Introduction
Innate immunity serves as the first line of defense
against microbial pathogens (1).
Toll-like receptors (TLRs) are essential sensor molecules of the
host innate immune system. They detect various pathogen-associated
molecular patterns and initiate innate immune responses via
distinct signaling pathways (2).
TLR9 is a member of the TLR family (3). It recognizes specific bacterial DNA
sequences containing unmethylated cytidine-phosphate-guanosine
(CpG) motifs, leading to activation of downstream signaling
pathways, including nuclear factor-κB (NF-κB) and mitogen-activated
protein kinase (MAPK). Activation of these signaling pathways
induces the production of type I interferon (IFN) and other
cytokines that influence the immune response against infectious
agents (4,5). Previous studies have demonstrated
that synthetic phosphorothioate oligodeoxynucleotides (ODN) bearing
unmethylated CpG motifs may mimic the TLR9-dependent
immune-stimulatory effects of bacterial DNA (6,7).
These studies permitted development of the
structure-immunostimulatory activity relationship of synthetic CpG
ODN. However, the features of natural bacterial sequences with
immunostimulatory potential remain to be elucidated.
Brucella species are gram-negative,
facultative intracellular bacteria (8). A total of 4 species of the genus
Brucella are pathogenic for humans, namely B.
melitensis, B. abortus, B. suis and B.
canis. B. melitensis is considered to be the most
pathogenic (9). Brucella is
able to survive and replicate inside host macrophages and dendritic
cells (10). In recent years it
has become apparent that Brucella DNA has an essential role
in the triggering of the innate immune system (11). Immunostimulatory effects of
Brucella DNA are exerted via TLR9 (12,13).
It is well-known that repetitive extragenic
palindromes (REPs) are small (20–40 bp) palindromic repeats,
present in high copies in gram-negative bacteria. They are
important for regulation of certain bacterial functions, including
DNA supercoiling, transcription termination and mRNA stabilization
(14). A previous study
demonstrated that REP sequences are natural TLR9 ligands (15). Studies with REP sequences have
determined the structural features of bacterial immunostimulatory
DNA sequences (15). To the best
of our knowledge, little work has been carried out concerning
Brucella REP candidates as natural TLR9 ligands.
The aim of the present study was to identify
Brucella REP sequences that have an important role in IFN-α
responses in macrophages by binding to TLR9. The interaction
between TLR9 and REPs was also investigated. Furthermore, it was
examined whether NF-κB, c-Jun N-terminal kinase (JNK),
extracellular signal-regulated kinase (ERK) 1/2 and p38 were
involved in TLR9 signaling stimulated by Brucella REPs in
macrophages. These data may be critical to the understanding of how
Brucella activates immune responses.
Materials and methods
Identification and synthesis of
REPs
The present study used a BLAST-based strategy
specifically designed to detect REPs as described previously
(15). The genome analyzed was
B. melitensis NI (chromosome 1: Genbank CP002931.1, RefSeq
NC_017248.1; chromosome 2: Genbank CP002932.1, RefSeq NC_017283.1).
The studied REPs (Table I) were
selected from the REPs identified in B. melitensis based on
the presence of self-complementary sequences and CpG motifs.
Purified phosphorothioate ODNs representing the selected REPs were
synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). For
methylated ODN synthesis, 5-methylcytidine was used (Sangon Biotech
Co., Ltd.).
 | Table I.REP and control sequences. |
Table I.
REP and control sequences.
| Study
designation | Name | Sequence, 5′→3′ |
|---|
| ODN 1 | B. melitensis
REP |
AGCGCAGTGATCCGCACCGCGCT |
| ODN 2 | B. melitensis
REPmethylCpG |
AGmCGCAGTGATCmCGCACmCGmCGCT |
| ODN 3 | B. melitensis
REP non-CpG |
AGGCCAGTGATCAGCACAGGCCT |
| Positive control | ODN2216 |
ggGGGACGATCGTCgggggg |
| Negative control |
ODN2216methylCpG |
ggGGGAGCATGCTAgggggg |
Cell culture
Murine RAW264.7 macrophages (Cell Resource Center,
IBMS, CAMS/PUMC, Beijing, China) were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum and 1%
penicillin/streptomycin at 37°C in a 5% CO2 atmosphere.
Following two passages, cells were seeded into 24-well plates at a
density of 2.0×106 cells/ml.
Treatment of macrophages with
ODNs
ODNs were diluted in Opti-MEM (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with lipofectin (10 µg/ml;
Invitrogen; Thermo Fisher Scientific, Inc.). The mixture was
incubated at room temperature for 5 min and then added to each well
of the 24-well plate. Culture supernatants were collected at 24 h.
A class stimulatory CpG ODN2216 (Sangon Biotech Co., Ltd.) was used
in the present study as a positive control. C-methylated ODN2216
(Sangon Biotech Co., Ltd.) was used as a negative control.
Transfection of macrophages with
anti-TLR9 small interfering RNA (siRNA)
The siRNA sequence used for silencing of murine TLR9
and the negative control siRNA sequence (scrambled siRNA) were
synthesized by Invitrogen (Thermo Fisher Scientific, Inc.).
Transfection compounds were prepared by adding siRNAs to 95 µl
Opti-MEM and 5 µl HiPerFect (Qiagen GmbH, Hilden, Germany) to give
a final concentration of 100 nM. The prepared transfection
compounds were placed at room temperature for 10 min. RAW264.7
macrophages were then transfected with these transfection mixes in
24-well plates. A total of 48 h after transfection, the silencing
effect of siRNA was detected by western blotting. RAW264.7 cells
were then stimulated with ODN1 for 24 h for use in subsequent
experiments.
Animals and ODN treatment
Female Wistar rats aged 6–8 weeks were purchased
from Vital River Lab Animal Technology Co., Ltd (Beijing, China).
Rats were housed in temperature (20±2°C) and humidity (55% ± 5)
controlled rooms, with a 12-h light/dark cycle. Animal studies were
performed under conditions approved by the Local Animal Care and
Use Committee. Rats were injected subcutaneously with 20 mg/kg
ODNs. Serum was collected for ELISA by retro-orbital bleeding 2 h
after injection.
Cytokine ELISA
Concentrations of cytokines in the culture
supernatants or serum were measured using ELISA kits according to
the manufacturer's protocol (R&D Systems Inc., Minneapolis, MN,
USA). Samples were tested, in duplicate, a total of three
times.
Reverse transcription-polymerase chain
reaction (RT-PCR) of RNA expression
Total RNA was isolated from RAW264.7 cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RNA was reverse
transcribed into complementary DNA using 10 µl PrimeScript RT
Enzyme Mix I (Takara Biotechnology Co., Ltd., Dalian, China) under
the following conditions: 37°C for 15 min, followed by 85°C for 5
sec. One-step semiquantitative RT-PCR (Takara Biotechnology Co.,
Ltd.) was performed at 95°C for 30 sec, followed by 40 cycles of
95°C for 5 sec and 60°C for 30 sec. Melting curve analysis of
amplification products was performed at the conclusion of each PCR.
GAPDH served as an internal control. The sequences of primers were
as follows: TLR9 forward, 5′-TTGGTCGCACCTCCAACAGT-3′ and reverse,
5′-TGGGCCCATTGTGATGAAC-3′; GAPDH forward, 5′-TCAACGGCACAGTCAAGG-3′
and reverse, 5′-ACTCCACGACATACTCAGC-3′ (Sangon Biotech Co.
Ltd.).
Western blot analysis
RAW264.7 cells treated with ODNs were harvested and
treated with radioimmunoprecipitation assay buffer (BioVision,
Inc., Milpitas, CA, USA) supplemented with protease inhibitor
cocktail (cOmplete™ Mini protease inhibitors; Roche Diagnostics,
Basel, Switzerland) for 15 min at 4°C. The cell lysates (20 µg of
protein) were subjected to 10% SDS-PAGE and then transferred onto a
nitrocellulose membrane (Sangon Biotech Co., Ltd.). The membrane
was blocked with 5% nonfat milk in TBST (Triton X-100 0.5% and
Tris-buffered saline), and incubated with primary antibodies,
including rabbit anti-mouse phosphorylated (p)-inhibitor of κB
(IκB)α monoclonal antibody (dilution, 1:1,000; catalog no., #2859),
rabbit anti-mouse IκBα polyclonal antibody (dilution, 1:1,000;
catalog no., #9242), rabbit anti-mouse p-NF-κB polyclonal antibody
(dilution, 1:1,000; catalog no., #3031), rabbit anti-mouse NF-κB
monoclonal antibody (dilution, 1:1,000; catalog no., #8242), rabbit
anti-mouse p-JNK polyclonal antibody (dilution, 1:1,000; catalog
no., #9251), rabbit anti-mouse JNK polyclonal antibody (dilution,
1:1,000; catalog no., #9252), rabbit anti-mouse p-ERK1/2 polyclonal
antibody (dilution, 1:1,000; catalog no., #9101), rabbit anti-mouse
ERK1/2 polyclonal antibody (dilution, 1:1,000; catalog no., #9102),
rabbit anti-mouse p-p38 polyclonal antibody (dilution, 1:1,000;
catalog no., #9211), rabbit anti-mouse p38 polyclonal antibody
(dilution, 1:1,000; catalog no., #9212), rabbit anti-mouse
α-tubulin polyclonal antibody (dilution, 1:1,000; catalog no.,
#2144; all from Cell Signaling Technology, Inc., Danvers, MA, USA),
and rabbit anti-mouse TLR9 polyclonal antibody (dilution, 1:500;
catalog no., #TA306291; OriGene Technologies, Rockville, MD, USA),
followed by a horseradish peroxidase-conjugated goat anti-rabbit
IgG secondary antibody (dilution, 1:5,000; catalog no., #D110058;
Sangon Biotech Co., Ltd.). Immunoreactivity was visualized by
enhanced chemiluminescence (ECL kit; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) with the ChemiDoc™ XRS+ system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Molecular modeling
The crystal structure of mouse TLR9 was obtained
from the Protein Data Bank (PDB Code: 3WPG) (16). The CpG ODN structure was built
using Biopolymer tools in Discovery Studio version 2.5 (Accelrys,
San Diego, CA, USA). The program Hex 6.1 (hex.loria.fr/dist61) was
used to perform a rigid-body docking search as previously described
(17).
Statistical analysis
Multiple groups were compared using one-way analysis
of variance, followed by the Fisher's least significant difference
test. All data were expressed as the mean ± standard deviation and
P<0.05 was considered to indicate a statistically significant
difference.
Results
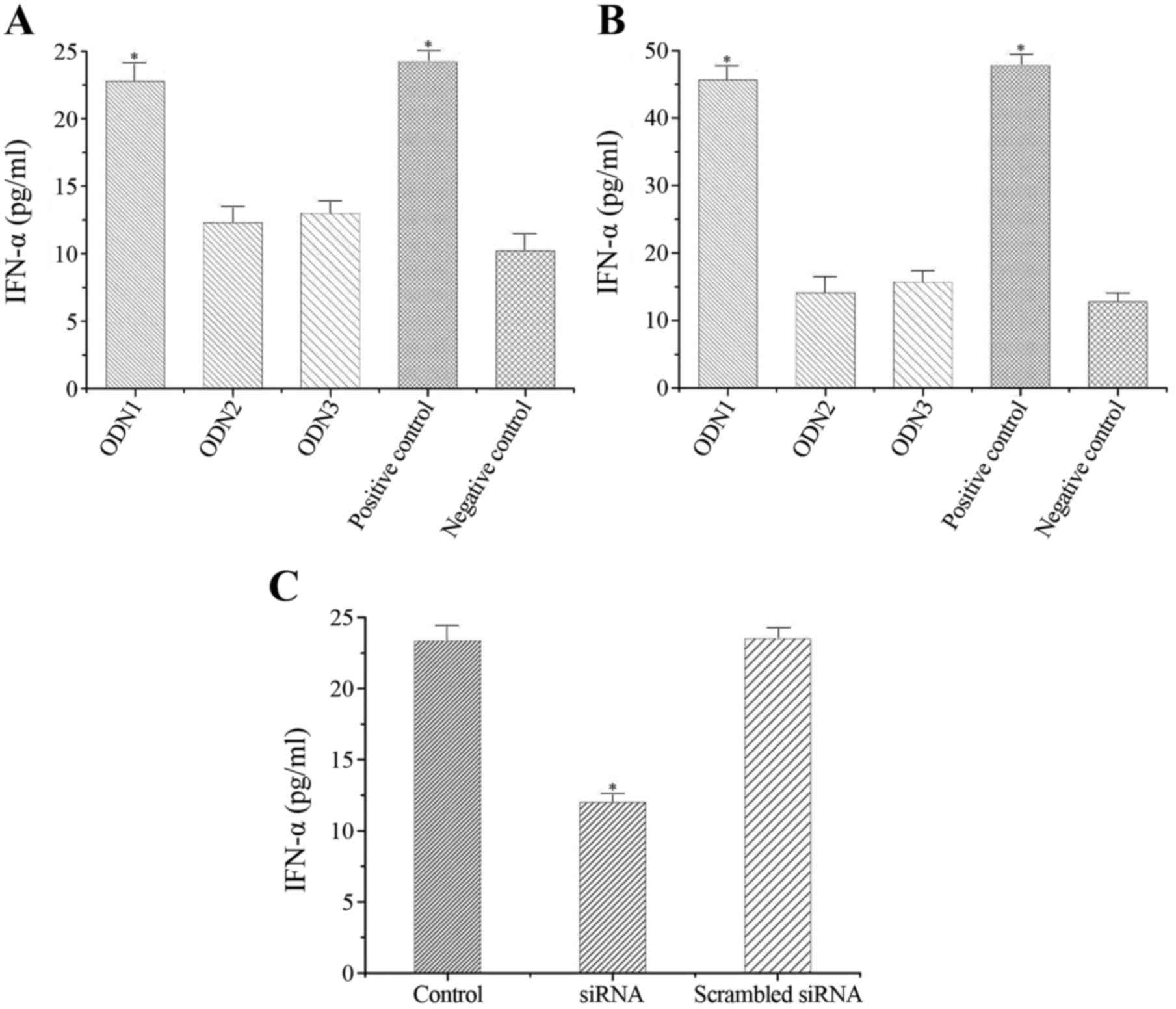
ODN1 induces IFN-α production in
vitro
The effects of ODNs on IFN-α secretion by
macrophages in vitro were determined (Fig. 1). As presented in Fig. 1A, ODN1 significantly enhanced the
release of IFN-α at 24 h (22.706±1.305 pg/ml) as compared to the
negative control (10.032±1.251 pg/ml) (P<0.001). However, ODN2
and ODN3 induced only low levels of IFN-α (10.627±1.237 and
10.831±1.012 pg/ml, respectively) compared with the negative
control group (P=0.589 and P=0.438, respectively; Fig. 1A).
ODN1 induces IFN-α production in
vivo
The effects of ODNs on serum IFN-α levels in
vivo were also determined. Significantly higher serum IFN-α
levels were observed in the ODN1 group (45.839±2.103 pg/ml)
compared with the negative control group (12.901±1.227 pg/ml) at 2
h (P<0.01; Fig. 1B). However,
production of IFN-α remained low in the ODN2 and ODN3 groups
(13.303±2.478 and 14.167±1.221 pg/ml, respectively) compared with
the negative control group (P=0.815 and P=0.274, respectively;
Fig. 1B).
Effects of anti-TLR9 siRNA on IFN-α
response to ODN1 in macrophages
RAW264.7 cells transfected with anti-TLR9 siRNA or
control siRNA were stimulated by ODN1. As presented in Fig. 1C, significantly lower IFN-α levels
were observed in the anti-TLR9 siRNA group (12.253±0.873 pg/ml)
compared to the untransfected (23.819±1.012 pg/ml) and control
siRNA-transfected (23.107±1.238 pg/ml) groups (P<0.001).
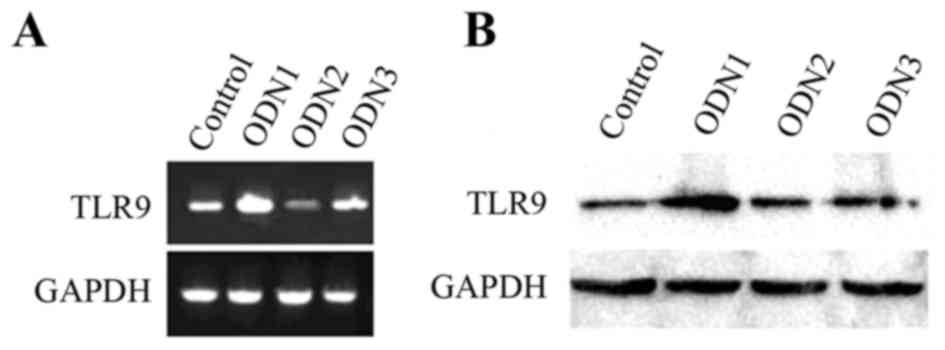
Effects of ODN1 on TLR9
expression
As presented in Fig.
2A, ODN1 enhanced the mRNA expression of TLR9. In addition, the
protein expression levels of TLR9 were increased in the ODN1 group
(Fig. 2B). However, TLR9 mRNA and
protein expression levels were not increased following stimulation
with ODN2 and ODN3 (Fig. 2B).
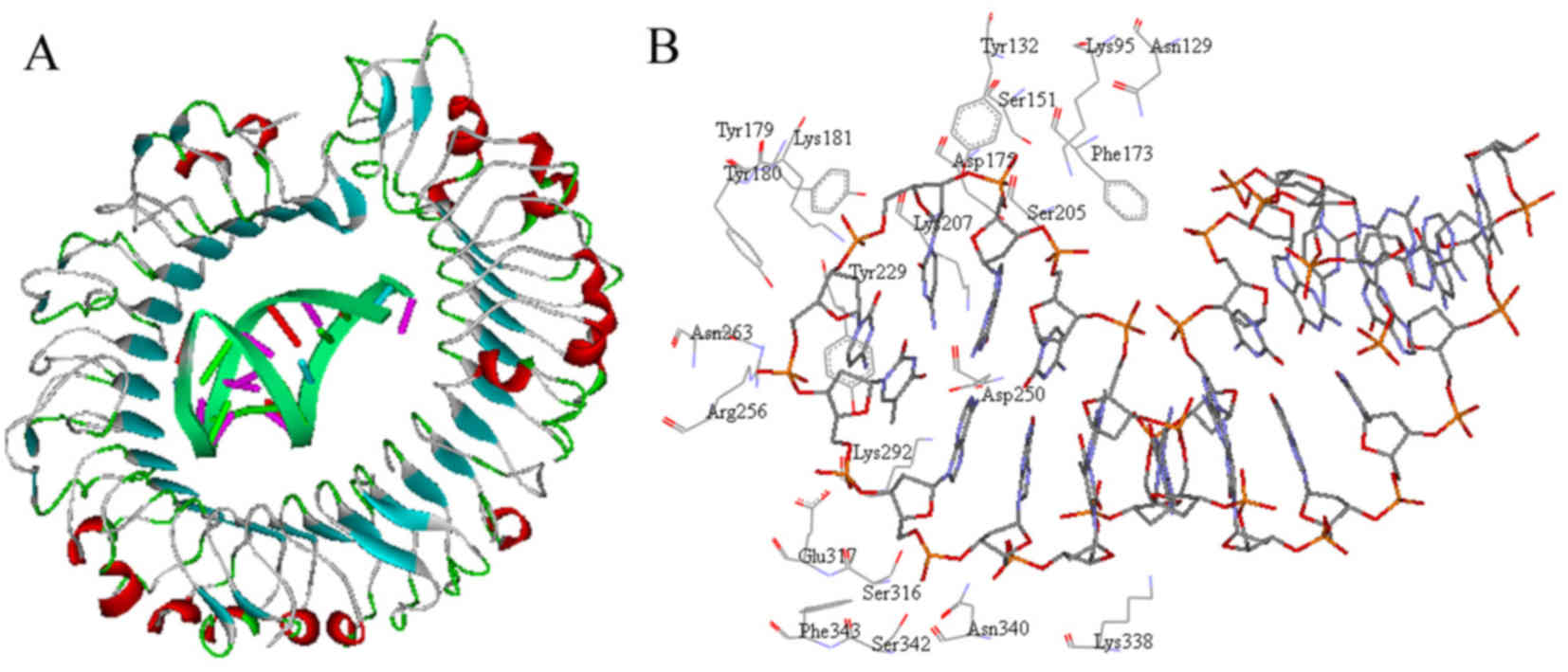
TLR9-ODN1 interaction analysis
The interaction pattern demonstrated that the
interface between TLR9 and ODN1 was geometrically complementary
(Fig. 3A). Analysis revealed that
>20 residues in TLR9 constituted the active site where ODN1
binds (Fig. 3B). Most of these
amino acid residues are basic and have an essential role in
TLR9-ODN1 interaction, including Lys95, Lys181, Lys207, Arg256,
Lys292 and Lys338.
Effects of ODN1 on the IκB/NF-κB
signaling pathway
To investigate whether the IκB/NF-κB signaling
pathway in RAW264.7 cells was affected by ODN1, the phosphorylation
levels of NF-κB and IκB-α in ODN1-treated macrophage cultures were
assessed by western blot analysis. As presented in Fig. 4A, the protein expression of p-NF-κB
p65 was upregulated following 60 and 120 min stimulation with ODN1.
However, the expression of p-IκB-α was reduced (Fig. 4A).
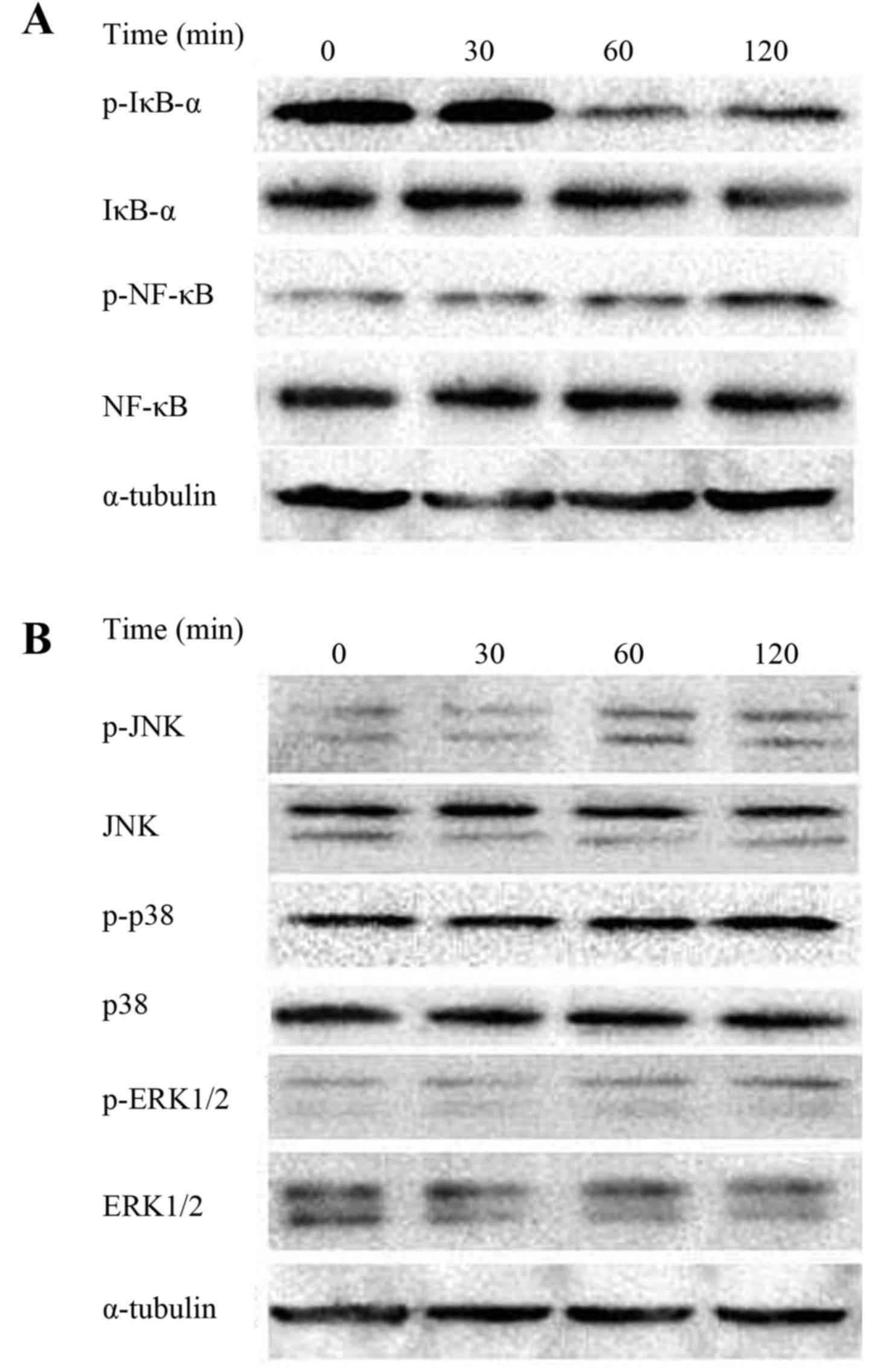 | Figure 4.ODN 1-induced IκB/NF-κB and MAPK
signaling pathways in macrophages. (A) The protein expression
levels of p-IκB-a, IκB-a, p-NF-κB and NF-κB. (B) The protein
expression levels of p-JNK, JNK, p-p38, p38, p-ERK1/2 and ERK1/2.
Increased phosphorylation of JNK, p38 and ERK1/2 was observed.
α-tubulin served as the loading control. ODN, oligodeoxynucleotide;
IκB, inhibitor of κB; NF-κB, nuclear factor-κB; MAPK,
mitogen-activated protein kinase; p, phosphorylated; JNK, c-Jun
N-terminal kinase; ERK, extracellular signal-regulated kinase. |
Effects of ODN1 on the MAPK signaling
pathway
The expression of p-JNK, p-p38 and p-ERK1/2 in
ODN1-stimulated RAW264.7 cells was also assessed. As presented in
Fig. 4B, an increased
phosphorylation of JNK was induced by ODN1 treatment. Furthermore,
phosphorylation of p38 and ERK1/2 was observed. The levels of p-p38
and p-ERK1/2 were further increased at 120 min in ODN1-treated
macrophages.
Discussion
REP sequences, which are palindromic and rich in
unmethylated CpG motifs, are important for the immune recognition
of bacteria (18). However,
whether REP sequences derived from the Brucella genome exert
immunostimulatory effects remains to be elucidated.
In the present study, synthetic ODNs with a natural
phosphodiester backbone were utilized to mimic natural REPs. ODNs
representing Brucella REP sequences were tested for their
ability to stimulate production of the cytokine IFN-α in
vitro and in vivo. It was observed that ODN1 with
unmethylated CpG motifs had immune stimulatory ability. However,
induction of IFN-α by ODN1 could be abrogated when ODN1 was
modified into non-CpG sequences or C-methylated variations. The
results of the present study confirmed that the immune stimulatory
ability of Brucella REP is CpG dependent.
It is well-established that CpG ODNs may be
recognized by TLR9 (19). The
effects of ODN1 on the TLR9 signaling pathway were thus analyzed in
the present study. It was observed that ODN1 significantly enhanced
the expression of TLR9 at mRNA and protein levels. In addition,
knockdown of TLR9 expression by siRNA in macrophages led to a
reduction of IFN-α production following REP stimulation. These
results support the notion that, similar to other CpG ODNs,
Brucella REP is able to activate macrophages through TLR9,
consequently upregulating IFN-α.
To improve our understanding of the mechanism of
TLR9-Brucella REP recognition, the interacting features
between TLR9 and ODN1 were examined using molecular modeling
approaches. The interaction pattern demonstrated that >20
residues in TLR9 were important for ligand recognition, and these
residues established direct contact with ODN1. This is in agreement
with the results of previous studies (16,17).
Furthermore, the role of NF-κB and MAPK signaling
pathways in Brucella REP-mediated innate immune responses
was determined. The present results revealed that the levels of the
NF-κB p65 phosphorylation were increased in macrophages stimulated
with ODN1. A simultaneous decrease in the phosphorylation of IκB-α
was also observed. In addition, the phosphorylation levels of p38,
ERK1/2 and JNK were increased in ODN1-treated cells. These data
suggest that the activation of the NF-κB and MAPK signaling
pathways may in part contribute to the immunological response
induced by Brucella REP.
In conclusion, the present study suggests that TLR9
recognizes and responds to Brucella REP, leading to the
activation of downstream signaling pathways, including NF-κB and
MAPK, which then induce IFN-α biosynthesis. The finding that
Brucella REPs are natural TLR9 agonists may be useful for
the development of novel therapeutic applications.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no., 81460049), the
Natural Science Foundation of Inner Mongolia Autonomous Region of
China (grant nos., 2014JQ04 and 2015MS0810) and the Program for
Young Talents of Science and Technology in Universities of Inner
Mongolia Autonomous Region of China (grant no., NJYT-14-A13).
References
|
1
|
Hoffmann J and Akira S: Innate immunity.
Curr Opin Immunol. 25:1–3. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
O'Neill LA, Golenbock D and Bowie AG: The
history of Toll-like receptors-redefining innate immunity. Nat Rev
Immunol. 13:453–460. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Riad A, Westermann D, Escher F, Becher PM,
Savvatis K, Lettau O, Heimesaat MM, Bereswill S, Volk HD,
Schultheiss HP and Tschöpe C: Myeloid differentiation factor-88
contributes to TLR9-mediated modulation of acute coxsackievirus
B3-induced myocarditis in vivo. Am J Physiol Heart Circ Physiol.
298:H2024–H2031. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bauer S, Kirschning CJ, Häcker H, Redecke
V, Hausmann S, Akira S, Wagner H and Lipford GB: Human TLR9 confers
responsiveness to bacterial DNA via species-specific CpG motif
recognition. Proc Natl Acad Sci USA. 98:9237–9242. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El Kebir D, József L, Pan W, Wang L and
Filep JG: Bacterial DNA activates endothelial cells and promotes
neutrophil adherence through TLR9 signaling. J Immunol.
182:4386–4394. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jurk M, Kritzler A, Debelak H, Vollmer J,
Krieg AM and Uhlmann E: Structure-activity relationship studies on
the immune stimulatory effects of base-modified CpG toll-like
receptor 9 agonists. ChemMedChem. 1:1007–1014. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu D, Putta MR, Bhagat L, Li Y, Zhu F,
Wang D, Tang JX, Kandimalla ER and Agrawal S: Agonists of Toll-like
receptor 9 containing synthetic dinucleotide motifs. J Med Chem.
50:6411–6418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moreno E, Cloeckaert A and Moriyón I:
Brucella evolution and taxonomy. Vet Microbiol. 90:209–227. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Galińska EM and Zagórski J: Brucellosis in
humans-etiology, diagnostics, clinical forms. Ann Agric Environ
Med. 20:233–238. 2013.PubMed/NCBI
|
|
10
|
Deghelt M, Mullier C, Sternon JF, Francis
N, Laloux G, Dotreppe D, Van der Henst C, Jacobs-Wagner C, Letesson
JJ and De Bolle X: G1-arrested newborn cells are the predominant
infectious form of the pathogen Brucella abortus. Nat Commun.
5:43662014.PubMed/NCBI
|
|
11
|
Campos PC, Gomes MT, Guimarães G, Franco
MM Costa, Marim FM and Oliveira SC: Brucella abortus DNA is a major
bacterial agonist to activate the host innate immune system.
Microbes Infect. 16:979–984. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gomes MT, Campos PC, de Almeida LA,
Oliveira FS, Costa MM, Marim FM, Pereira GS and Oliveira SC: The
role of innate immune signals in immunity to Brucella abortus.
Front Cell Infect Microbiol. 2:1302012.PubMed/NCBI
|
|
13
|
Huang LY, Ishii KJ, Akira S, Aliberti J
and Golding B: Th1-like cytokine induction by heat-killed Brucella
abortus is dependent on triggering of TLR9. J Immunol.
175:3964–3970. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Messing SA, Ton-Hoang B, Hickman AB,
McCubbin AJ, Peaslee GF, Ghirlando R, Chandler M and Dyda F: The
processing of repetitive extragenic palindromes: The structure of a
repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res. 40:9964–9979. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Magnusson M, Tobes R, Sancho J and Pareja
E: Cutting edge: Natural DNA repetitive extragenic sequences from
gram-negative pathogens strongly stimulate TLR9. J Immunol.
179:31–35. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ohto U, Shibata T, Tanji H, Ishida H,
Krayukhina E, Uchiyama S, Miyake K and Shimizu T: Structural basis
of CpG and inhibitory DNA recognition by Toll-like receptor 9.
Nature. 520:702–705. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou W, Li Y, Pan X, Gao Y, Li B, Qiu Z,
Liang L, Zhou H and Yue J: Toll-like receptor 9 interaction with
CpG ODN-an in silico analysis approach. Theor Biol Med Model.
10:182013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nunvar J, Huckova T and Licha I:
Identification and characterization of repetitive extragenic
palindromes (REP)-associated tyrosine transposases: Implications
for REP evolution and dynamics in bacterial genomes. BMC Genomics.
11:442010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Suwarti S, Yamazaki T, Svetlana C and
Hanagata N: Recognition of CpG oligodeoxynucleotides by human
Toll-like receptor 9 and subsequent cytokine induction. Biochem
Biophys Res Commun. 430:1234–1239. 2013. View Article : Google Scholar : PubMed/NCBI
|