Introduction
Activated leukocyte cell adhesion molecule
(ALCAM/CD166) is a 105 kDa transmembrane glycoprotein and a member
of the cell surface immunoglobulin superfamily. It was initially
identified in thymic epithelial cells and activated leukocytes
(1,2). ALCAM mediates cell-cell homophilic
(ALCAM-ALCAM) and heterophilic (ALCAM-CD166) interactions.
Deregulation of ALCAM is associated with the development and
malignant progression of various types of cancer, including colon,
gastric, liver, lung, prostate, pancreatic cancer, breast
carcinomas and melanoma (3–11).
Furthermore, it is a marker for cancer stem cells in colon and
prostate cancer (12,13). However, the exact role of ALCAM
during the tumorigenesis of different types of malignant tumor
remains to be elucidated. It was proposed that ALCAM may promote
the survival of breast cancer cells by inhibiting apoptosis and
autophagy (14). Evidence has also
suggested that ALCAM may suppress the migration and invasion of
cancer cells by controlling the activity of matrix
metalloproteinases (15). For
example, the expression of ALCAM is associated with the suppression
of breast cancer cell invasion (16). Furthermore, ALCAM-negative
pancreatic cancer cells demonstrated stronger invasive and
migratory activities compared with ALCAM-positive cancer cells
(17). Our previous study
demonstrated that silencing ALCAM caused no affect on cell growth
or invasion in Su86.86 pancreatic cancer cells, but significantly
reduced cell adhesion and increased pancreatic cancer cell
resistance towards chemotherapeutic agents (4). However, the underlying mechanism
remains to be elucidated.
In the present study, the expression of ALCAM in
pancreatic cancer cells and pancreatic stellate cells (PSCs) was
analyzed, and the role of ALCAM in the growth, proliferation,
invasion and cell-cell interaction of pancreatic cancer cells and
PSCs was further investigated.
Materials and methods
Tissue specimens and cell cultures
Tissue specimens were obtained from patients who
underwent pancreatic resection. The patients underwent surgery for
a range of pancreatic diseases, including pancreatic ductal
adenocarcinoma (PDAC; n=56) and chronic pancreatitis (CP; n=10).
Normal pancreatic tissue samples (n=10) were obtained during
resection for tumor infiltration of the periampullary area by
another malignancy (i.e. colon cancer) or metastasis to the
pancreas by kidney tumors that required resection of healthy
pancreatic tissue. Tissue collection was approved by the Ethics
Committees of the Technical University of Munich (Munich, Germany)
and the University of Heidelberg (Heidelberg, Germany). Written
informed consent was obtained from all patients.
Freshly removed tissues were fixed in 4%
paraformaldehyde for 24 h and embedded in paraffin for histological
analysis. A portion of the tissue samples was preserved in RNAlater
(Ambion Europe Ltd., Huntingdon, Cambridgeshire, UK) or snap-frozen
in liquid nitrogen immediately upon surgical removal, and
maintained at −80°C until use. Pancreatic cancer cell lines (Panc-1
and T3M4) were obtained from American Type Culture Collection
(Manassas, VA, USA) and grown in RPMI-1640 medium (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal calf serum (FCS; Invitrogen; Thermo Fisher Scientific,
Inc.) and 100 U/ml penicillin and streptomycin (complete medium) at
37°C in a 5% CO2 humidified atmosphere. PSCs between
passages 3 and 6 were cultured in a 1:1 (vol/vol) mixture of
low-glucose (1,000 mg/l) Dulbecco's modified Eagle's medium with
Ham's F12 medium (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% FCS, L-glutamine (2 mmol/l),
penicillin/streptomycin and amphotericin, as previously described
(18–21).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
All reagents and equipment for mRNA and cDNA
preparation were purchased from Roche (Mannheim, Germany). The
forward and reverse primer sequences were as follows: ALCAM sense,
5′-TAG CAG GAA TGC AAC TGT GG-3′; ALCAM anti-sense, 5′-CGC AGA CAT
AGT TTC CAG-3′. mRNA was prepared by automated isolation using the
MagNA Pure LC instrument and isolation kit I (for cells) and kit II
(for tissues). RNA was reverse-transcribed into cDNA using the cDNA
synthesis kit for RT-PCR (AMV), according to the manufacturer's
protocol. qPCR was performed on a LightCycler 480 Real-Time PCR
system (Roche Diagnostics, Basel, Switzerland) with the Light
Cycler Fast Start DNA SYBR Green kit. The PCR program consisted of
an initial denaturation cycle (10 min at 95°C) followed by 40
cycles of denaturation (15 sec at 95°C) and annealing and
elongation (60 sec at 60°C). The relative number of specific
transcripts was normalized against the levels of cyclophilin B and
hypoxanthine guanine phosphoribosyltransferase, and the data were
analyzed using the 2−ΔΔCq method (22).
Immunohistochemistry
Immunohistochemistry was performed using the Dako
Envision System (Dako Cytomation GmbH, Hamburg, Germany).
Consecutive paraffin-embedded tissue sections (3–5 µm thick)
were deparaffinized and rehydrated using routine methods (23). Antigen retrieval was performed by
pretreatment of the slides in citrate buffer (pH 6.0) in a
microwave oven for 10 min. Endogenous peroxidase activity was
quenched by incubation in deionized water containing 3% hydrogen
peroxide at room temperature for 10 min. Following blocking of
non-specific reactivity with diluted normal goat serum, the tissue
sections were incubated with rabbit anti-human ALCAM polyclonal
antibodies (4 µg/ml; 1:1,000, Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) at 4°C overnight. The tissue sections
were subsequently incubated with horseradish peroxidase
(HRP)-linked goat anti-rabbit antibodies (1:5,000; Dako GmbH,
Hamburg, Germany), followed by reaction with diaminobenzidine and
counterstaining with Mayer's hematoxylin. In addition, to confirm
the specificity of the primary antibodies, tissue sections were
incubated in the absence of the primary antibodies and with
negative control rabbit immunoglobulin G (1:50; Santa Cruz
Biotechnology, Inc.).
Immunocytochemistry
The cells (Panc-1, T3M4 and PSCs) were transfected
with control or ALCAM small interfering (si)RNA for 48 h,
trypsinized and seeded onto slides for 12 h. Following washing
three times with phosphate-buffered saline (PBS), the cells were
incubated with 4% paraformaldehyde for 10 min, 30 mM glycine/PBS
for 5 min and 0.1% Triton X-100 for 5 min. After washing three
times with PBS, the cells were incubated with 3%
H2O2 for 10 min, followed by incubation with
the primary antibody diluted in universal block DAKO (ALCAM, 1:50;
keratin, 1:500, Santa Cruz Biotechnology, Inc.) for 1 h. Following
washing with Tris-buffered saline (TBS)/bovine serum albumin (BSA;
Carl Roth GmbH & Co. KG, Karlsruhe, Germany) and Tween 20
(0.05%), the secondary antibody labeled with HRP (1:1,000,
Chemicon, Hofheim, Germany) was added, followed by color reaction
and counterstaining as in immunohistochemistry.
Enzyme-linked immunosorbent assay
(ELISA)
The ALCAM ELISA kit was used (R&D Systems,
Wiesbaden-Nordenstadt, Germany) to detect ALCAM levels in the serum
and tissue culture medium at room temperature. Briefly, 96-well
Nunc Immuno plates (Nunc, Roskilde, Denmark) were coated overnight
with 100 µl (2 µg/ml) of ALCAM capture antibodies
(1:500; R&D Systems, Inc., Wiesbaden-Nordenstadt, Germany) in
PBS (pH 7.0). PBS with 0.05% Tween 20 was used as the washing
solution. Non-specific binding sites were blocked with 300
µl blocking buffer (1% BSA in PBS) for 1 h at 37°C. Either
recombinant human ALCAM or serum/cell culture supernatant (100
µl/well) were added and incubated for 2 h at 37°C. Following
washing, 100 µl of biotin-conjugated goat anti-human ALCAM
detection antibodies (50 ng/ml) were added into each well and
incubated for 2 h at room temperature. HRP-conjugated streptavidin
(100 µl) 1:200 diluted in PBS was added to each well and
incubated for 20 min at 37°C. Following washing with PBS with Tween
20 three times, 100 µl of 1:1 mixed TMB substrate reagent A
and reagent B (BD Biosciences, San Diego, CA, USA) were added for
20 min at 37°C. Colorimetric reactions were stopped by adding 50
µl of 2 N H2SO4 and analyzed using a
microplate reader at 450 and 570 nm for correction.
Immunoblotting
Cultured pancreatic cancer cells and PSCs were lysed
in ice-cold lysis buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM
NaCl, 1% Triton X-100, 2.5 mM sodium pyrophosphate and one tablet
EDTA-free protease inhibitor cocktail (Roche) for 30 min. Cell
lysates were collected following centrifugation at 15,000 × g for
10 min at 4°C. The total protein (20 µg) was loaded onto 10%
polyacrylamide gels and transferred onto polyvinylidene difluoride
membranes. The membranes were blocked with 20 ml TBS, 5% skim milk
and 0.05% Tween-20 for 1 h, and then incubated with rabbit
anti-human ALCAM polyclonal antibodies (1:200; Santa Cruz
Biotechnology, Inc.) or anti-Erk2 (1:2,500; Santa Cruz
Biotechnology, Inc.) overnight at 4°C. The membranes were washed
three times with 0.05% Tween-20-TBS and incubated with
HRP-conjugated anti-rabbit antibody (1:2,500; Chemicon) for 1 h at
room temperature. Signals were detected using the enhanced
chemiluminescence system (Amersham Life Science, Buckinghamshire,
UK).
Hypoxia
Panc-1, T3M4 and PSC cells were exposed to hypoxic
conditions of 0.75% O2, 10% CO2 and 89.25%
N2, as described previously (24). The medium was switched to
serum-free medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) prior to subjecting the cells to hypoxia. Control
cultures were grown in serum-free medium under normoxia in a 5%
CO2 incubator (Forma Scientific Co., Marietta, OH, USA).
After incubation for 48 h, supernatants were collected for ALCAM
assay using ELISA.
siRNA transfection
Human ALCAM specific siRNA (sense, GCC CGA UGG CUC
CCC AGU A; antisense, UAC UGG GGA GCC AUC GGG C) (14) was purchased from Qiagen (Hilden,
Germany). The cells were grown to 50–70% confluence. siRNA
transfection was performed with HiPerFect transfection reagent
(Qiagen), according to the manufacturer's instructions. The final
concentration of the control and specific siRNA was 5 nM. The
efficacy of the siRNA transfection was ascertained by immunoblot
analysis and ELISA after 72 h of transfection.
Invasion assay
To assess cell migration in vitro, Transwell
migration chambers with an 8 µm pore size (BD Biosciences)
were used and reconstituted with 600 µl serum-free RPMI-1640
medium in the top and bottom chambers for 24 h. The Panc 1, T3M4
and pancreatic stellate cells were trypsinized and seeded into the
top chamber at a density of 2.5×104 cells/well in 600
µl RPMI-1640 containing 10% FCS. Following incubation at
37°C for 20 h, the cells remaining attached to the upper surface of
the filters were carefully removed with cotton swabs, while cells
that reached the underside of the chamber were stained with
hematoxylin and eosin and counted under a microscope in five random
fields at a magnification of ×200.
Cell interaction assay
Panc-1 or T3M4 cells in RPMI-1640 supplemented with
10% FCS and PSCs in a 1:1 (vol/vol) mixture of low glucose (1,000
mg/l) Dulbecco's modified Eagle's medium with Ham's F12 medium
containing with 10% FCS were transfected with ALCAM siRNA or
control siRNA for 2 days. The tumor cells and PSCs were
trypsinized, counted, mixed and seeded directly for co-culture at a
density of 4,000 cells/well in 24-well plates. After 2 days of
culture, immunocytochemistry using keratin as a specific marker for
cancer cells was performed to analyze the interaction between tumor
cells (Panc-1 and T3M4) and PSCs. Cell-cell interactions were
counted randomly in five areas under the microscope and calculated
as a percentage (interaction number/total number of cells). All
assays were performed in triplicate and repeated three times.
Statistical analysis
The data are presented as the mean ± standard error
of the mean for in vitro assays, and median and individual
data for the RT-qPCR and ELISA results, unless indicated otherwise.
Statistical analysis was performed using SPSS 17.0 software (SPSS
Inc., Chicago, IL, USA). The Mann-Whitney U test and the
Kruskal-Wallis test were utilized, and groups were compared using
Dunn's multiple comparison test. P<0.05 was considered to
indicate a statistically significant difference. The mean
difference between groups was estimated with a 95% confidence
interval.
Results
ALCAM expression and localization in
pancreatic tissues
Our previous study (4) demonstrated that ALCAM was expressed
on the membrane of islet cells in the normal pancreas whereas
normal pancreatic ducts were negative for ALCAM. ALCAM was
expressed in ductal and acinar cells in CP tissues. Furthermore,
ALCAM expression was generally low in PDAC, while membranous or
cytoplasmic ALCAM expression was found in certain types of tumor
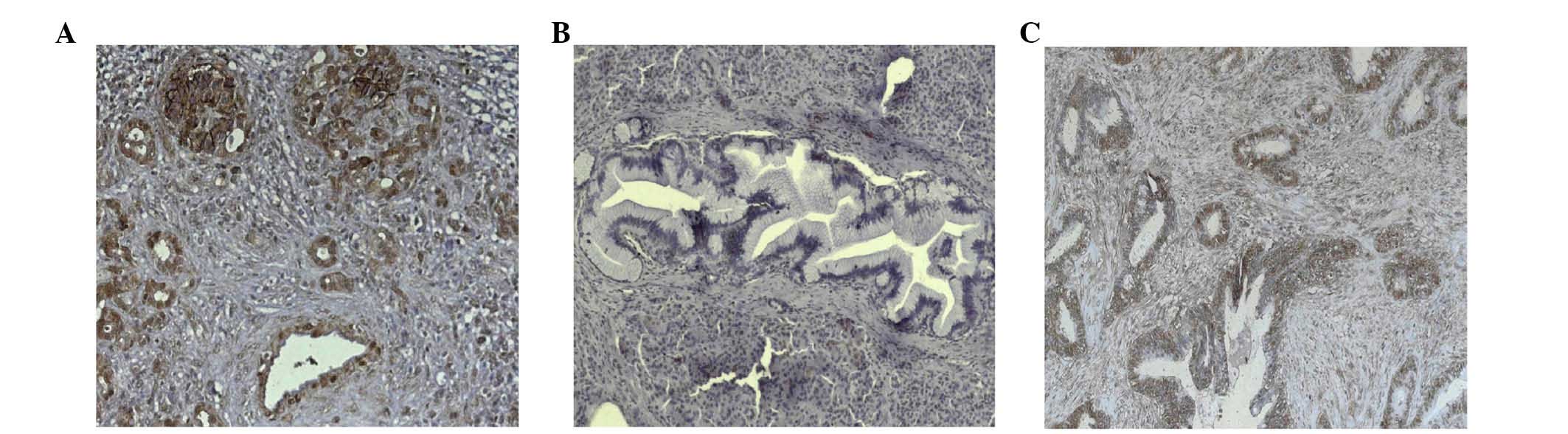
(9). The present study
demonstrated strong ALCAM expression in PSCs of CP tissues
(Fig. 1A), and PSCs surrounding
pancreatic intraepithelial neoplasias (Fig. 1B), as well as in pancreatic cancer
cells (Fig. 1C).
ALCAM expression in pancreatic cancer
cells and PSCs
A previous study demonstrated that ALCAM was
expressed in pancreatic cancer cell lines (9). The present study compared the
expression of ALCAM in pancreatic cancer Panc-1 and T3M4 cells with
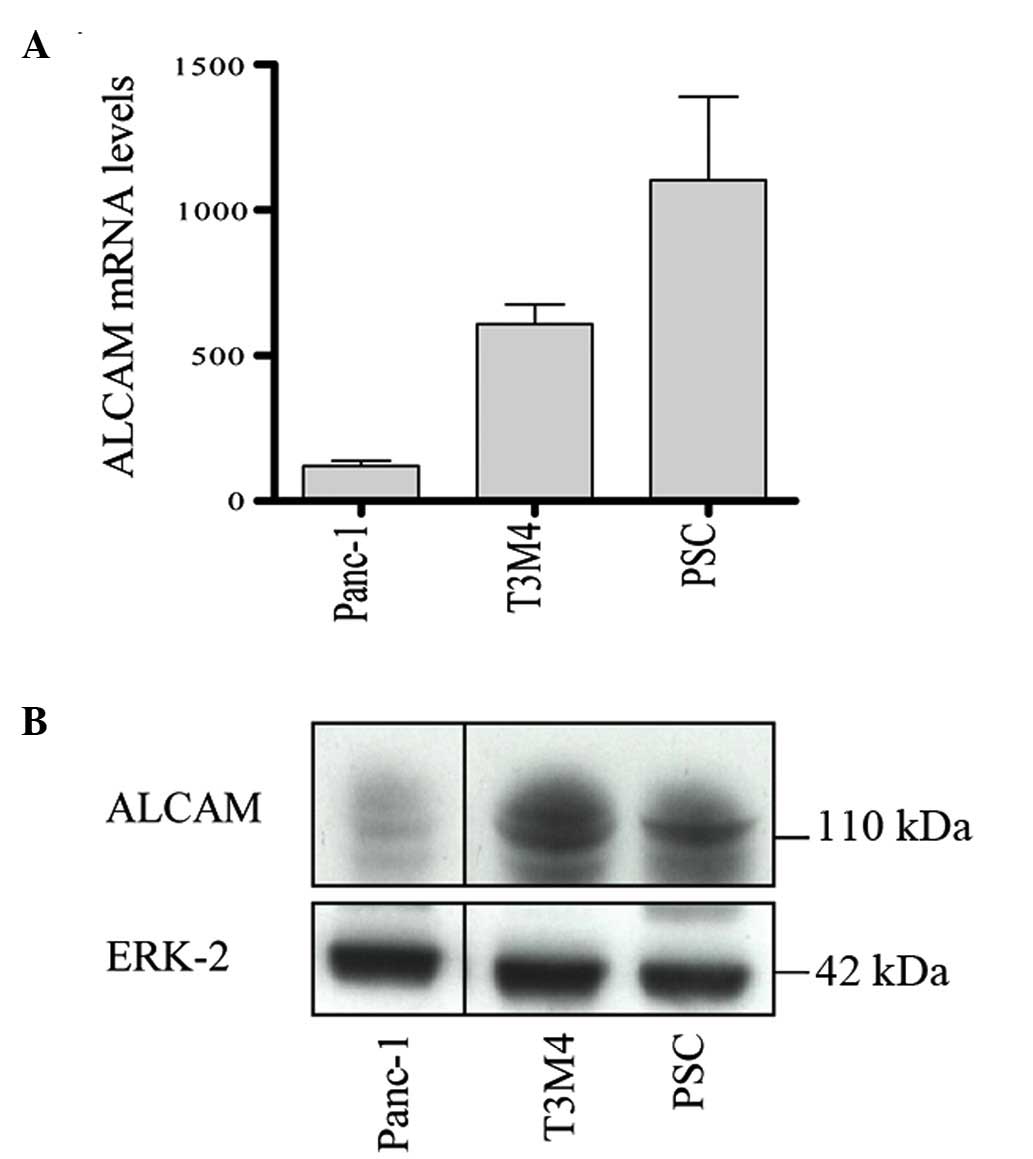
its expression in PSCs. As shown in Fig. 2A, ALCAM mRNA was highly expressed
in PSCs, while it was low to moderately expressed in T3M4 and
Panc-1 cells. Similar to mRNA expression, western blot analysis
demonstrated that ALCAM protein levels were high in PSCs and T3M4
cells, but low in Panc-1 cells (Fig.
2B).
Soluble levels of ALCAM are regulated by
tumor necrosis factor (TNF)-α, transforming growth factor (TGF)-β
and hypoxia
To assess the secretion of ALCAM following
stimulation in pancreatic cells, Panc-1 and T3M4 cells, and PSCs
were treated with TNF-α, TGF-β and hypoxia for 48 h, and ALCAM
protein levels were detected in cell culture supernatant by ELISA.
The results demonstrated that ALCAM levels were significantly
increased by TNF-α in Panc-1 (P<0.001), T3M4 (P=0.003) and PSCs
(P<0.001), while ALCAM levels were significantly decreased by
hypoxia in PSCs (P<0.001). Following treatment with TGF-β, ALCAM
levels did not change in Panc-1 cells, increased in T3M4 cells
(P=0.043) and decreased in PSCs (P=0.01; Fig. 3).
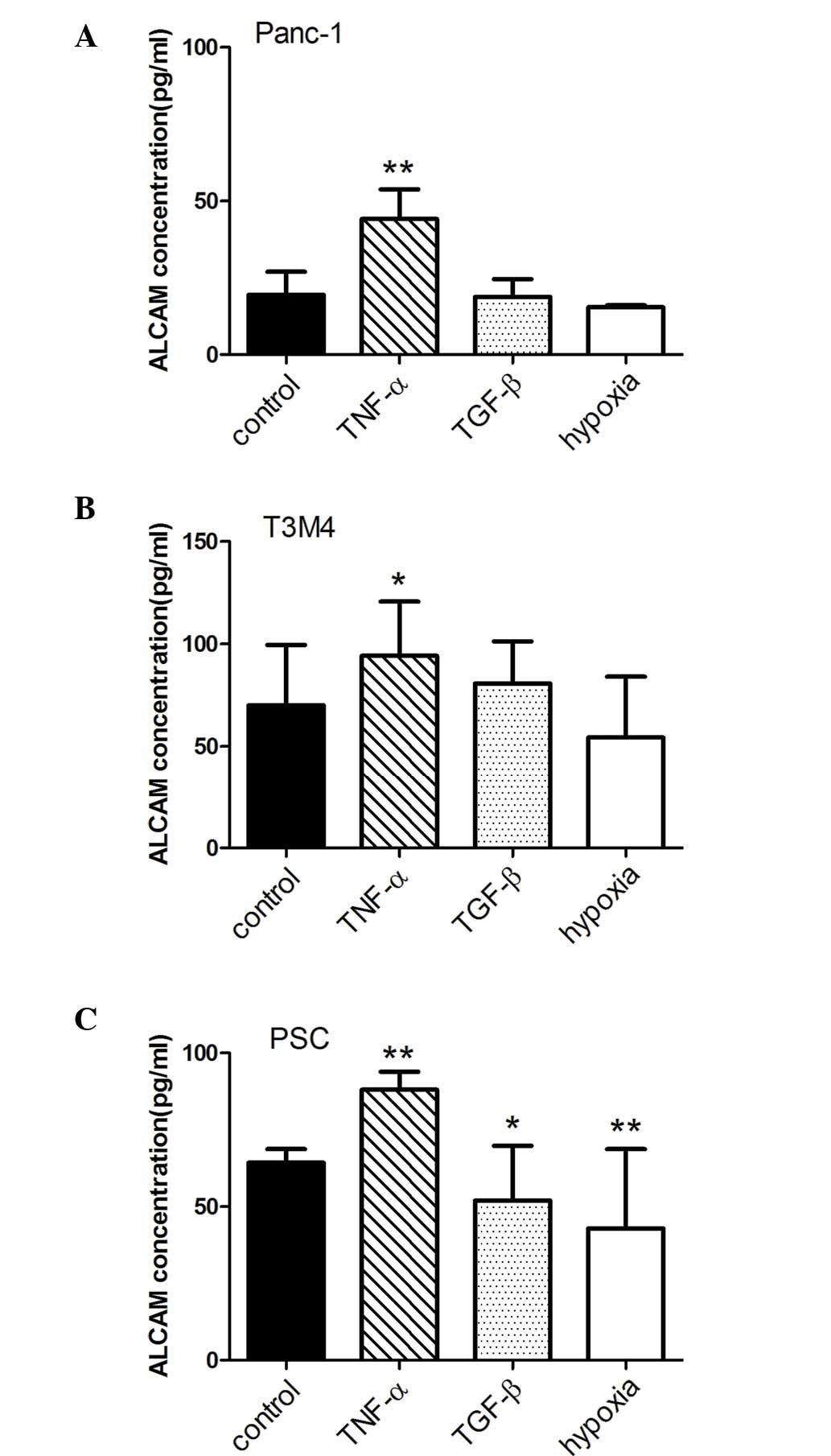 | Figure 3ALCAM levels following stimulation
with TNF-α, TGF-β and hypoxia. Enzyme-linked immunosorbent assay
analysis of the levels of ALCAM was performed, as described in the
Materials and methods. TNF-α, TGF-β and hypoxia were used to
stimulate (A) Panc-1 and (B) T3M4 cells, and also (C) PSCs. ALCAM
levels were significantly increased by TNF-α in (A) Panc-1
(P<0.001), (B) T3M4 (P=0.003) and (C) PSCs (P<0.001), whereas
ALCAM levels were significantly decreased by hypoxia in PSCs
(P<0.001). *P<0.05; **P<0.001.
TNF-α, tumor necrosis factor-α; TGF-β, transforming growth
factor-β; ALCAM, activated leukocyte cell adhesion molecule; PSCs,
pancreatic stellate cells. |
ALCAM promotes PSC invasion
As previously demonstrated by our group, ALCAM
silencing did not affect pancreatic cancer cell growth or invasion
but significantly reduced cell adhesion in Su86.86 pancreatic
cancer cells (4). To assess the
role of ALCAM in the regulation of invasion of pancreatic cells and
PSCs, ALCAM was knocked down using siRNA in Panc-1 and T3M4 cells,
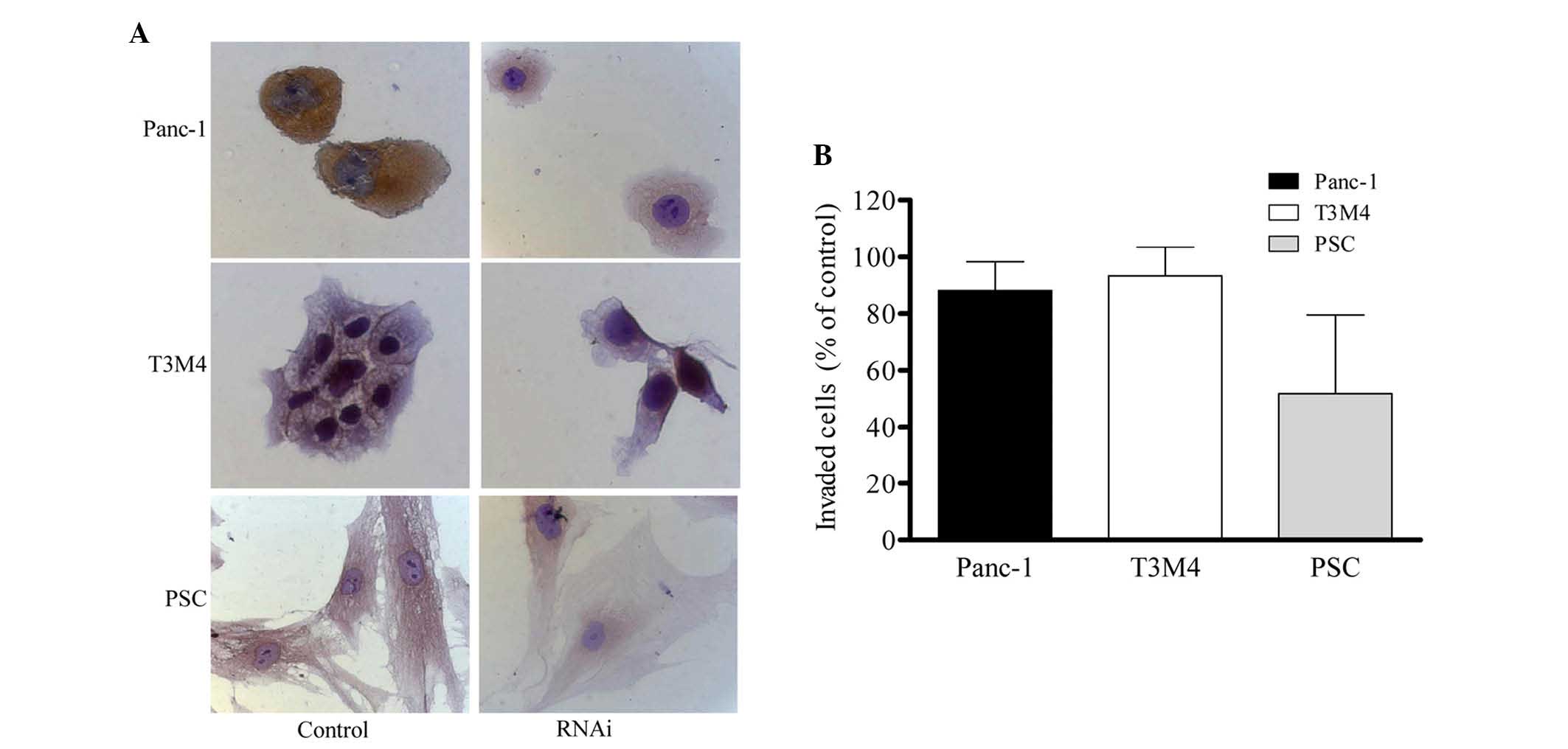
as well as in PSCs. Immunocytochemistry demonstrated that ALCAM was
predominantly expressed in the cytoplasm in Panc-1 and PSC cells,
and in the membrane of T3M4 cells. Transfection of these cells with
ALCAM siRNA for 48 h resulted in a decrease in ALCAM expression
(Fig. 4A). ALCAM silencing by
siRNA did not affect the invasion of Panc-1 and T3M4 cells, but
resulted in 50% inhibition of invasion of PSCs (P=0.047; Fig. 4B).
ALCAM silencing results in decreased
interaction between Panc-1 cells and PSCs
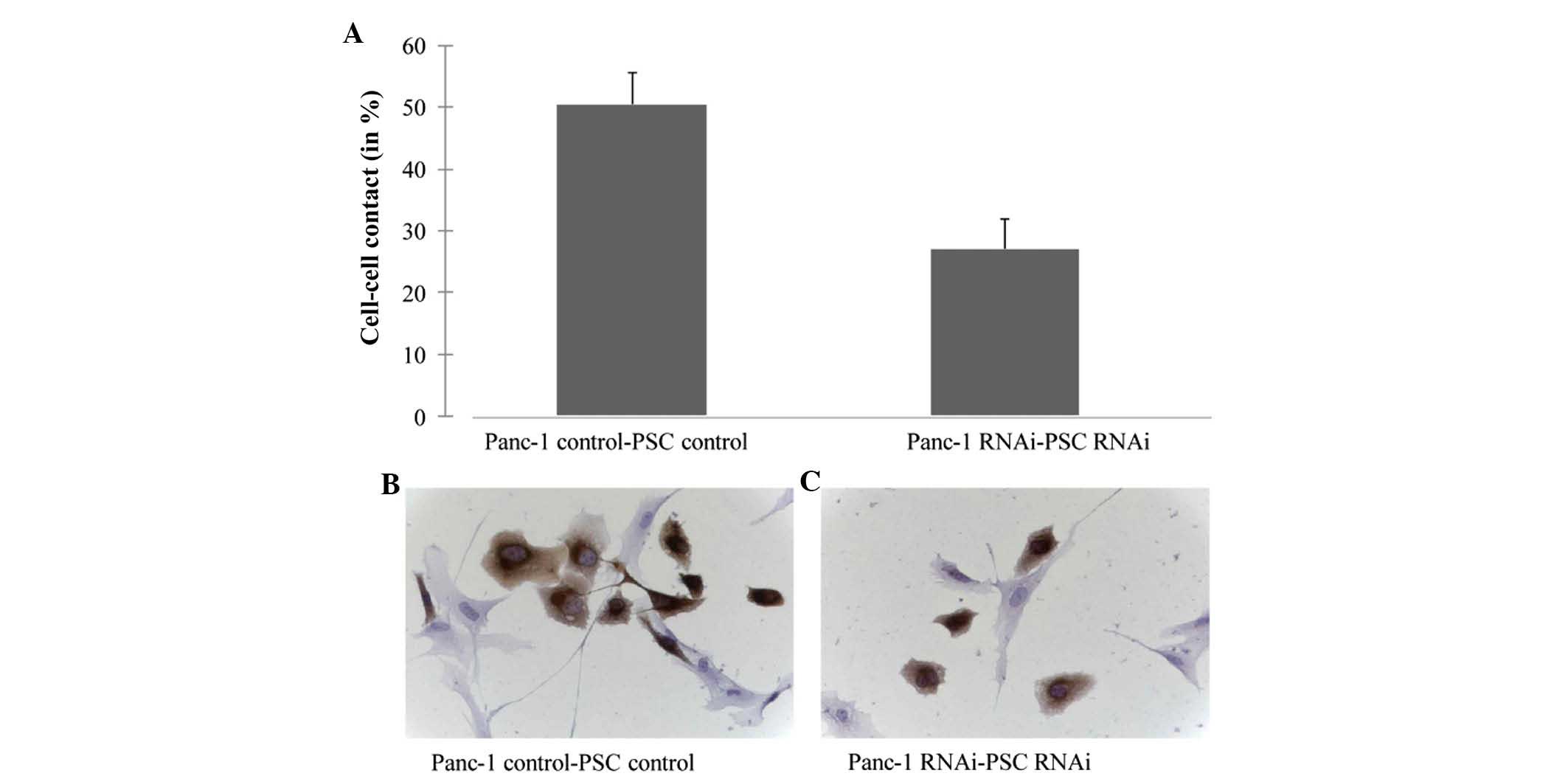
A previous study demonstrated that ALCAM is
important in the regulation of tumor cell-stromal cell interactions
(25). To assess whether silencing
of ALCAM alters the interaction between tumor and stromal cells,
Panc-1 and PSCs were co-cultured. Silencing of ALCAM by siRNA led
to a decreased interaction between Panc-1 cells and PSCs (Fig. 5).
Discussion
The molecular functions of ALCAM in the
tumorigenesis of different types of cancer are largely unknown. In
the present study, immunohistochemistry demonstrated that ALCAM was
only partially expressed in the membrane and cytoplasm of
pancreatic cancer cells, which was consistent with the results of a
previous study by Hong et al (4). ALCAM expression is generally variable
in pancreatic cancer cells (17).
In the present study, RT-qPCR and immunoblot analyses revealed that
the expression of ALCAM was higher in PSCs compared with that in
Panc-1 and T3M4 pancreatic cancer cells, suggesting that ALCAM may
be more important in PSCs compared with in pancreatic cancer
cells.
It was reported that ALCAM protected breast cancer
cells against apoptosis and autophagy, suggesting that ALCAM may
promote tumorigenesis (14).
However, another study demonstrated that ALCAM suppressed breast
cancer cell invasion, suggesting inhibition of tumorigenesis at the
late stages (16). In addition,
CD166+ pancreatic cancer cells are strongly tumorigenic,
while CD166− pancreatic cancer cells exhibit
comparatively stronger invasive and migratory activities (16). These data suggested that ALCAM may
either promote or suppress tumorigenesis. However, the effect of
ALCAM on PSCs is unknown. Notably, the results of the present study
demonstrated that ALCAM silencing by siRNA decreased the invasive
ability of PSCs, which are a component of the pancreatic tumor
microenvironment. ALCAM shedding would release ALCAM into the tumor
environment and circulation to exert its function. When the
malignant environment was mimicked in vitro using TNF-α,
TGF-β and hypoxia, ALCAM shedding was significantly induced by
TNF-α from pancreatic cancer cells and PSCs, while it was decreased
by hypoxia and TGF-β, particularly in PSCs. The results are
consistent with the observation that ALCAM is a cytokine-regulated
cell adhesion molecule (26). This
indicated that the expression of ALCAM is regulated by an altered
tumor microenvironment.
The tumor microenvironment is important in the
progression, invasion and metastasis of cancer cells (27,28).
ALCAM is variably expressed in stromal and tumor cells, and its
expression is affected by the tumor microenvironment. Hong et
al reported an association between CD166 and adhesiveness
(4). Adhesiveness may cause the
functional differences between CD166+ and
CD166− cells. Thus, it may be hypothesized that
ALCAM-ALCAM interactions facilitate stromal-cancer cell
adhesiveness. In support of this hypothesis, in the present study,
ALCAM silencing by siRNA did not significantly alter the
proliferation of pancreatic cancer cells, but decreased the
invasive ability of PSCs. Furthermore, co-culture experiments of
PSCs with pancreatic cancer cells demonstrated that silencing of
ALCAM in PSCs and Panc-1 cells resulted in decreased tumor cell-PSC
adhesiveness. It is well known that once tumor growth has reached a
critical mass, the metastatic spread of tumor cells is dependent on
their dissociation from the primary tumor and migration towards the
systemic circulation. Primary tumors with invasive properties
usually exhibit reduced intercellular adhesion, which allows cells
to break away from the parental cell mass. Thus, it is possible
that ALCAM may indirectly regulate the metastatic potential of
pancreatic cancer cells by modulating tumor-stroma
interactions.
In conclusion, ALCAM is upregulated in PSCs of
pancreatic cancer tissues, promotes PSC invasion and increases the
interaction between Panc-1 cells and PSCs, suggesting a potential
role of ALCAM in regulating pancreatic cancer cell-PSC
interactions.
Acknowledgments
The present study was supported by the Qingdao
Municipal Health Science and Technology Program.
References
|
1
|
Bowen MA, Patel DD, Li X, Modrell B,
Malacko AR, Wang WC, Marquardt H, Neubauer M, Pesando JM, Francke
U, et al: Cloning, mapping and characterization of activated
leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J Exp Med.
181:2213–2220. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patel DD, Wee SF, Whichard LP, Bowen MA,
Pesando JM, Aruffo A and Haynes BF: Identification and
characterization of a 100-kD ligand for CD6 on human thymic
epithelial cells. J Exp Med. 181:1563–1568. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hansen AG, Arnold SA, Jiang M, Palmer TD,
Ketova T, Merkel A, Pickup M, Samaras S, Shyr Y, Moses HL, et al:
ALCAM/CD166 is a TGF-β-responsive marker and functional regulator
of prostate cancer metastasis to bone. Cancer Res. 74:1404–1415.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hong X, Michalski CW, Kong B, Zhang W,
Raggi MC, Sauliunaite D, De Oliveira T, Friess H and Kleeff J:
ALCAM is associated with chemoresistance and tumor cell adhesion in
pancreatic cancer. J Surg Oncol. 101:564–569. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kozovska Z, Gabrisova V and Kucerova L:
Colon cancer: Cancer stem cells markers, drug resistance and
treatment. Biomed Pharmacother. 68:911–916. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Penna E, Orso F, Cimino D, Vercellino I,
Grassi E, Quaglino E, Turco E and Taverna D: miR-214 coordinates
melanoma progression by upregulating ALCAM through TFAP2 and
miR-148b downmodulation. Cancer Res. 73:4098–4111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Piao D, Jiang T, Liu G, Wang B, Xu J and
Zhu A: Clinical implications of activated leukocyte cell adhesion
molecule expression in breast cancer. Mol Biol Rep. 39:661–668.
2012. View Article : Google Scholar
|
|
8
|
Tachezy M, Zander H, Wolters-Eisfeld G,
Müller J, Wicklein D, Gebauer F, Izbicki JR and Bockhorn M:
Activated leukocyte cell adhesion molecule (CD166): An 'inert'
cancer stem cell marker for non-small cell lung cancer? Stem Cells.
32:1429–1436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye M, Du YL, Nie YQ, Zhou ZW, Cao J and Li
YF: Overexpression of activated leukocute cell adhesion molecule in
gastric cancer is associated with advanced stages and poor
prognosis and miR-9 deregulation. Mol Med Rep. 11:2004–2012.
2015.
|
|
10
|
Yu W, Wang J, Ma L, Tang X, Qiao Y, Pan Q,
Yu Y and Sun F: CD166 plays a pro-carcinogenic role in liver cancer
cells via inhibition of FOXO proteins through AKT. Oncol Rep.
32:677–683. 2014.PubMed/NCBI
|
|
11
|
Zhao Z, Lu P, Zhang H, Xu H, Gao N, Li M
and Liu C: Nestin positively regulates the Wnt/β-catenin pathway
and the proliferation, survival and invasiveness of breast cancer
stem cells. Breast Cancer Res. 16:4082014. View Article : Google Scholar
|
|
12
|
Dalerba P, Dylla SJ, Park IK, Liu R, Wang
X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rajasekhar VK, Studer L, Gerald W, Socci
ND and Scher HI: Tumour-initiating stem-like cells in human
prostate cancer exhibit increased NF-κB signalling. Nat Commun.
2:1622011. View Article : Google Scholar
|
|
14
|
Jezierska A, Matysiak W and Motyl T:
ALCAM/CD166 protects breast cancer cells against apoptosis and
autophagy. Med Sci Monit. 12:BR263–BR273. 2006.PubMed/NCBI
|
|
15
|
Lunter PC, van Kilsdonk JW, van Beek H,
Cornelissen IM, Bergers M, Willems PH, van Muijen GN and Swart GW:
Activated leukocyte cell adhesion molecule (ALCAM/CD166/MEMD), a
novel actor in invasive growth, controls matrix metalloproteinase
activity. Cancer Res. 65:8801–8808. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jezierska A, Olszewski WP, Pietruszkiewicz
J, Olszewski W, Matysiak W and Motyl T: Activated Leukocyte Cell
Adhesion Molecule (ALCAM) is associated with suppression of breast
cancer cells invasion. Med Sci Monit. 12:BR245–BR256.
2006.PubMed/NCBI
|
|
17
|
Fujiwara K, Ohuchida K, Sada M, Horioka K,
Ulrich CD III, Shindo K, Ohtsuka T, Takahata S, Mizumoto K, Oda Y
and Tanaka M: CD166/ALCAM expression is characteristic of
tumorigenicity and invasive and migratory activities of pancreatic
cancer cells. PLoS One. 9:e1072472014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Erkan M, Adler G, Apte MV, Bachem MG,
Buchholz M, Detlefsen S, Esposito I, Friess H, Gress TM, Habisch
HJ, et al: StellaTUM: Current consensus and discussion on
pancreatic stellate cell research. Gut. 61:172–178. 2012.
View Article : Google Scholar
|
|
19
|
Erkan M, Kleeff J, Gorbachevski A, Reiser
C, Mitkus T, Esposito I, Giese T, Büchler MW, Giese NA and Friess
H: Periostin creates a tumor-supportive microenvironment in the
pancreas by sustaining fibrogenic stellate cell activity.
Gastroenterology. 132:1447–1464. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Erkan M, Reiser-Erkan C, Michalski CW,
Deucker S, Sauliunaite D, Streit S, Esposito I, Friess H and Kleeff
J: Cancer-stellate cell interactions perpetuate the
hypoxia-fibrosis cycle in pancreatic ductal adenocarcinoma.
Neoplasia. 11:497–508. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang W, Erkan M, Abiatari I, Giese NA,
Felix K, Kayed H, Büchler MW, Friess H and Kleeff J: Expression of
extracellular matrix metalloproteinase inducer (EMMPRIN/CD147) in
pancreatic neoplasm and pancreatic stellate cells. Cancer Biol
Ther. 6:218–227. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Abiatari I, Esposito I, Oliveira TD, Felix
K, Xin H, Penzel R, Giese T, Friess H and Kleeff J: Moesin
dependent cytoskeleton remodeling is associated with an anaplastic
phenotype of pancreatic cancer. J Cell Mol Med. 14:1166–1179.
2009.
|
|
23
|
Kayed H, Kleeff J, Kolb A, Ketterer K,
Keleg S, Felix K, Giese T, Penzel R, Zentgraf H, Büchler MW, et al:
FXYD3 is overexpressed in pancreatic ductal adenocarcinoma and
influences pancreatic cancer cell growth. Int J Cancer. 118:43–54.
2006. View Article : Google Scholar
|
|
24
|
Erkan M, Kleeff J, Esposito I, Giese T,
Ketterer K, Büchler MW, Giese NA and Friess H: Loss of BNIP3
expression is a late event in pancreatic cancer contributing to
chemoresistance and worsened prognosis. Oncogene. 24:4421–4432.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Behnan J, Isakson P, Joel M, Cilio C,
Langmoen IA, Vik-Mo EO and Badn W: Recruited brain tumor-derived
mesenchymal stem cells contribute to brain tumor progression. Stem
Cells. 32:1110–1123. 2014. View Article : Google Scholar
|
|
26
|
Levesque MC, Heinly CS, Whichard LP and
Patel DD: Cytokine-regulated expression of activated leukocyte cell
adhesion molecule (CD166) on monocyte-lineage cells and in
rheumatoid arthritis synovium. Arthritis Rheum. 41:2221–2229. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Habisch H, Zhou S, Siech M and Bachem MG:
Interaction of stellate cells with pancreatic carcinoma cells.
Cancers (Basel). 2:1661–1682. 2010. View Article : Google Scholar
|
|
28
|
Wilson JS, Pirola RC and Apte MV: Stars
and stripes in pancreatic cancer: Role of stellate cells and stroma
in cancer progression. Front Physiol. 5:522014. View Article : Google Scholar : PubMed/NCBI
|