Introduction
β-crystallin B2 (CRYBB2) expression was initially
reported in the eye lens where it functions to maintain lens
transparency and refractive index. CRYBB2 deficiency was
demonstrated to result in the generation of age-associated
cataracts (1). However, a number
of previous studies demonstrated that CRYBB2 is also expressed in
the retina, brain, testis, and ovary (2–5). Our
previous study observed that CRYBB2 was expressed in human and
mouse ovaries, particularly in ovarian granulosa cells (6). CRYBB2 knockout mice exhibited
morphological and functional abnormalities in the ovary, including
reduced ovarian index (ratio of ovary weight to total body weight)
with increased follicle atresia, reduced mature follicles and
dysregulated estrous cycle (7).
Our data from a previous study also indicated a high level of
estrogen in the diestrus and metestrus, but a low level of
progesterone in the metestrus compared with wild-type (WT) mice
(6). At the genetic level,
expression of cell cycle and apoptosis-associated proteins,
including B-cell lymphoma 2, cyclin-dependent kinase 4, and cyclin
D2, were markedly lower in the CRYBB2 knockout mice compared
with WT mice (6). These data
suggest that CRYBB2 may be important in ovary development,
however, the underlying molecular mechanism by which CRYBB2
regulates ovarian development remains to be elucidated.
To identify and assess the role of CRYBB2 in
ovary development, differentially expressed long non-coding RNAs
(lncRNAs) were profiled in CRYBB2 knockout mice. lncRNAs are
a class of non-coding RNAs with nucleotides >200 bp and
transcribed by RNA polymerase II. Functionally, lncRNAs may
regulate gene transcription, protein translation, and epigenetic
modification of genomic DNA. Altered expression and regulation of
lncRNAs has been associated with human diseases and aging (8–12).
For example, previous studies have suggested that overexpression of
lncRNA HOX transcript antisense RNA associated with the recurrence
of hepatocellular carcinoma, poor prognosis in colorectal cancer,
and malignant behaviors of gastrointestinal stromal tumors
(13–15). lncRNAs modulate cell functions by
regulating expression of targeted downstream genes (16), which may in turn affect embryo
development (17), inactivate the
X chromosome, and regulate genomic imprinting (18). Thus, the current study assessed
whether these lncRNAs mediate the functions of CRYBB2 in ovary
development and investigated the underlying mechanisms. Microarray
profiling between ovarian tissues from CRYBB2 knockout and
WT mice was conducted and bioinformatic analysis of differentially
expressed lncRNAs and mRNAs was performed. A number of these
differentially expressed lncRNAs and mRNAs were verified using
quantitative reverse transcription-polymerase chain reaction
(RT-qPCR).
Materials and methods
Animals
Male and female C57BL/6 mice were obtained from the
Experimental Animal Center of the Second Military Medical
University (Shanghai, China). CRYBB2 knockout mice were
generated by the Ingenious Targeting Laboratory (Ronkonkoma, NY,
USA), as described previously (19). All mice were maintained on a 12 h
light/dark cycle with a temperature of 21±1°C and humidity of
50~70% in a pathogen-free facility with access to food and water
ad libitum. Bedding material and a plastic house or tube was
placed in the cage for environmental enrichment. Daily examinations
were performed on all animals throughout the experimental period.
Humane euthanasia of mice was performed under isoflurane anesthesia
using intracardiac injection of pentobarbitone (150 mg/kg). The
present study was conducted in accordance with institutional
guidelines and approved by the Animal Care and Use Committee of
Changhai Hospital (Shanghai, China).
A total of three 8-9-week-old (weight, 18.0±2.0 g)
female CRYBB2 knockout mice and three age-matched WT female
mice were obtained. Ovary tissues were collected from mice
following a 10-day experimental period.
RNA isolation, cDNA synthesis and
labeling, and hybridization
Ovarian tissues were homogenized on ice and total
cellular RNA was isolated using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol and then quantified using NanoDrop ND-1000
(Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and agarose
gel electrophoresis. RNA samples were further purified using an
RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) and reverse
transcribed into cDNA with fluorescent labeling for microarray
hybridization using using the AffinityScript QPCR cDNA Synthesis
kit (Agilent Technologies, Inc., Santa Clara, CA, USA). These
labeled cDNA probes were then hybridized to Agilent mouse
expression profiling (8×60K) microarray using the Gene Expression
Hybridization kit (Agilent Technologies, Inc.) according to the
manufacturer's protocol. The arrays were scanned into a file and
analyzed using Feature Extraction software, version v10.7.3.1
(Agilent Technologies, Inc.). The arrays were scanned at 5 µm/pixel
resolution using an Axon GenePix 4000B scanner (Molecular Devices,
LLC, Sunnyvale, CA, USA) piloted by GenePix Pro 6.0 software
(Molecular Devices, LLC) and then imported into NimbleScan software
(version 2.5; Roche NimbleGen, Inc., Madison, WI, USA) for grid
alignment and expression data analysis. Expression data were
normalized using quantile normalization and the Robust Multichip
Average algorithm included in the NimbleScan software. The probe
level files and mRNA level files were generated following
normalization. All mRNA level files were imported into Agilent
GeneSpringGX software (version 11.0; Agilent Technologies, Inc.)
for further analysis. Differentially expressed lncRNAs and mRNAs
were identified via fold change filtering.
Functional analysis of microarray
data
Gene Ontology (GO; www.geneontology.org) was performed to group
differentially expressed lncRNAs and their targeted genes into
biological processes, cellular components, and molecular features
of the biological functions. Kyoto Encyclopedia of Genes and
Genomes (KEGG; www.genome.jp/kegg) analysis was performed to identify
roles of the target genes and group them into gene pathways. The
correlation matrix method was used to establish a diagram of lncRNA
and mRNA co-expression regulatory networks. Prior to functional
analyses, the predicted potential lncRNA targets were integrated
with the differentially expressed mRNAs using a cut-off value of
≥2.0 fold change or P<0.05.
RT-qPCR
Total RNA was isolated from granulosa cells from the
ovaries of WT and CRYBB2 knockout mice using TRIzol reagent
and reverse transcribed into cDNA using the PrimeScript RT Reagent
Kit (Takara Biotechnology, Co., Ltd., Dalian, China) according to
the manufacturer's protocols. These cDNA samples were then
subjected to qPCR analysis using a Applied Biosystems 7300
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with the SYBR Green PCR Master Mix kit (Takara Biotechnology,
Co., Ltd.). qPCR amplification was conducted at 95°C for 2 min,
followed by 40 cycles of 95°C for 15 sec, and 60°C for 30 sec. The
relative expression of each target gene compared to β-actin was
calculated using the 2−ΔΔCq method (20). Specific primers used were as
follows: Forward, 5′-AGCCATGTACGTAGCCATCC-3′ and reverse,
5′-CTCTCAGCTGTGGTGGTGAA-3′ for β-actin; forward,
5′-CGAGTTGGTGCCAGTGTGGA-3′ and reverse, 5′-CCTGCTGTTGGTGGCCTCTT-3′
for P2rx7; and forward, 5′-TCCACTCAGGAAGAGCTGGT-3′ and
reverse, 5′-TAGCACCCTCGGGATATCTG-3′ for lncRNAA-30-P01019163.
Statistical analysis
All data were presented as the mean ± standard
deviation. The data were Log2-transformed and median centered by
genes using Cluster software, version 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm).
Statistical analyses were performed using SPSS 17.0 (SPSS, Inc.,
Chicago, IL, USA). The data were statistically analyzed using an
unpaired Student's t-test or by one-way analysis of variance
followed by Tukey's test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Microarray profiling of differentially
expressed lncRNAs and mRNA in ovary tissues from wild type and
CRYBB2 knockout mice
Microarray data on differential lncRNA and mRNA
expression levels were analyzed using Cluster software, version 3.0
with the cut-off values set as ≥2-fold difference or P<0.05. The
data indicated 157 differentially expressed lncRNAs (42
downregulated vs. 115 upregulated) and 1,085 differentially
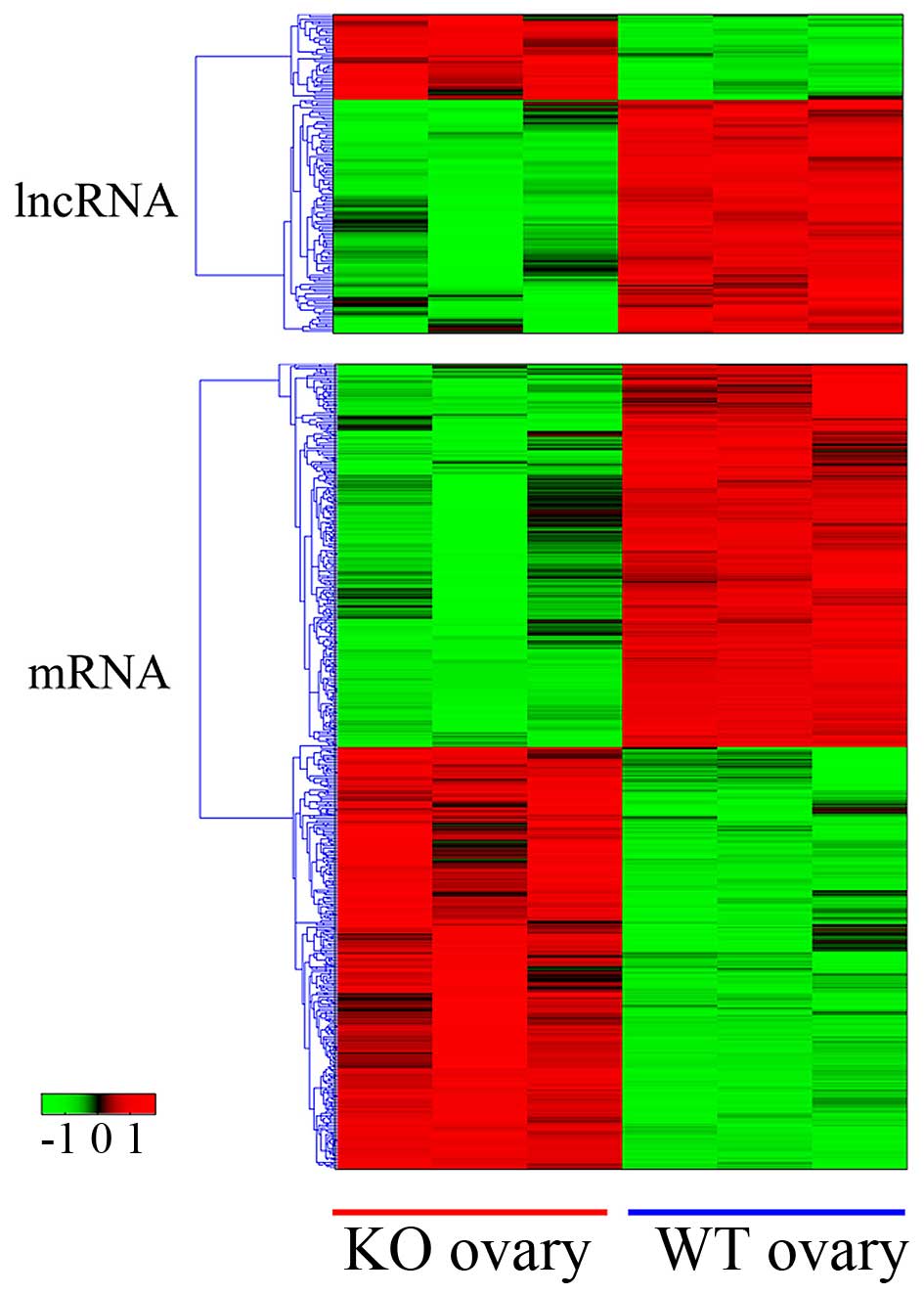
expressed mRNAs (570 downregulated vs. 515 upregulated; Fig. 1). The genes were selected according
to a software forecast associated with the processes of ovarian
development, and the lncRNAs associated with these genes were
selected (Tables I and II).
 | Table I.Differentially expressed long
non-coding RNAs in ovary tissues from β-crystallin B2 knockout
mice. |
Table I.
Differentially expressed long
non-coding RNAs in ovary tissues from β-crystallin B2 knockout
mice.
| Probe name | P-value | FC (abs) | Regulation |
|---|
| A_30_P01032506 | 0.001900622 | 2.244835 | Down |
| A_30_P01029732 | 0.026376737 | 2.3413253 | Down |
| A_30_P01020038 | 0.020382054 | 2.3550973 | Down |
| A_30_P01019163 | 0.008867065 | 2.3823233 | Down |
| A_30_P01017808 | 0.003141625 | 2.3902857 | Down |
| A_30_P01028465 | 0.026580842 | 2.3909771 | Down |
| A_30_P01024278 | 0.010205811 | 2.4727218 | Down |
| A_30_P01031572 | 0.036648612 | 2.5956807 | Down |
| A_30_P01027590 | 0.034441914 | 2.6343288 | Down |
| A_30_P01022135 | 0.04770927 | 3.120179 | Down |
| A_30_P01031631 | 0.001265911 | 3.135137 | Down |
| A_30_P01032372 | 0.009654216 | 3.4001627 | Down |
| A_30_P01032133 | 0.012271924 | 3.434039 | Down |
| A_30_P01024342 |
2.97×10−4 | 3.7296832 | Down |
| A_30_P01024108 | 0.004697592 | 3.8198798 | Down |
| A_30_P01022445 |
2.66×10−5 | 3.8623033 | Down |
| A_30_P01022656 |
2.08×10−4 | 4.069888 | Down |
| A_30_P01023636 | 0.02164327 | 4.4273615 | Down |
| A_30_P01022538 |
9.77×10−4 | 5.492365 | Down |
| A_30_P01033546 | 0.004913332 | 5.9940124 | Down |
| A_30_P01019023 | 0.024659809 | 3.7894382 | Up |
| A_30_P01024108 | 0.004697592 | 3.8198798 | Up |
| A_30_P01017880 | 0.034943227 | 3.8256395 | Up |
| A_30_P01022445 |
2.66×10−5 | 3.8623033 | Up |
| A_30_P01026472 | 0.022304738 | 3.8728366 | Up |
| A_30_P01030900 | 0.023211088 | 3.9997435 | Up |
| A_30_P01022656 |
2.08×10−4 | 4.069888 | Up |
| A_30_P01018745 |
1.84×10−5 | 4.266393 | Up |
| A_30_P01033353 | 0.03583063 | 4.398616 | Up |
| A_30_P01021636 | 0.01934708 | 4.4207296 | Up |
| A_30_P01023636 | 0.02164327 | 4.4273615 | Up |
| A_30_P01022538 |
9.77×10−4 | 5.492365 | Up |
| A_30_P01019825 | 0.006051722 | 5.5902176 | Up |
| A_30_P01033546 | 0.004913332 | 5.9940124 | Up |
| A_30_P01024788 |
1.11×10−5 | 7.118162 | Up |
| A_30_P01024270 |
6.38×10−5 | 8.226261 | Up |
| A_30_P01027087 | 0.003292078 | 8.981714 | Up |
| A_30_P01031162 | 0.011685452 | 9.579193 | Up |
| A_30_P01021698 |
1.31×10−5 | 10.52238 | Up |
| A_30_P01020936 |
4.52×10−4 | 22.788074 | Up |
 | Table II.Differentially expressed mRNAs in
ovary tissues from β-crystallin B2 knockout mice. |
Table II.
Differentially expressed mRNAs in
ovary tissues from β-crystallin B2 knockout mice.
| Gene | P-value | FC (abs) | Regulation |
|---|
| Prkar2b | 0.040216 | 2.1857524 | Down |
| Lrp11 | 0.006975 | 2.9040618 | Down |
| P2rx7 |
4.11×10−4 | 4.6213336 | Down |
| Calml3 | 0.011701 | 5.126569 | Down |
| Dclre1c |
3.39×10−4 | 5.1453366 | Down |
| Lpcat2 | 0.002695 | 5.828531 | Down |
| Stk32a | 0.001242 | 5.833555 | Down |
| Cyp19a1 | 0.034041 | 6.1799 | Down |
| Megf10 | 0.045621 | 6.1827664 | Down |
| Plcxd1 |
1.04×10−4 | 6.82413 | Down |
| Gm5103 | 0.005622 | 8.389297 | Down |
| Fermt1 | 0.001363 | 8.656997 | Down |
| Mlxip |
3.10×10−4 | 8.746344 | Down |
| Slc6a2 | 0.046581 | 8.750824 | Down |
| Gm7969 | 0.012795 | 9.970471 | Down |
| Ces2a | 0.028129 | 10.472376 | Down |
| Dclre1c |
1.23×10−4 | 10.645219 | Down |
| Ostn |
9.07×10−6 | 11.019576 | Down |
| Jph4 | 0.001674 | 12.722166 | Down |
| Onecut2 | 0.048542 | 13.894124 | Down |
| Ces2a | 0.028128909 | 10.472376 | Up |
| Dclre1c |
1.23×10−4 | 10.645219 | Up |
| Jph4 | 0.001673621 | 12.722166 | Up |
| Onecut2 | 0.04854157 | 13.894124 | Up |
| Itln1 |
5.97×10−5 | 14.339393 | Up |
| Arsk |
7.13×10−5 | 16.06716 | Up |
| Sfrp4 | 0.023781205 | 17.065975 | Up |
| Plekhg4 | 0.047409292 | 17.067673 | Up |
| Nuf2 |
6.05×10−5 | 17.872303 | Up |
| Adh7 | 0.0064019 | 18.950857 | Up |
| Itln1 |
4.63×10−5 | 19.05549 | Up |
| Ptgfr | 0.032550577 | 20.059986 | Up |
| Pou6f1 |
1.71×10−4 | 23.766891 | Up |
| Saa2 | 0.023944996 | 25.063555 | Up |
| Wnt10b | 0.02046059 | 27.648937 | Up |
| Ifi202b | 0.001051159 | 28.72067 | Up |
| Dcpp1 | 0.014649089 | 40.37458 | Up |
| Cd5l |
4.12×10−4 | 42.74245 | Up |
| Dcpp2 | 0.012848383 | 45.13117 | Up |
Bioinformatic analysis of
differentially expressed lncRNAs and mRNA in ovary tissues from WT
and CRYBB2 knockout mice
GO analysis was performed for functional annotation
of differentially expressed lncRNAs and mRNAs and it was observed
that they were predominantly involved in cell cycle regulation,
cell proliferation, metabolism, and signal transduction (Table III). One particular gene, P2rx7,
localized in the cytoplasm, functions to regulate cell cycle
progression and proliferation, and the signal transduction
process.
 | Table III.Gene ontology analysis of
differentially expressed genes. |
Table III.
Gene ontology analysis of
differentially expressed genes.
| Term | Number of genes
(%) | P-value |
|---|
| Biological
process |
|
| Cell
cycle and proliferation | 3
(4) | 0.479979 |
| Stress
response | 4
(6) | 0.205102 |
|
Transport | 5
(7) | 0.563981 |
|
Developmental processes | 6
(9) | 0.459856 |
| RNA
metabolism | 7
(10) | 0.315857 |
| Other
metabolic processes | 12 (17) | 0.007676 |
| Cell
organization and biogenesis | 6
(9) | 0.193590 |
|
Cell-cell signaling | 1
(1) | 0.502761 |
| Signal
transduction | 11
(16) | 0.120754 |
| Protein
metabolism | 6
(9) | 0.502818 |
|
Death | 1
(1) | 0.827556 |
| Other
biological processes | 8
(11) | 0.990425 |
| Cellular
component |
|
|
Cytosol | 1
(2) | 0.505969 |
|
Mitochondrion | 1
(2) | 0.862146 |
|
Endoplasmic
reticulum/golgi | 3
(5) | 0.460760 |
| Other
cytoplasmic organelle | 1
(2) | 0.604395 |
|
Nucleus | 10 (18) | 0.306385 |
| Plasma
membrane | 6
(1) | 0.438812 |
| Other
membranes | 14 (26) | 0.661329 |
|
Translational apparatus | 1
(2) | 0.330956 |
|
Non-structural
extracellular | 2
(4) | 0.830334 |
| Other
cellular component | 15 (28) | 0.354726 |
| Molecular
function |
|
| Enzyme
regulator activity | 3
(5) | 0.141417 |
|
Transcription regulatory
activity | 4
(7) | 0.174629 |
|
Transporter activity | 2
(4) | 0.623057 |
| Signal
transduction activity | 11 (20) | 0.082302 |
| Nucleic
acid binding activity | 7
(13) | 0.252320 |
| Kinase
activity | 2
(4) | 0.503159 |
| Other
molecular function | 26 (47) | 0.535279 |
Subsequently, KEGG analysis was conducted to group
differentially expressed lncRNAs and mRNA into gene pathways. The
data from the present study indicated that the genes predominantly
formed signaling pathways associated with the regulation of
Ca2+ signaling, ligand and receptor interactions, and
other cell signaling pathways (Table
IV). For example, P2rx7 was identified to be predominantly
involved in Ca2 + signaling.
 | Table IV.KEGG pathways analysis of
differentially expressed genes. |
Table IV.
KEGG pathways analysis of
differentially expressed genes.
| Term | Count | P-value | Genes | Up | Down |
|---|
| Glutathione
metabolism | 1 | 0.106032 | Gsta2 | 0 | 1 |
| Metabolism of
xenobiotics by cytochrome P450 | 1 | 0.143567 | Gsta2 | 0 | 1 |
| Drug
metabolism-cytochrome P450 | 1 | 0.147434 | Gsta2 | 0 | 1 |
| Ribosome | 1 | 0.172182 | Rps3a | 0 | 1 |
| PPAR signaling
pathway | 1 | 0.143567 | Adipoq | 0 | 1 |
| Calcium signaling
pathway | 3 | 0.00614 | P2rx7, Ptger1,
Ptafr | 3 | 0 |
| Cytokine-cytokine
receptor interaction | 1 | 0.45143 | Cxcl1 | 0 | 1 |
| Chemokine signaling
pathway | 1 | 0.353888 | Cxcl1 | 0 | 1 |
| Neuroactive
ligand-receptor interaction | 4 | 0.001745 | P2rx7, Ptger1,
Ptafr, Vipr1 | 4 | 0 |
| Cell cycle | 1 | 0.226819 | Hdac1 | 0 | 1 |
| Apoptosis | 1 | 0.179664 | Prkar2b | 1 | 0 |
| Notch signaling
pathway | 1 | 0.099971 | Hdac1 | 0 | 1 |
| Insulin signaling
pathway | 1 | 0.268146 | Prkar2b | 1 | 0 |
| Adipocytokine
signaling pathway | 1 | 0.139685 | Adipoq | 0 | 1 |
| Type II diabetes
mellitus | 1 | 0.099971 | Adipoq | 0 | 1 |
| Huntington's
disease | 1 | 0.334126 | Hdac1 | 0 | 1 |
| Pathways in
cancer | 1 | 0.530933 | Hdac1 | 0 | 1 |
| Chronic myeloid
leukemia | 1 | 0.155119 | Hdac1 | 0 | 1 |
Building the lncRNA-mRNA regulatory
network
The lncRNA target predictions were superimposed into
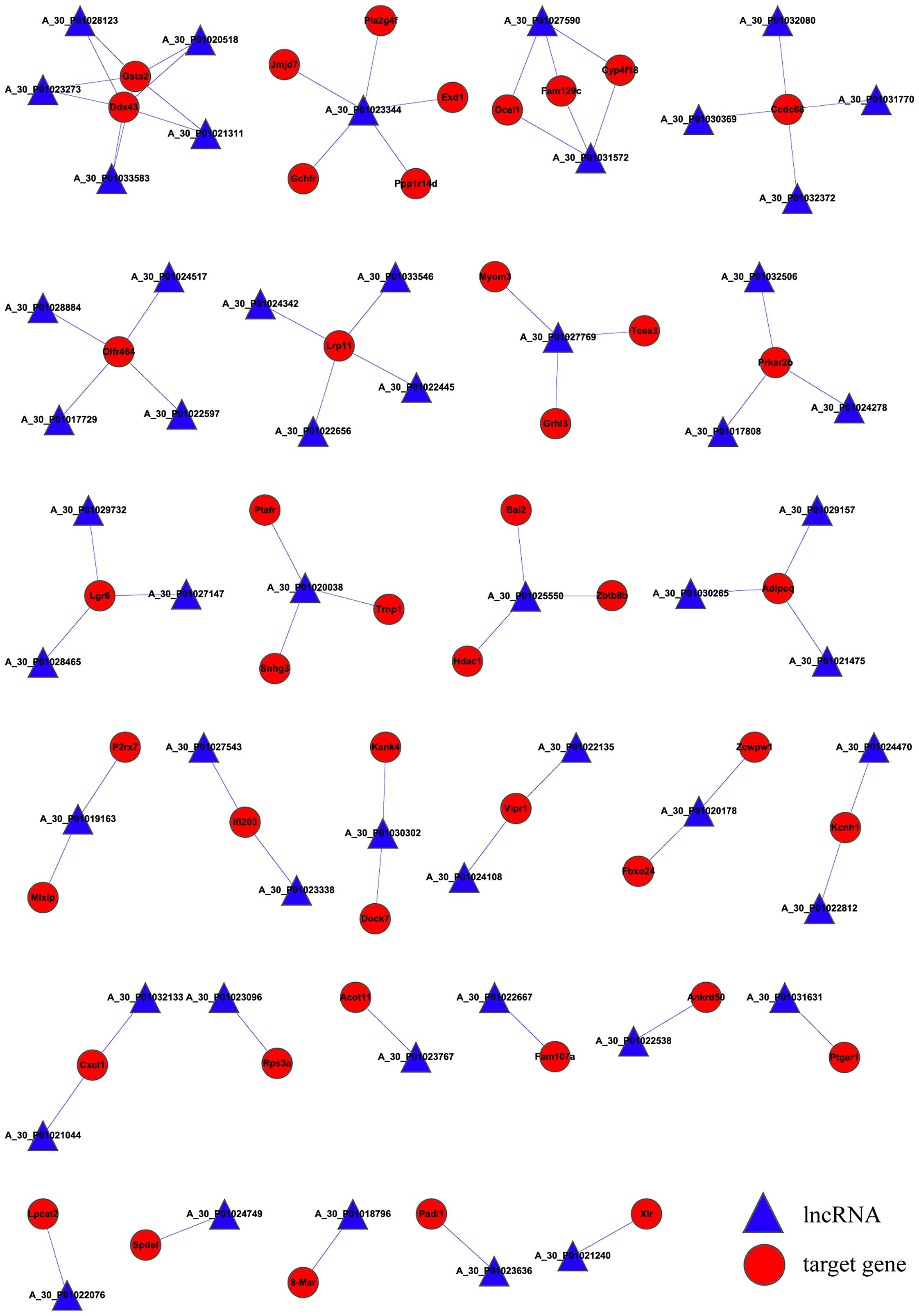
the lncRNA-mRNA correlation network. As presented in Fig. 2, the network has a total of 98
nodes, 75 connections, 53 lncRNAs and 45 mRNAs. Expression of
glutathione S-transferase α 2 (Gsta2) and DEAD (Asp-Glu-Ala-Asp)
box polypeptide 43 (Ddx43) was regulated by a total of five
lncRNAs, while expression of coiled-coil domain containing 68
(Ccdc68), olfactory receptor 464 (Olfr464) and low density
lipoprotein receptor-related protein 11 (Lrp11) were regulated by
four lncRNAs. In addition, expression levels of protein kinase,
cAMP dependent regulatory, type II β (Prkar2b), leucine-rich
repeat-containing G protein-coupled receptor 6 (Lgr6) and
adiponectin, C1Q and collagen domain containing (Adipoq) were
regulated by a total of three lncRNAs.
Validation of differentially expressed
lncRNAs and mRNAs in ovary tissues of WT and CRYBB2 knockout
mice
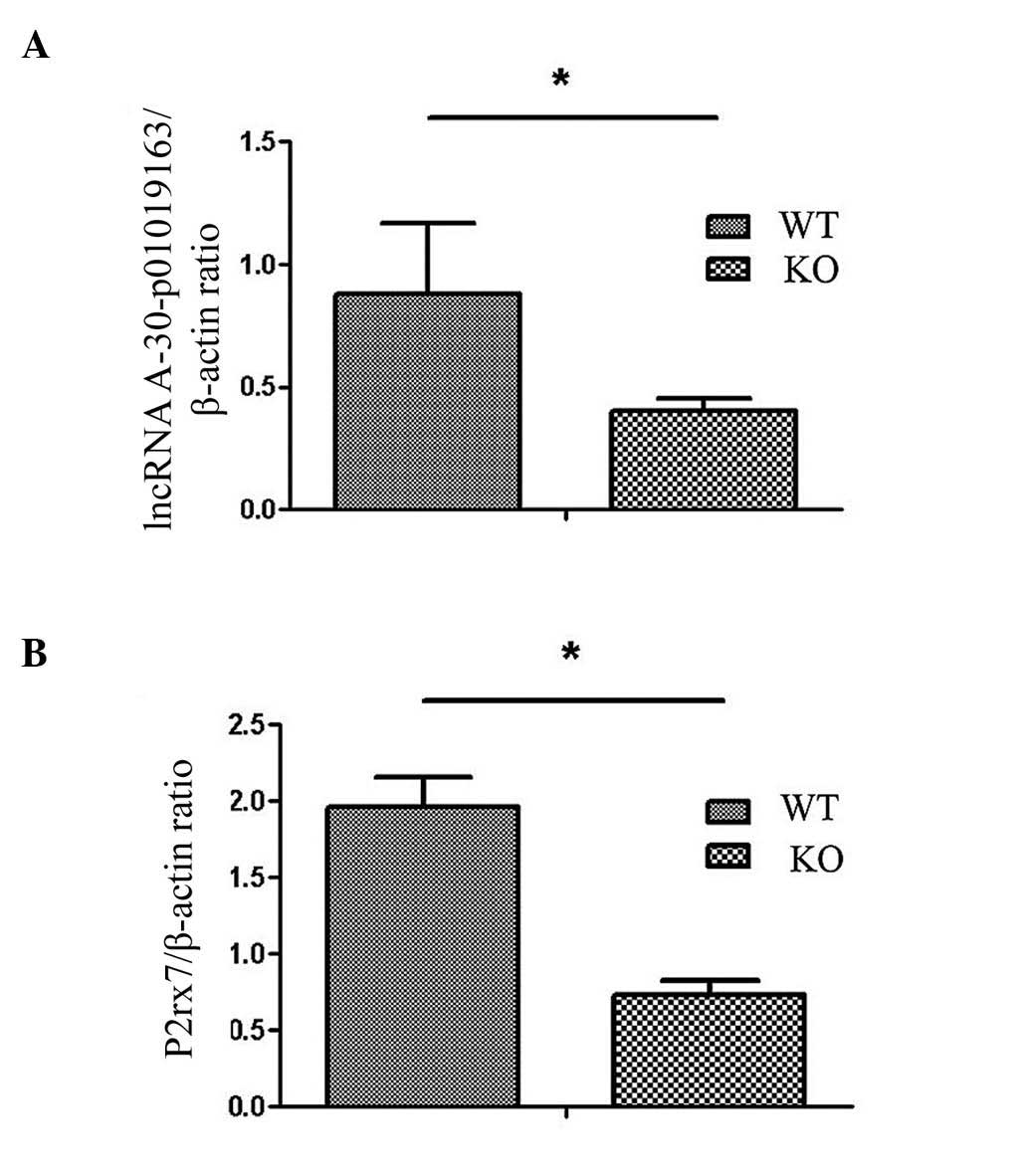
RT-qPCR analysis of different lncRNAs and mRNAs was
performed and it was demonstrated that expression levels of lncRNA
A-30-P01019163 and P2rx7 were significantly different between10
cases of ovary tissues from WT and CRYBB2 knockout mice
(P<0.05; Fig. 3), whereas other
lncRNAs and mRNAs did not indicate any difference between WT and
CRYBB2 knockout mouse ovary tissues (data not shown).
Discussion
CRYBB2 knockout mice exhibit abnormal
morphological and functional mouse ovaries. The current study
investigated the underlying molecular mechanisms by which
CRYBB2 affects expression of lncRNAs to alter mouse ovary
structure and function. Certain aberrant lncRNA expression in the
regulation of other genes was assessed. A total of 157
differentially expressed lncRNAs and 1,085 mRNAs were observed. The
GO database analysis indicated that these altered gene expressions
and were predominantly distributed in the biological process of
metabolism, immune system, and signal transduction. The KEGG
database pathway analysis suggested that the predominant signaling
pathways were associated with Ca2+ signaling and
ligand-receptor interaction.
Subsequent to establishing a correlation matrix
lncRNA and mRNA co-expression network map, a total of 98 nodes, 75
connections, 53 lncRNAs, and 45 mRNAs were identified. Gsta2 and
Ddx43 were regulated by five lncRNAs, Ccdc68, Olfr464 and Lrp11 by
four lncRNAs, Prkar2b, Lgr6, and Adipoq by three lncRNAs. Gsta2
participates in cytochrome P450 metabolism and cytochrome P450 is
important in the conversion of androgens to estrogens. P2rx7,
Olfr464, Lrp11, Prkar2b, Lgr6, and Adipoq are involved in cellular
signal transduction and P2rx7, Prkar2b, and Hdac1 are involved in
regulation of the cell cycle and cell proliferation, while Prkar2b,
Hdac1, and Dock7 are important in cell development. lncRNA
A-30-P01019163 and P2rx7 were differentially expressed in ovarian
tissues of CRYBB2 knockout mice.
Crybb2 not only functions to maintain lens
transparency and refractive index, but also affects ovary
development in mice. At the gene level, Crybb2 may mediate
Ca2+ signal transduction (21,22).
The current study demonstrated that Crybb2 also regulates
expression of lncRNAs in ovarian tissues, which may be a novel area
of research on Crybb2-regulation of gene expression. Previous
studies using large scale cDNA library sequencing and next
generation sequencing demonstrated abundant lncRNAs in mammals,
however, not all lncRNAs are functional and only a relatively small
proportion have been demonstrated to be biologically relevant
(11,23). For example, as of mid-2014, 197
lncRNAs have been functionally annotated in lncRNA databases
(24). However, other lncRNAs may
be translated into proteins (25).
In addition, lncRNAs, unlike miRNAs, can down- or upregulate gene
expression by targeting transcriptional activators or repressors
(10), in addition to
post-transcriptional regulation of protein expression.
In the current study differentially expressed
lncRNAs in ovary tissues of CRYBB2 knockout mice were
profiled and a total of 157 differentially expressed lncRNAs were
observed. Furthermore, differentially expressed mRNAs were profiled
and a total of 1,085 differentially expressed mRNAs in ovary
tissues of CRYBB2 knockout mice were observed. GO and KEGG
pathway analyses were performed to determine associations between
these lncRNAs and mRNAs, and a number of these were subsequently
verified using RT-qPCR. It was observed that lncRNA A-30-P01019163
and P2rx7 were significantly differentially expressed between10
cases of ovary tissues from WT and CRYBB2 knockout mice.
Thus, CRYBB2 knockout could downregulate expression of
lncRNA A-30-P01019163 and, subsequently, suppress expression of the
downstream gene P2rx7 and affect ovarian cell signal transduction,
cell cycle, and ultimately ovarian development.
The current study is a proof-of-principle study and
there are certain limitations. For example, it was not confirmed
how lncRNA A-30-P01019163 regulates P2rx7 expression and the
mechanistic investigation into how lncRNA A-30-P01019163-regulated
P2rx7 expression mediates the effects of CRYBB2 on ovary
development could be expanded on.
In conclusion, CRYBB2 regulates expression of
different lncRNAs to influence ovary development. lncRNA
A-30-P01019163 may affect ovarian cell cycle and proliferation by
regulating P2rx7 expression in the ovary.
Acknowledgements
The authors would like to thank Shanghai Sensichip
Infotech Co., Ltd. (Shanghai, China) for assistance with
bioinformatic analysis and Medjaden Bioscience Ltd. for assisting
in the preparation of the manuscript. The present study was
supported in part by grants from the National Natural Science
Foundation of China (grant nos. 81170834 and 81300748).
References
|
1
|
Zhang J, Li J, Huang C, Xue L, Peng Y, Fu
Q, Gao L, Zhang J and Li W: Targeted knockout of the mouse
betaB2-crystallin gene (Crybb2) induces age-related cataract.
Invest Ophthalmol Vis Sci. 49:5476–5483. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ganguly K, Favor J, Neuhüuser-Klaus A,
Sandulache R, Puk O, Beckers J, Horsch M, Schädler S, Weisenhorn D
Vogt, Wurst W and Graw J: Novel allele of crybb2 in the mouse and
its expression in the brain. Invest Ophthalmol Vis Sci.
49:1533–1541. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liedtke T, Schwamborn JC, Schröer U and
Thanos S: Elongation of axons during regeneration involves retinal
crystallin beta b2 (crybb2). Mol Cell Proteomics. 6:895–907. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Magabo KS, Horwitz J, Piatigorsky J and
Kantorow M: Expression of betaB(2)-crystallin mRNA and protein in
retina, brain, and testis. Invest Ophthalmol Vis Sci. 41:3056–3060.
2000.PubMed/NCBI
|
|
5
|
Duprey KM, Robinson KM, Wang Y, Taube JR
and Duncan MK: Subfertility in mice harboring a mutation in
betaB2-crystallin. Mol Vis. 13:366–373. 2007.PubMed/NCBI
|
|
6
|
Gao Q, Sun LL, Xiang FF, Gao L, Jia Y,
Zhang JR, Tao HB, Zhang JJ and Li WJ: Crybb2 deficiency impairs
fertility in female mice. Biochem Biophys Res Commun. 453:37–42.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao Q, Yang XY, Gu ML, Zhu RR, Zhang JJ
and Li WJ: Effect of beta-B2 crystallin on ovary development and
estrous cycle of mouse. Reprod Contracept. 33:217–223. 2013.
|
|
8
|
Hangauer MJ, Vaughn IW and McManus MT:
Pervasive transcription of the human genome produces thousands of
previously unidentified long intergenic noncoding RNAs. PLoS Genet.
9:e10035692013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Faghihi MA, Modarresi F, Khalil AM, Wood
DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ and
Wahlestedt C: Expression of a noncoding RNA is elevated in
Alzheimer's disease and drives rapid feed-forward regulation of
beta-secretase. Nat Med. 14:723–730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goodrich JA and Kugel JF: Non-coding-RNA
regulators of RNA polymerase II transcription. Nat Rev Mol Cell
Biol. 7:612–616. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Reinius B, Shi C, Hengshuo L, Sandhu KS,
Radomska KJ, Rosen GD, Lu L, Kullander K, Williams RW and Jazin E:
Female-biased expression of long non-coding RNAs in domains that
escape X-inactivation in mouse. BMC Genomics. 11:6142010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Niinuma T, Suzuki H, Nojima M, Nosho K,
Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki
Y, et al: Upregulation of miR-196a and HOTAIR drive malignant
character in gastrointestinal stromal tumors. Cancer Res.
72:1126–1136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu
TP, Yin L, Zhang EB, De W and Shu YQ: Long non-coding RNA ANRIL is
upregulated in hepatocellular carcinoma and regulates cell
apoptosis by epigenetic silencing of KLF2. J Hematol Oncol.
8:502015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kiefer JC: Epigenetics in development. Dev
Dyn. 236:1144–1156. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee JT: Gracefully ageing at 50,
X-chromosome inactivation becomes a paradigm for RNA and chromatin
control. Nat Rev Mol Cell Biol. 12:815–826. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Huang CG, Li WJ, Weng W and Wang
JQ: Establishment of a βB2crystallin gene knockout mice model. Acad
J Sec Mil Med Univ. 27:1246–1248. 2006.
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jobby MK and Sharma Y: Calcium-binding to
lens betaB2- and betaA3-crystallins suggests that all
beta-crystallins are calcium-binding proteins. FEBS J.
274:4135–4147. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kang D, Hur CG, Park JY, Han J and Hong
SG: Acetylcholine increases Ca2+ influx by activation of CaMKII in
mouse oocytes. Biochem Biophys Res Commun. 360:476–482. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dinger ME, Amaral PP, Mercer TR and
Mattick JS: Pervasive transcription of the eukaryotic genome:
Functional indices and conceptual implications. Brief Funct Genomic
Proteomic. 8:407–423. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Amaral PP, Clark MB, Gascoigne DK, Dinger
ME and Mattick JS: lncRNAdb: A reference database for long
noncoding RNAs. Nucleic Acids Res. 39:(Database Issue). D146–D151.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Smith JE, Alvarez-Dominguez JR, Kline N,
Huynh NJ, Geisler S, Hu W, Coller J and Baker KE: Translation of
small open reading frames within unannotated RNA transcripts in
Saccharomyces cerevisiae. Cell Rep. 7:1858–1866. 2014. View Article : Google Scholar : PubMed/NCBI
|