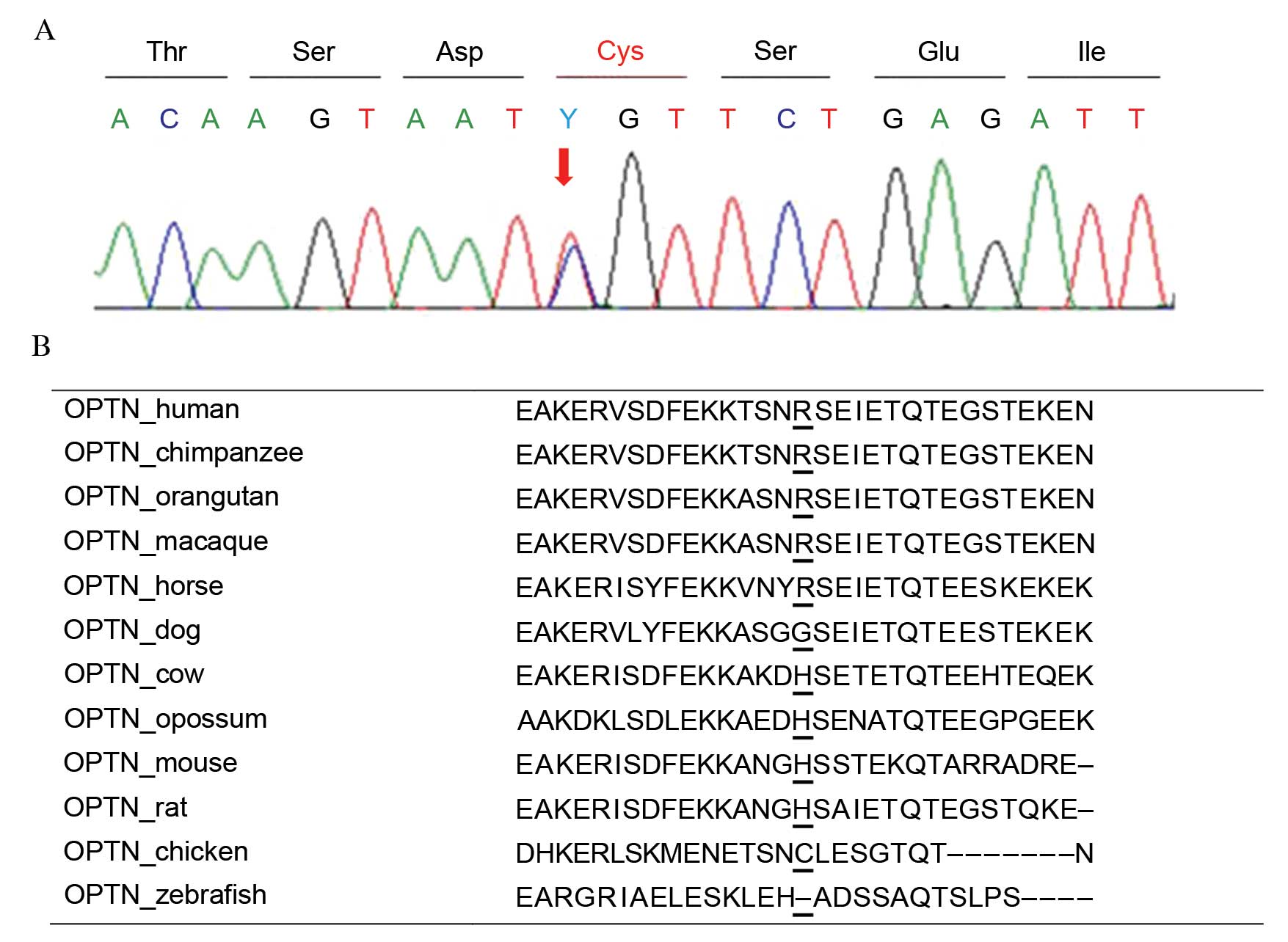
Molecular analysis of OPTN
OPTN variants were identified in 18 patients
with primary glaucoma. One patient (P086) harbored a novel variant,
p.Arg271Cys as a heterozygous state (Fig. 2A). This variant was predicted to be
'benign' with a score of 0.047 by PolyPhen-2 and was expected to be
'neutral' with a score of −1.726 by PROVEAN. This protein sequence
was not highly conserved across the compared species, and the
sequence in chicken had a cysteine in that position (Fig. 2B). One novel synonymous variant,
p.Leu568=; c.1704A>G was identified in heterozygosity.
These two variants were not detected in healthy controls.
In addition, three reported variants associated with
primary glaucoma (10,27), including
p.Thr34=(rs2234968), p.Met98Lys (rs11258194) and
p.Arg545Gln (rs75654767) were identified (Table V). The p.Thr34=,
p.Met98Lys and p.Arg545Gln mutations were detected in
40 (36%), 19 (17%), and 10 (9%) patients with primary glaucoma as
well as in 25 (25%), 18 (18%), and 2 (2%) healthy controls,
respectively, in a heterozygous or homozygous state. The odds ratio
(OR) was not statistically different for p.Thr34= and
p.Met98Lys by the χ2 test (P=0.061 and P=0.492,
respectively); however, it was significant for p.Arg545Gln
by Fisher's exact test [OR=4.804, 95% confidence interval
(CI)=1.026–22.482; relative risk=1.634, 95% CI=1.226–2.178;
P=0.037]. The presence of one of homozygous
p.[Thr34=;Thr34=] (4/112; 4%), homozygous
p.[Met98Lys;Met98Lys] (4/112; 4%) or combined heterozygous
p.[Thr34=;Arg545Gln] (9/112, 8%) was significantly
associated with the development of primary glaucoma (OR=8.768, 95%
CI=1.972–38.988; relative risk=1.818, 95% CI=1.473–2.244; P=0.001).
Clinical manifestations of primary glaucoma with OPTN
variants are summarized in Table
VI.
 | Table VGenotype frequencies of OPTN
gene identified in Korean patients with primary glaucoma. |
Table V
Genotype frequencies of OPTN
gene identified in Korean patients with primary glaucoma.
| Location | cDNA change | Protein change | Genotype
frequencies (%)
| rs IDs |
|---|
| Primary glaucoma,
n=112 | Normal control,
n=100 |
|---|
| Exon4 | c.102G>A |
p.Thr34= | GG 72/112
(64.3) | GA 36/112
(32.1) | AA 4/112 (3.6) | GG 75/100 (75) | GA 25/100 (25) | AA 0/100 (0) | rs2234968 |
| Exon4 | c.147C>T |
p.Thr49= | CC 111/112
(99.1) | CT 1/112 (0.9) | TT 0/112 (0) | CC 95/100 (95) | CT 5/100 (5) | TT 0/100 (0) | rs187734249 |
| Exon5 | c.293T>A |
p.Met98Lys | TT 93/112
(83.1) | TA 15/112
(13.4) | AA 4/112 (3.5) | TT 82/100 (82) | TA 18/100 (18) | AA 0/100 (0) | rs11258194 |
| Intron6 | c.552+63C>T | – | CC 112/112
(100) | CT 0/112 (0) | TT 0/112 (0) | CC 99/100 (99) | CT 1/100 (1) | TT 0/100 (0) | rs184333348 |
| Intron6 | c.553-10G>A | – | GG 84/112 (75) | GA 20/112
(17.9) | AA 8/112 (7.1) | GG 66/100 (66) | GA 34/100 (34) | AA 0/100 (0) | rs11258210 |
| Intron6 | c.553-5C>T | – | CC 20/112
(17.9) | CT 39/112
(34.8) | TT 53/112
(47.3) | CC 7/100 (7) | CT 45/100 (45) | TT 48/100 (48) | rs2244380 |
| Intron7 | c.626+24G>A | – | GG 103/112
(92) | GA 7/112 (6.3) | AA 2/112 (1.8) | GG 90/100 (90) | GA 10/100 (10) | AA 0/100 (0) | rs11258211 |
| Intron8 | c.780-53T>C | – | TT 88/112
(78.6) | TC 22/112
(19.6) | CC 2/112 (1.8) | TT 89/100 (92) | TC 10/100 (10) | CC 1/100 (1) | rs765884 |
| Exon9 | c.811C>T |
p.Arg271Cys | TT 111/112
(99.1) | CT 1/112 (0.9) | TT 0/112 (0) | TT 100/100
(100) | CT 0/100 (0) | TT 0/100 (0) | – |
| Intron9 | c.882+19C>T | – | CC 111/112
(99.1) | CT 1/112 (0.9) | TT 0/112 (0) | CC 99/100 (99) | CT 1/100 (1) | TT 0/100 (0) | rs2277219 |
| Exon10 | c.964G>A |
p.Glu322= | GG 111/112
(99.1) | GA 0/112 (0) | AA 1/112 (0.9) | GG 100/100
(100) | GA 0/100 (0) | AA 0/100 (0) | rs523747 |
| Intron11 |
c.1149-86G>T | – | GG 28/112 (25) | GT 33/112
(29.5) | TT 51/112
(45.5) | GG 10/100 (10) | GT 41/100 (41) | TT 49/100 (49) | rs676302 |
| Intron15 |
c.1613-48C>A | – | CC 64/112
(57.1) | CA 30/112
(26.8) | AA 18/112
(16.1) | CC 34/100 (34) | CA 50/100 (50) | AA 16/100 (16) | rs10906310 |
| Exon16 | c.1634G>A |
p.Arg545Gln | GG 102/112
(91.1) | GA 10/112
(8.9) | AA 0/112 (0) | GG 98/100 (98) | GA 2/100 (2) | AA 0/100 (0) | rs75654767 |
| Exon16 | c.1704A>G |
p.Leu568= | AA 111/112
(99.1) | AG 1/112 (0.9) | GG 0/112 (0) | AA 100/100
(100) | AG 0/100 (0) | GG 0/100 (0) | – |
 | Table VIClinical manifestations in 18 primary
glaucomas with OPTN variants. |
Table VI
Clinical manifestations in 18 primary
glaucomas with OPTN variants.
| Patient | Diagnosis | Variant 1 | Variant 2 | Sex | Age at Dx
(year) | BCVA
| Max IOP (mmHg)
| VCDR
| VFMD
|
|---|
| OD | OS | OD | OS | OD | OS | OD | OS |
|---|
| P059 | JOAG |
c.102G>A;p.Thr34= |
c.102G>A;p.Thr34= | M | 7 | 1 | 1 | 28 | 28 | 0.8 | 0.7 | −1.52 | −1.81 |
| P060 | POAG |
c.102G>A;p.Thr34= |
c.102G>A;p.Thr34= | F | 47 | 0.1 | 0.1 | 25 | 25 | 0.6 | 0.6 | −10.47 | −12.93 |
| P098 | NTG |
c.102G>A;p.Thr34= |
c.102G>A;p.Thr34= | M | 25 | 1 | 1 | 13 | 13 | 0.7 | 0.8 | −4.08 | −3.09 |
| P109 | NTG |
c.102G>A;p.Thr34= |
c.102G>A;p.Thr34= | M | 34 | 1 | 1 | 16 | 17 | 0.7 | 0.6 | −0.69 | −1.38 |
| P014 | POAG |
c.293T>A;p.Met98Lys |
c.293T>A;p.Met98Lys | M | 40 | 1 | 1 | 24 | 22 | 0.8 | 0.8 | −12.88 | −32.71 |
| P029 | JOAG |
c.293T>A;p.Met98Lys |
c.293T>A;p.Met98Lys | M | 7 | 0.4 | 1 | 24 | 27 | 0.7 | 0.7 | −8.32 | −6.63 |
| P054 | NTG |
c.293T>A;p.Met98Lys |
c.293T>A;p.Met98Lys | M | 29 | 0.8 | 0.8 | 19 | 19 | 0.6 | 0.4 | −16.36 | −2.2 |
| P058 | NTG |
c.293T>A;p.Met98Lys |
c.293T>A;p.Met98Lys | M | 26 | 1 | 1 | 21 | 21 | 0.5 | 0.6 | −0.04 | −1.93 |
| P086 | NTG |
c.811C>T;p.Arg271Cys | None | M | 17 | 1 | 1 | 19 | 19 | 0.7 | 0.6 | −1.46 | −1.25 |
| P003 | POAG |
c.102G>A;p.Thr34= |
c.1634G>A;p.Arg545Gln | M | 49 | 0.8 | 1 | 23 | 22 | 0.8 | 0.7 | −3.42 | −2.48 |
| P010 | NTG |
c.102G>A;p.Thr34= |
c.1634G>A;p.Arg545Gln | M | 36 | 1 | 1 | 16 | 16 | 0.6 | 0.6 | 0.23 | −0.95 |
| P023 | NTG |
c.102G>A;p.Thr34= |
c.1634G>A;p.Arg545Gln | F | 28 | 0.8 | 1 | 17 | 18 | 0.7 | 0.5 | −1.18 | −3.25 |
| P034 | JOAG |
c.102G>A;p.Thr34= |
c.1634G>A;p.Arg545Gln | F | 29 | 1 | 1 | 29 | 28 | 0.4 | 0.8 | NA | NA |
| P049 | NTG |
c.102G>A;p.Thr34= |
c.1634G>A;p.Arg545Gln | F | 29 | 1 | 1 | 20 | 20 | 0.7 | 0.7 | −3.75 | −3.3 |
| P066 | NTG |
c.102G>A;p.Thr34= |
c.1634G>A;p.Arg545Gln | F | 32 | 1 | 1 | 15 | 15 | 0.2 | 0.2 | −4.66 | −3.77 |
| P081 | JOAG |
c.102G>A;p.Thr34= |
c.1634G>A;p.Arg545Gln | M | 15 | 1 | 1 | 23 | 22 | 0.5 | 0.5 | −0.08 | −0.54 |
| P095 | POAG |
c.102G>A;p.Thr34= |
c.1634G>A;p.Arg545Gln | M | 43 | 1 | 1 | 31 | 31 | 0.6 | 0.7 | −6.48 | −4.35 |
| P105 | NTG |
c.102G>A;p.Thr34= |
c.1634G>A;p.Arg545Gln | F | 43 | 1 | 1 | 21 | 21 | 0.6 | 0.6 | −0.34 | −0.18 |
Discussion
The present study reports a molecular analysis of
MYOC and OPTN genes in 112 patients with primary
glaucoma. One novel MYOC variant, p.Leu255Pro, in
patient P107 with a family history of glaucoma was predicted to
have a deleterious effect by in silico analysis. This
variant is a possible pathogenic mutation associated with the
development of primary glaucoma. The second MYOC variant,
p.Thr353Ile, in patient P044 with a family history of
glaucoma has been previously reported as a pathologic mutation for
POAG and resides in the olfactomedin-homology region. It may be
involved in the regulation of IOP in trabecular-meshwork cells
(26). The presence of
heterozygous p.Thr123= and p.Ile288= was observed
only in patients with JOAG and NTG. Thus,
p.[Thr123=;Ile288=] may be a part of the haplotype variant
associated with the development of primary glaucoma. Further
extended haplotype analysis is required to confirm the association
of MYOC haplotype variants. The variant in the 5′-UTR
region, c.-83G>A, which was initially reported in Western
countries (20,28), and subsequently observed in Hong
Kong (29) and the Philippines
(30), was identified only in
primary glaucoma patients in the present study. The association of
c.-83G>A and the risk of primary glaucoma remains controversial
due to the variable frequency and the non-significant differences
between POAG patients and controls observed in previous studies
(28,31,32).
Genetic analysis of the OPTN coding region
was performed in all patients with primary glaucoma. Notably,
OPTN variants (10/76, 13.2%) were identified more frequently
than MYOC variants (1/76, 1.3%) in NTG patients, which is
consistent with previous studies (33,34).
The three OPTN variants p.Thr34=, p.Leu40= and
p.Glu89His were identified in 7 of 53 Korean NTG patients
(33), whereas only one patient
with the MYOC variant p.Leu411= was reported from 32
separate Korean NTG patients (34). Supporting the findings of the
present study, Rezaie et al (10) suggested that mutations in
OPTN may be responsible for 16.7% of the hereditary forms of
NTG and that there is an additional risk factor of 13.6% in
familial and sporadic cases. Furthermore, Sohn et al
(35) demonstrated that the
MYOC gene itself was not associated with OAG, including
POAG, NTG and SIG. Their results do not support the hypothesis that
MYOC induction may be linked to IOP variation and that promoter
variants of MYOC may be a risk factor for the pathogenesis of
OAG.
A novel OPTN variant, p.Arg271Cys, of
unknown significance was identified in patient P086 with NTG
(1/112, 1%). This variant led to replacement of arginine with
cysteine at codon 271, however, the protein sequence was not highly
conserved across species. Although this variant was predicted to be
'benign' or 'neutral', it is potentially pathogenic as it was not
present in the healthy controls. Segregation analysis and/or
functional studies are required to verify the pathogenicity or
neutrality of this variant.
Of the OPTN haplotype variants,
p.Thr34=, p.Met98Lys and p.Arg545Gln have been
previously reported as possible glaucoma-causing mutations
(10,27). However, they were present at
reduced frequencies as a heterozygous state in healthy
controls.
The p.Thr34= variant was detected in 40 of
112 (36%) patients with primary glaucoma and in 25 of 100 (25%)
healthy controls in a heterozygous or homozygous state in the
present study. Homozygosity for p.Thr34= was observed only
in four patients with primary glaucoma (P059, P060, P098 and P109).
Funayama et al (27)
reported that p.Thr34= was weakly associated with patients
with OAG with elevated IOP, whereas p.Met98Lys was weakly
associated with patients with OAG with normal IOP. In interaction
analysis between olfactomedin 2 (OLFM2) and OPTN
genes in patients with OAG, the c.317G>A; p.Arg106Gln of
OLFM2 and c.412G>A; p.Thr34= of OPTN and
the c.1281C>T; p.Arg427Arg of OLFM2 and
c.412G>A; p.Thr34= of OPTN were significantly
associated with OAG with elevated IOP. These results suggest that
these variants in OLFM2 and OPTN contribute
interactively to OAG, indicating a polygenic etiology with
different properties for p.Thr34= and p.Met98Lys
variants of OPTN.
The p.Met98Lys variant was identified in 19
of 112 (17%) patients with primary glaucoma as well as in 18 of 100
(18%) healthy controls as a heterozygous or homozygous state in the
present study. Alward et al (36) and Fuse et al (37) reported a significant association
between p.Met98Lys and glaucoma in Japanese patients. By
contrast, Toda et al (38)
identified similar frequencies of p.Met98Lys in Japanese
glaucoma patients and controls. Notably, p.Met98Lys has been
demonstrated to be a polymorphic variant in German, French and
Moroccan patients (39,40). By contrast to previous studies
(10,41,42),
the frequency difference in the present study (4 vs. 0%) between
the patients with homozygous p.Met98Lys (P014, P029, P054
and P058) and healthy controls was highly significant.
p.Met98Lys is located within a putative basic leucine zipper
domain and is conserved in macaques; it may represent a risk
associated factor or a dominant susceptibility allele (10). Wild-type OPTN protein, operating
through the tumor necrosis factor α pathway, may have a
neuroprotective role in the eye and optic nerve; however when
defective, it produces visual loss and optic neuropathy as
typically observed in normal and high-pressure glaucoma (10).
The p.Arg545Gln variant was reported
previously in POAG families with normal IOP (10). Although the p.Arg545Gln
variant is not part of a known protein domain, it is situated near
the only zinc finger motif within OPTN. This motif is typically
observed in transcription factors. p.Arg545Gln has been
detected in similar frequencies in Japanese glaucoma patients and
healthy controls (38). Alward
et al (36) suggested that
p.Arg545Gln may not be a disease-causing polymorphism.
Results concerning the effect of OPTN on glaucoma have been
equivocal (10,32,43,44);
however, the expression of MYOC may be regulated by
OPTN (45). Notably,
results from the present study support the view that the three
variants of OPTN may be involved in the development of
primary glaucoma, as the OR for the haplotype harboring homozygous
p.Thr34= or p.Met98Lys as well as a simultaneous
presence of heterozygous p.Thr34= and p.Arg545Gln was
statistically significant by the Fisher's exact test (OR=8.768,
95%, CI=1.972–38.988; relative risk=1.818, 95% CI=1.473–2.244;
P=0.001).
In conclusion, haplotype variants identified in the
present study may be regarded as potential contributing factors for
primary glaucoma in Korea. The present study provides insight into
the genetic or haplotype variants contributing to primary glaucoma.
Further studies, including on additional genes, are required to
elucidate the underlying pathogenic mechanism using larger cohorts
to provide additional statistical power.
Acknowledgments
The authors would like to thank The Catholic
Genetic Laboratory Center (Seoul, Korea) for its assistance in
conducting the present study and compiling this report. The present
study was supported by a grant from the Basic Science Research
Program through the National Research Foundation of Korea (Daejeon,
Korea; grant no. NRF-2013R1A1A2006801) funded by the Ministry of
Education (Sejong, Korea).
References
|
1
|
Foster PJ, Buhrmann R, Quigley HA and
Johnson GJ: The definition and classification of glaucoma in
prevalence surveys. Br J Ophthalmol. 86:238–242. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pakravan M, Yazdani S, Javadi MA, Amini H,
Behroozi Z, Ziaei H, Katibeh M, Solaimanizad R, Ghahari E and
Yaseri M: A population-based survey of the prevalence and types of
glaucoma in central Iran: The Yazd eye study. Ophthalmology.
120:1977–1984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Casson RJ, Chidlow G, Wood JP, Crowston JG
and Goldberg I: Definition of glaucoma: Clinical and experimental
concepts. Clin Experiment Ophthalmol. 40:341–349. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weinreb RN, Aung T and Medeiros FA: The
pathophysiology and treatment of glaucoma: A review. JAMA.
311:1901–1911. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goldwyn R, Waltman SR and Becker B:
Primary open-angle glaucoma in adolescents and young adults. Arch
Ophthalmol. 84:579–582. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Booth A, Churchill A, Anwar R, Menage M
and Markham A: The genetics of primary open angle glaucoma. Br J
Ophthalmol. 81:409–414. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wiggs JL: Genetic etiologies of glaucoma.
Arch Ophthalmol. 125:30–37. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khan AO: Genetics of primary glaucoma.
Curr Opin Ophthalmol. 22:347–355. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stone EM, Fingert JH, Alward WL, Nguyen
TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A,
Nichols BE, et al: Identification of a gene that causes primary
open angle glaucoma. Science. 275:668–670. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rezaie T, Child A, Hitchings R, Brice G,
Miller L, Coca-Prados M, Héon E, Krupin T, Ritch R, Kreutzer D, et
al: Adult onset primary open-angle glaucoma caused by mutations in
optineurin. Science. 295:1077–1079. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Monemi S, Spaeth G, DaSilva A, Popinchalk
S, Ilitchev E, Liebmann J, Ritch R, Héon E, Crick RP, Child A and
Sarfarazi M: Identification of a novel adult-onset primary
open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet.
14:725–733. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Footz TK, Johnson JL, Dubois S, Boivin N,
Raymond V and Walter MA: Glaucoma-associated WDR36 variants encode
functional defects in a yeast model system. Hum Mol Genet.
18:1276–1287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pasutto F, Matsumoto T, Mardin CY, Sticht
H, Brandstätter JH, Michels-Rautenstrauss K, Weisschuh N, Gramer E,
Ramdas WD, van Koolwijk LM, et al: Heterozygous NTF4 mutations
impairing neurotrophin-4 signaling in patients with primary
open-angle glaucoma. Am J Hum Genet. 85:447–456. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aung T, Ocaka L, Ebenezer ND, Morris AG,
Krawczak M, Thiselton DL, Alexander C, Votruba M, Brice G, Child
AH, et al: A major marker for normal tension glaucoma: Association
with polymorphisms in the OPA1 gene. Hum Genet. 110:52–56. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo Y, Chen X, Zhang H, Li N, Yang X,
Cheng W and Zhao K: Association of OPA1 polymorphisms with NTG and
HTG: A meta-analysis. PLoS One. 7:e423872012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaur K, Mandal AK and Chakrabarti S:
Primary congenital glaucoma and the involvement of CYP1B1. Middle
East Afr J Ophthalmol. 18:7–16. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jelodari-Mamaghani S, Haji-Seyed-Javadi R,
Suri F, Nilforushan N, Yazdani S, Kamyab K and Elahi E:
Contribution of the latent transforming growth factor-β binding
protein 2 gene to etiology of primary open angle glaucoma and
pseudoexfoliation syndrome. Mol Vis. 19:333–347. 2013.
|
|
18
|
Fingert J: Primary open-angle glaucoma
genes. Eye (Lond). 25:587–595. 2011. View Article : Google Scholar
|
|
19
|
Shastry BS: Genetic susceptibility to
primary angle closure glaucoma (PACG). Discov Med. 15:17–22.
2013.PubMed/NCBI
|
|
20
|
Fingert JH, Héon E, Liebmann JM, Yamamoto
T, Craig JE, Rait J, Kawase K, Hoh ST, Buys YM, Dickinson J, et al:
Analysis of myocilin mutations in 1703 glaucoma patients from five
different populations. Hum Mol Genet. 8:899–905. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Menaa F, Braghini CA, Vasconcellos JP,
Menaa B, Costa VP, Figueiredo ES and Melo MB: Keeping an eye on
myocilin: A complex molecule associated with primary open-angle
glaucoma susceptibility. Molecules. 16:5402–5421. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Turalba AV and Chen TC: Clinical and
genetic characteristics of primary juvenile-onset open-angle
glaucoma (JOAG). Semin Ophthalmol. 23:19–25. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Choi Y, Sims GE, Murphy S, Miller JR and
Chan AP: Predicting the functional effect of amino acid
substitutions and indels. PLoS One. 7:e466882012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sievers F, Wilm A, Dineen D, Gibson TJ,
Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al:
Fast, scalable generation of high-quality protein multiple sequence
alignments using Clustal Omega. Mol Syst Biol. 7:5392011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoon SJ, Kim HS, Moon JI, Lim JM and Joo
CK: Mutations of the TIGR/MYOC gene in primary open-angle glaucoma
in Korea. Am J Hum Genet. 64:1775–1778. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Funayama T, Mashima Y, Ohtake Y, Ishikawa
K, Fuse N, Yasuda N, Fukuchi T, Murakami A, Hotta Y and Shimada N;
Glaucoma Gene Research Group: SNPs and interaction analyses of
noelin 2, myocilin, and optineurin genes in Japanese patients with
open-angle glaucoma. Invest Ophthalmol Vis Sci. 47:5368–5375. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alward WL, Fingert JH, Coote MA, Johnson
AT, Lerner SF, Junqua D, Durcan FJ, McCartney PJ, Mackey DA,
Sheffield VC and Stone EM: Clinical features associated with
mutations in the chromosome 1 open-angle glaucoma gene (GLC1A). N
Engl J Med. 338:1022–1027. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fan BJ, Leung YF, Pang CP, Baum L, Tam OS,
Wang N and Lam SC: Single nucleotide polymorphisms of the myocilin
gene in primary open-angle glaucoma patients. Zhonghua Yi Xue Yi
Chuan Xue Za Zhi. 21:70–73. 2004.PubMed/NCBI
|
|
30
|
Wang DY, Fan BJ, Canlas O, Tam PO, Ritch
R, Lam DS, Fan DS and Pang CP: Absence of myocilin and optineurin
mutations in a large Philippine family with juvenile onset primary
open angle glaucoma. Mol Vis. 10:851–856. 2004.PubMed/NCBI
|
|
31
|
Mukhopadhyay A, Acharya M, Mukherjee S,
Ray J, Choudhury S, Khan M and Ray K: Mutations in MYOC gene of
Indian primary open angle glaucoma patients. Mol Vis. 8:442–448.
2002.PubMed/NCBI
|
|
32
|
Kumar A, Basavaraj MG, Gupta SK, Qamar I,
Ali AM, Bajaj V, Ramesh TK, Prakash DR, Shetty JS and Dorairaj SK:
Role of CYP1B1, MYOC, OPTN, and OPTC genes in adult-onset primary
open-angle glaucoma: Predominance of CYP1B1 mutations in Indian
patients. Mol Vis. 13:667–676. 2007.PubMed/NCBI
|
|
33
|
Jeong DH, Kim MR, Mun YK and Kee C: OPTN
gene mutation in normal-tension glaucoma. J Korean Ophthalmol Soc.
44:1903–1907. 2003.
|
|
34
|
Lee SJ, Hur WH and Kee C: Analysis of TIGR
gene mutation in glaucoma. J Korean Ophthalmol Soc. 41:1095–1101.
2000.
|
|
35
|
Sohn S, Hur W, Choi YR, Chung YS, Ki CS
and Kee C: Little evidence for association of the glaucoma gene
MYOC with open-angle glaucoma. Br J Ophthalmol. 94:639–642. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alward WL, Kwon YH, Kawase K, Craig JE,
Hayreh SS, Johnson AT, Khanna CL, Yamamoto T, Mackey DA, Roos BR,
et al: Evaluation of optineurin sequence variations in 1,048
patients with open-angle glaucoma. Am J Ophthalmol. 136:904–910.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fuse N, Takahashi K, Akiyama H, Nakazawa
T, Seimiya M, Kuwahara S and Tamai M: Molecular genetic analysis of
optineurin gene for primary open-angle and normal tension glaucoma
in the Japanese population. J Glaucoma. 13:299–303. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Toda Y, Tang S, Kashiwagi K, Mabuchi F,
Iijima H, Tsukahara S and Yamagata Z: Mutations in the optineurin
gene in Japanese patients with primary open-angle glaucoma and
normal tension glaucoma. Am J Med Genet A. 125A:1–4. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Melki R, Belmouden A, Akhayat O, Brézin A
and Garchon HJ: The M98K variant of the OPTINEURIN (OPTN) gene
modifies initial intraocular pressure in patients with primary open
angle glaucoma. J Med Genet. 40:842–844. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Weisschuh N, Neumann D, Wolf C, Wissinger
B and Gramer E: Prevalence of myocilin and optineurin sequence
variants in German normal tension glaucoma patients. Mol Vis.
11:284–287. 2005.PubMed/NCBI
|
|
41
|
Mukhopadhyay A, Komatireddy S, Acharya M,
Bhattacharjee A, Mandal AK, Thakur SK, Chandrasekhar G, Banerjee A,
Thomas R, Chakrabarti S and Ray K: Evaluation of Optineurin as a
candidate gene in Indian patients with primary open angle glaucoma.
Mol Vis. 11:792–797. 2005.PubMed/NCBI
|
|
42
|
Sripriya S, Nirmaladevi J, George R,
Hemamalini A, Baskaran M, Prema R, Ve Ramesh S, Karthiyayini T,
Amali J, Job S, et al: OPTN gene: Profile of patients with glaucoma
from India. Mol Vis. 12:816–820. 2006.PubMed/NCBI
|
|
43
|
Tektas OY and Lütjen-Drecoll E: Structural
changes of the trabecular meshwork in different kinds of glaucoma.
Exp Eye Res. 88:769–775. 2009. View Article : Google Scholar
|
|
44
|
Cheng JW, Cheng SW, Ma XY, Cai JP, Li Y
and Wei RL: The prevalence of primary glaucoma in mainland China: A
systematic review and meta-analysis. J Glaucoma. 22:301–306. 2013.
View Article : Google Scholar
|
|
45
|
Huang X, Li M, Guo X, Li S, Xiao X, Jia X,
Liu X and Zhang Q: Mutation analysis of seven known
glaucoma-associated genes in Chinese patients with glaucoma. Invest
Ophthalmol Vis Sci. 55:3594–3602. 2014. View Article : Google Scholar : PubMed/NCBI
|