Introduction
The abnormal proliferation of vascular smooth muscle
cells (VSMCs) is critical in several types of vascular disorders,
including essential hypertension and atherosclerosis (1). The remodeled structure of the artery,
with increased media and a smaller lumen, leads to cardiovascular
complications, which are the predominant causes of
hypertension-associated mortality (1). A number of regulators have been
reported to be involved in the control of proliferation of VSMCs
(1,2). Investigations in this field are
increasingly focused on the function of microRNAs (miRNAs) in VSMCs
proliferation.
miRNAs are comprised of ~22 nucleotides, and are
considered to be involved in the post-transcriptional regulation of
mRNAs of its target genes by binding to sequences in the
3′-untranslated regions (3′UTRs) in the target mRNAs, leading to
accelerated degradation of the target mRNA or repressed
translation, and thereby causing a reduction in protein synthesis
(3,4). It has been reported that miRNAs are
involved in several human pathophysiological and disease processes
by controlling biological processes (4). Studies have demonstrated that miRNAs,
including miRNA (miR)-143/145 (5–9),
miR-221/222 (10,11) and miR-21 (12), are crucial in modulating the
proliferation of VSMCs during the process of atherosclerosis and
vascular injury. However, whether miRNAs are involved in modulating
the proliferation of VSMCs in hypertension remains to be fully
elucidated.
Accumulating evidence has shown that several
specific miRNAs target the mRNAs of multiple genes, and function as
regulators in the differentiation, apoptosis and proliferation of
VSMCs (11,13–15).
For example, miR-145 has been reported to target Krüppel-like
factor 5, which is associated with the proliferation of VSMCs and
has a regulatory effect on the phenotypic modulation of VSMCs
(14). Other miRNAs, including
miR-222 and miR-221, have been identified as novel regulators for
neointimal hyperplasia and VSMC proliferation, by suppressing the
expression of p57 (Kip2) and p27 (Kip1) (11). The findings may provide novel
therapeutic targets to develop promising treatments for numerous
proliferative vascular diseases, including hypertension and
atherosclerosis.
Previously, high-throughput screening has been used
to identify the candidate miRNAs with potential functional
involvement in the pathogenesis of hypertension by comparing the
miRNA expression profiles between Wistar Kyoto (WKY) rats and
spontaneously hypertensive rats (SHRs) (16). Genetically, WKY rats and SHRs are
from the same lineage, however, they have a notable difference in
vascular phenotype (17). As the
proliferation activity is markedly higher in the VSMCs of SHRs,
compared with that of the VSMCs of WKY rats (17), the present study hypothesized that
the different proliferation characteristics between SHR VSMCs and
WKY rat VSMCs may be the major cause of the presence of
hypertension in SHRs and the absence of hypertension in WKY rats.
To confirm this hypothesis, the present study screened miRNAs,
which have been reported to be aberrantly expressed in SHR VSMCs
(16) and performed further
investigations to identify the possible genes associated with these
miRNAs.
Materials and methods
Cell culture
VSMCs were obtained from the medial layer of the
thoracic aorta, which were collected from a total of 36 female SHRs
and WKY rats (10-week-old), purchased from the Animal Centre of
Zhengzhou University (Zhengzhou, China). The rats were maintained
in a 12 h light/dark cycle at 25°C with food and water ad
libitum. All animals were sacrificed using cervical dislocation
The cultures were maintained at 37°C and a humidified atmosphere of
5% CO2 in Dulbecco's modified Eagle's medium (DMEM)
containing 10 U/ml streptomycin (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), 10 U/ml penicillin (Gibco; Thermo Fisher
Scientific, Inc.) and 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA). Cells in passages 3–6
were selected and used for experiments. The study protocol was
approved by the Animal Ethical Committee at Zhengzhou University
(Zhengzhou, China).
Cell culture and transfection
Human smooth muscle cells were obtained from the
Chinese Academy of Sciences Cell Bank (Shanghai, China) and
cultured in DMEM medium with 10% FBS, 100 µg/ml streptomycin and
100 U/ml penicillin in a humidified atmosphere with 5%
CO2 at 37°C, with cells in passages 3–8 used for further
experiments. The inhibitor and mimic of miR-34b, and CDK6 small
interfering (si)RNA were obtained from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China) with the following sequence:
5′-TACTTCTGAAGTGTTTGACATTT-3′. Cell transfection was performed
using HiPerFect transfection reagent (Qiagen China Co., Ltd,
Shanghai, China). The transfection complexes were added to the
culture plates, cells were seeded at a density of
1.0×105, and incubated for 4 h at room temperature,
following which the medium was replaced with fresh medium in
accordance with the manufacturer's protocol.
RNA preparation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was isolated from the cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The Prime
Script reverse transcription reagent kit (Takara Biotechnology Co.,
Ltd., Dalian, China) was used to perform the RT reaction with
0.2–0.5 µg RNA in order to detect miRNAs and mRNA, in accordance
with the manufacturer's protocol. SYBR Premix Ex Taq (cat. no.
DRR041A; Takara Biotechnology Co., Ltd.) was used to perform
quantitative analysis to identify the expression levels of miR-1,
miR-34b, miR-500, miR-98, miR-72, and let-7a, and the mRNA
expression levels of CCNG1 and CDK6, in an Applied Biosystems 7900
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The cDNA was amplified using qPCR using a SYBR green qPCR
kit (Takara Bio, Inc., Otsu, Japan). The DNA polymerase was
purchased from Takara Bio, Inc. The sequences of the primers were
as follows: CDK6 sense, 5′-TGCACAGTGTCACGAACAGA-3′; antisense,
5′-ACCTCGGAGAAGCTGAAACA-3′; GAPDH sense,
5′-GATATTGTTGCCATCAATGAC-3′; U6 sense, 5′-CTCGCTTCGGCAGCACA-3′ and
antisense, 5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH sense,
5′-TGCACCACCAACTGCTTAGC-3′ and antisense,
5′-GGCATGGACTGTGGTCATGAG-3′. The qPCR thermal cycling was performed
as follows: Initial incubated for 15 sec at 95°C, followed by
denaturing in 40 cycles at 95°C for 5 sec and annealing for 31 sec
at 60°C. The 2-ΔΔCq method (18) was applied to analyze the data. U6
was used to normalize the expression levels of mRNA and miRNAs as
an internal control.
Western blot analysis
Cold phosphate-buffered saline (PBS) was used to
wash the VSCMs three times, following which
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Jiangsu, China) supplemented with phosphatase
inhibitor cocktail and protease (Merck Millipore, Darmstadt,
Germany) was used to lyse the cells. A Bradford assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was performed to determine
protein content. Subsequently, equal quantities of protein (30 µg)
were separated using 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and then transferred onto a nitrocellulose membrane
(EMD Millipore, Billerica, MA, USA). Membranes were blocked using
PBS containing 3% bovine serum albumin (Invitrogen; Thermo Fisher
Scientific, Inc.) at room temperature for 60 min. Primary
antibodies were used to incubate the membranes at 4°C overnight
following blocking. The antibodies comprised β-actin at a dilution
of 1:1,000 (Santa Cruz Biotechnology, Inc., Dallas, TX, USA), and
CCHG1 (cat. no. 3978) and CDK6 (cat. no. 13331) antibodies (Cell
Signaling Technology, Inc., Danvers, MA, USA) at a dilution of
1:500. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit
secondary antibodies (cat. no. 7072; 1:1,000; Cell Signaling
Technology, Inc.) were used to incubate the membranes for 1 h at
room temperature. A chemiluminescence system (Cell Signaling
Technology, Inc.) and ImageJ software (National Institutes of
Health, Bethesda, MD, USA) were used to perform detection and
relative density quantification.
Proliferation assay
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Rockville, MD, USA) was used to count cell
numbers according to the manufacturer's protocol. At 48 h
post-transfection, the cells were collected and washed with PBS.
Cells were seeded at a density of 2×106 and CCK-8
reagent (10 µl) was added into each plate, followed by incubation
at room temperature for 4 h. A microplate reader was used to
measure the absorbance at 450–540 nm (Multiskan Spectrum; Thermo
Fisher Scientific, Inc.).
Luciferase reporter assay
The 3′UTRs of CDK6 and CCNG1 were amplified by qPCR
using a SYBR green qPCR kit. The amplification was performed at
98°C for 60 sec, 30 cycles of 98°C for 30 sec, 58°C for 30 sec,
72°C for 2 min and 72°C for 5 min. The PCR products were
subsequently subcloned into a pGL3-basic vector (Invitrogen; Thermo
Fisher Scientific, Inc.) carrying a firefly reporter gene. The
reaction was performed in an 8 μl mixture containing 100 ng pGL3
basic vector, 200 ng PCR products and placed in a 45°C water bath
for 5 min and an ice bath immediately. Subsequently, 1 µl 10xT4
ligase buffer and 1µl 2 mmol/l ATP were added, and incubated at
12°C overnight. The accuracy of the insert sequence was confirmed
using direct Sanger sequencing. Furthermore, mutations were
introduced into the constructs using site-directed mutagenesis
(Stratagene, La Jolla, CA, USA).
The VSMCs were seeded into 24-well plates
(2×105/well) and, when the confluence reached 70–80%,
the cells were cotransfected with wild-type or mutant vectors and
miR-34b mimics (Thermo Fisher Scientific, Inc.). After 48 h, the
cells were assayed for luciferase activity using a Dual-Luciferase
Reporter Assay system (Promega Corporation, Madison, WI, USA)
according to the manufacturer's protocol. The Dual-Luciferase
Reporter Assay system and GloMax 40/40 luminometer (Promega
Corporation) were used to measure firefly luciferase activity
according to the manufacturer's protocol.
Statistical analysis
MiRanda (www.microrna.org), TargetScan (www.targetscan.org) were used in the present study.
All data are expressed as the mean ± standard error of the mean.
SPSS 16.0 (SPSS, Inc, Chicago, IL, USA) was used to perform one-way
analysis of variance or Student's independent t-test to compare the
parameters among groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-34b is differentially expressed in
SHRs
To investigate the molecular mechanism of
hypertension, the present study investigated miRNAs (miR-1, let-7a,
miR-34b, miR-500, miR-98 and miR-72), and their downstream
mediators and signaling pathways, as evidenced by observations that
they are differentially expressed between SHRs and WKY rats. As
shown in Fig. 1, the expression
level of miR-34b in the SHRs was downregulated, compared with that
in the WKY rats (P<0.05; Fig.
1A), whereas the other mRNAs showed comparable levels of
expression in the SHRs and WKY rats.
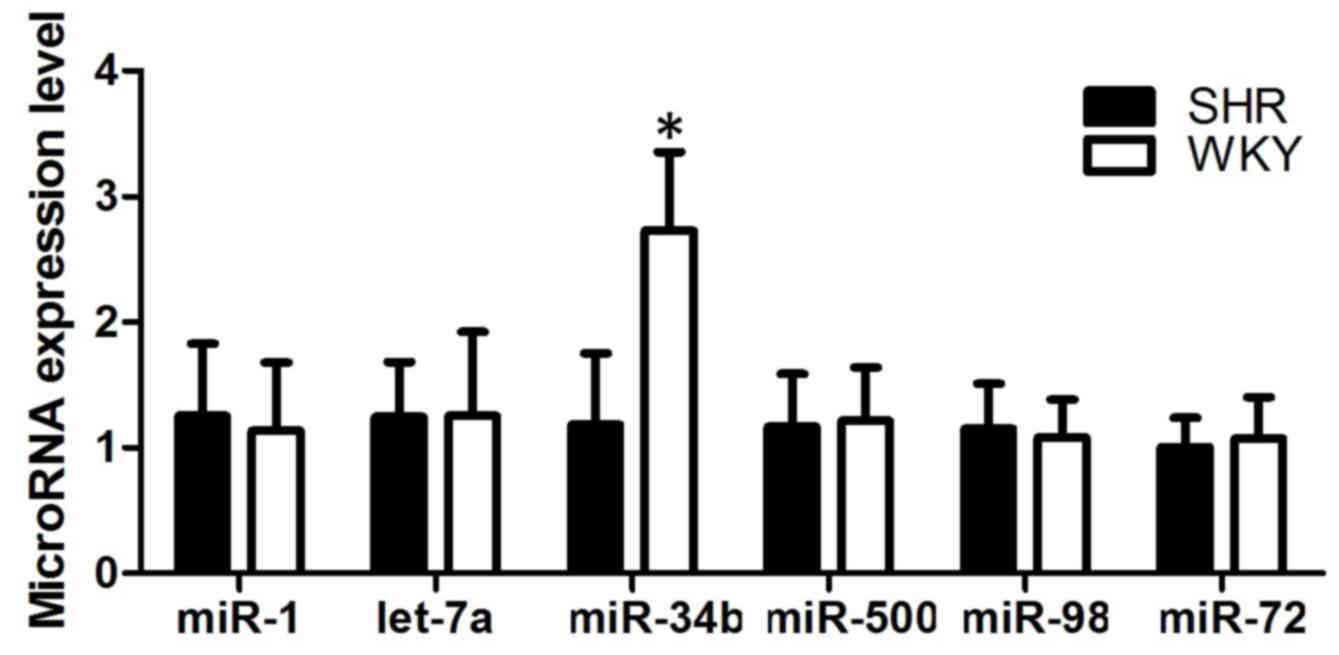 | Figure 1.Expression levels of miR-1, let-7a,
miR-34b, miR-500, miR-98 and miR-72 in SHR and WKY rat cell
samples. Expression of miR-34b was upregulated in the SHRs,
compared with the WKY rats, whereas the other microRNAs showed
comparable expression levels in the SHRs and WKY rats, indicating
miR-34b was involved in hypertensive disease. *P<0.05 vs. WKY
group. miR, microRNA; SHR, spontaneously hypertensive rats; WKY,
Wistar Kyoto. |
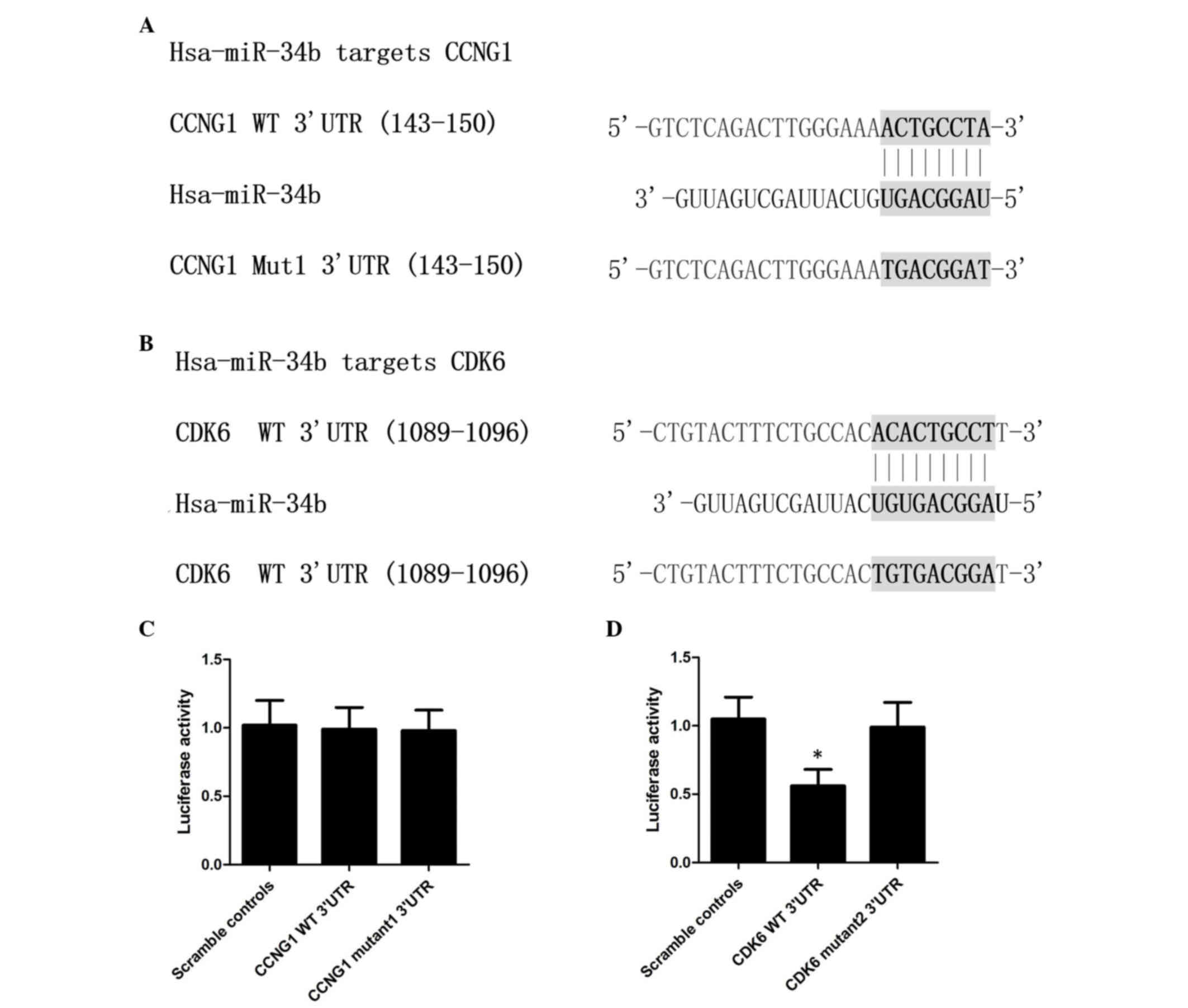
CDK6 is a target of miR-34b
The present study used online miRNA target
prediction tools to predict the candidate target genes of miR-34b
in the database, from which CCNG1 and CDK6 were identified as its
possible target gene with the ‘seed sequence’ in the 3′UTR of CCNG1
(Fig. 2A) or CDK6 (Fig. 2B) separately. To identify the
direct target gene of miR-34b, a luciferase reporter assay was
performed in VSMCs, transfected with wild-type and mutant candidate
target genes, respectively, and compared with scramble controls. As
shown in Fig. 2C, the wild-type
and mutant CCNG1 cells showed similar relative luciferase activity
to that of the scramble control cells, indicating the mutant CCNG1
had no effect on the VSMCs. As shown in Fig. 2D, wild-type CDK6 shows showed lower
relative luciferase activity, compared with the scramble controls
(P<0.05), whereas the relative luciferase activity in the mutant
CDK-6 group was comparable to that of the scramble control,
indicating CDK6 to be the target gene of miR-34b.
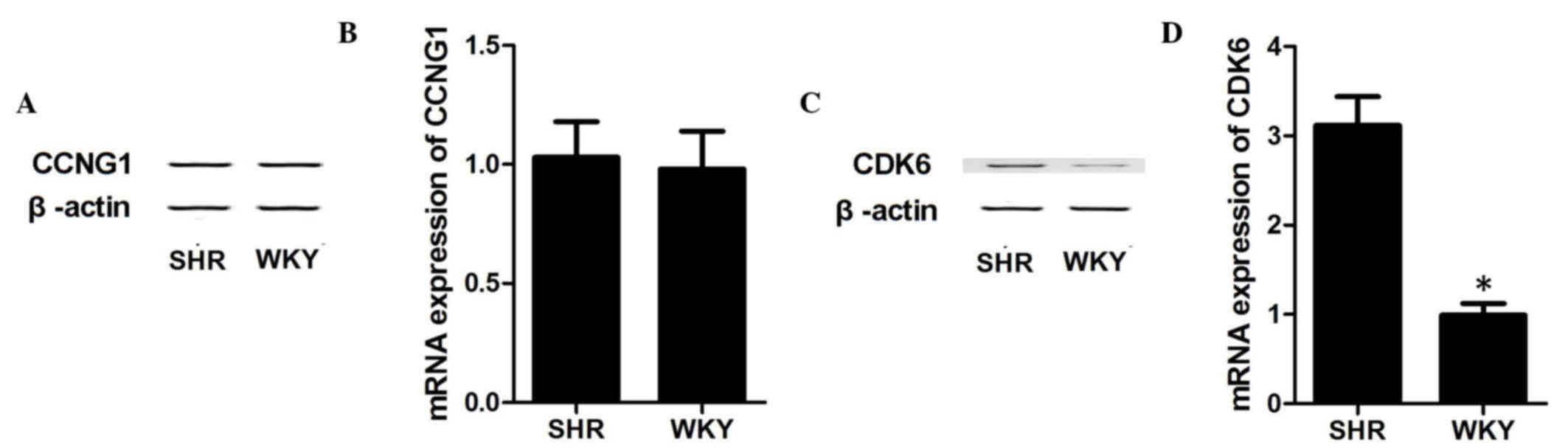
To further confirm CDK6 as the target gene of
miR-34b, the present study investigated the mRNA and protein
expression levels of CDK6 and CCNG1 in VSMCs collected from SHRs
and WKY rats, respectively. As shown in Fig. 3, in the WKY rat VSMCs with
overexpression of miR-34b, the mRNA expression levels of CCNG1
(Fig. 3B) and protein expression
levels of CCNG1 (Fig. 3A) were
similar between the two groups. By contrast, the mRNA (Fig. 3D) and protein (Fig. 3C) levels of CDK6 were downregulated
in the WKY rats, compared with the SHRs (P<0.05), which
confirmed CDK6 as the direct target gene of miR-34b, and indicate a
possible negative regulatory association between miR-34b and
CDK6.
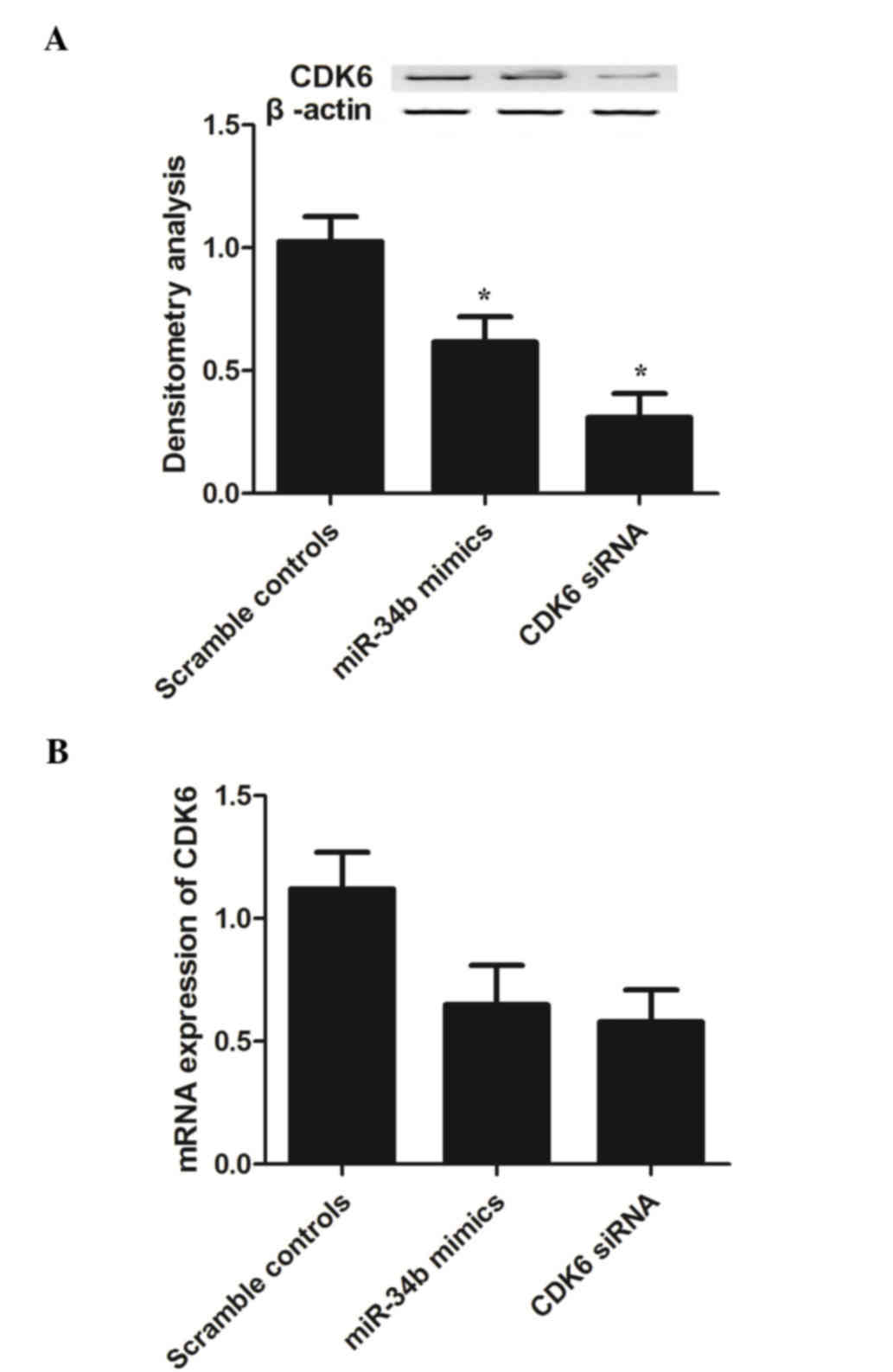
Negative regulatory association
between miR-34b and its target, CDK6
To investigate the signaling pathways between
miR-34b and CDK6, the present study examined the mRNA and protein
expression levels of CDK6 in VSMCs transfected with CDK6 siRNA or
miR-34b mimics. As shown in Fig.
4, the mRNA levels of CDK6 (Fig.
4B) in the VSMCs transfected with the CDK6 siRNA or miR-34b
mimics were comparably lower, compared with the scramble controls,
indicating that the miR-34b mimics inhibited the expression of CDK6
in addition to CDK6 siRNA. The protein band of CDK6 (Fig. 4A; P<0.05) in the miR-34b mimic
group was lower in density, compared with that of the scramble
control group, whereas the band of the CDK6 siRNA group showed
comparable density with that of the miR-34b mimic group, indicating
that the miR-34b mimics exhibited the same effect on the expression
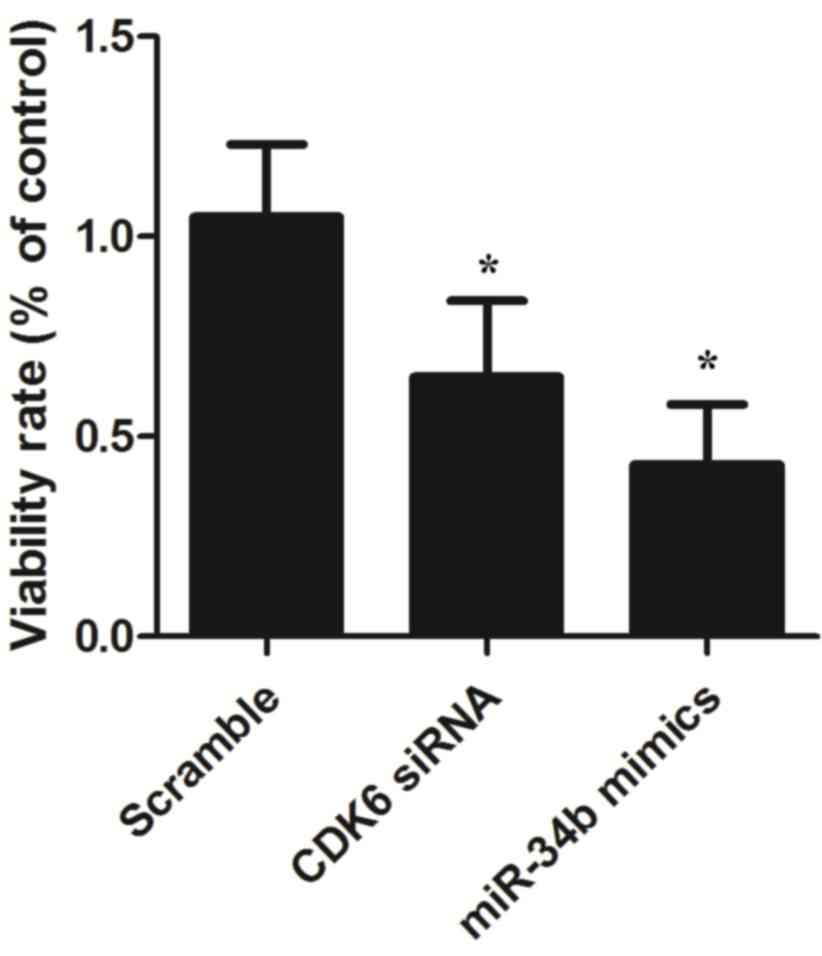
of CDK6 as CDK6 siRNA. The present study also examined the
viabilities of the VSMCs in the groups. As shown in Fig. 5, the relative viability of the
VSMCs transfected with miR-34b mimics was lower, compared with that
of VSMCs in the scramble control group, and similar results were
observed in the VSMCs transfected with CDK6 siRNA, confirming the
negative regulatory association between miR-34b and its target,
CDK6 (P<0.05).
Discussion
Accumulating evidence indicates that miRNAs are
involved in vascular remodeling in cardiovascular diseases and in
modulating VSMC proliferation. miR-146a, miR-26a, miR-24,
miR-221/222 and miR-21 have been reported to directly induce the
proliferation of VSMCs, which has been found to be mediated via
platelet-derived growth factor and bone morphogenetic proteins in
in vitro experiments (3,12,13,19).
However, the proliferation of VSMCs is inhibited by miR-145,
miR-143 and miR-1 (12,20). The expression levels of miR-146a,
miR-221/222 and miR-21 have been reported to be substantially
elevated in balloon-injured carotid arteries in animal experiments,
indicating the injury was attenuated by overexpression of these
miRNAs (3,19). Neointimal lesions in the femoral
arteries of mice with hypertension or of old age can be induced by
the deficiency of miR-143/145 (12). Other studies have reported that
there is an upregulation of miR-125b in the VSMCs of diabetic mice
(21), and there is abnormal
vascularization of the retina when miR-218 is knocked down
(22). Clinical studies have shown
that miR-145, which is abundant in smooth muscle, and miR-92a,
miR-17 and miR-126, which are abundant in the endothelium, are
significantly decreased in patients suffering from coronary artery
disease, compared with healthy individuals (23). In the present study, it was found
that the expression level of miR-34b in SHRs was upregulated,
compared with that in WKY rats. CDK6, rather than CCNG1, was
identified as the target gene of miR-34b using computational
analysis and a luciferase assay.
miR-34b is a member of the miR-34 family, which is
comprised of miR-34c, miR-34b and miR-34a. The physiological and
pathophysiological importance of this family in human diseases,
particularly in cancer, has been well documented; for example, the
miR-34 family acts as a tumor suppressor by inducing cell-cycle
arrest and apoptosis (24,25). In particular, miR-34a represses the
expression of pluripotency genes, including Mycn, (sex determining
region Y)-box 2 and Nanog, which leads to the restriction of
somatic reprogramming (26). The
late steps of spermatogenesis are associated with miR-34c (27), and sperm-borne miR-34c is involved
in modulating the expression of B cell lymphoma-2, which is
critical for the control of cell division (28). The miR-34 family has been shown to
be involved in the nervous system. miR-34c can act as a repressor
of stress-induced anxiety by targeting stress-related
corticotrophin releasing factor receptor type 1 and has a
physiological function in regulating the central stress response
(29). miR-34a targets silent
information regulator 1, which leads to regulation of the
differentiation of neural stem cells from mice (30). In the present study, it was found
that the mRNA and protein levels of CDK6 were downregulated in WKY
rats, compared with SHRs, which confirmed CDK6 as the direct target
gene of miR-34b and indicated the possible negative regulatory
association between miR-34b and CDK6. In addition, the present
study found that the mRNA and protein expression levels of CDK6 in
VSMCs transfected with CDK6 siRNA or miR-34b mimics were comparably
lower, compared with levels in the scramble control cells,
indicating that the miR-34b mimics, in addition to CDK6 siRNA,
inhibited the expression of CDK6.
Cell proliferation is primarily controlled by the
cell cycle, and progression of the cell cycle is predominantly
regulated by cyclin-dependent kinases (CDKs) and cyclins. Cyclin D1
is a key protein in regulating the G1 phase and the cell cycle is
more sensitive to alterations in cyclin D1, compared with other
cyclins (31). CDK6 acts as a
binding partner of cyclin D1, and its expression is critical for
the entry of cells into the S phase, which can be induced by the
activated cyclin D1/CDK6 complex (32). In the present study, it was shown
that the viability of VSMCs transfected with miR-34b mimics was
lower, compared with the scramble controls, similar to the results
of the VSMCs transfected with CDK6 siRNA. This confirmed the
negative regulatory association between miR-34b and its target,
CDK6.
Taken together, the present study demonstrated that
miR-34b regulated the proliferation of VSMCs by inhibiting the
expression of CDK6. The results of the present study, focused on
miR-34b, provide further insight into the molecular mechanism of
the development of hypertension, and improve current understanding
of the pathogenesis of vascular remodeling in hypertension.
However, further investigations are warranted to confirm the exact
role of miR-34b in the pathogenesis of hypertension in other
models, including transgenic mice in which miR-34b is knocked down
or overexpressed.
References
|
1
|
Owens GK, Kumar MS and Wamhoff BR:
Molecular regulation of vascular smooth muscle cell differentiation
in development and disease. Physiol Rev. 84:767–801. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rzucidlo EM: Signaling pathways regulating
vascular smooth muscle cell differentiation. Vascular. 17:(Suppl
1). S15–S20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moazed D: Small RNAs in transcriptional
gene silencing and genome defence. Nature. 457:413–420. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
5
|
Boettger T, Beetz N, Kostin S, Schneider
J, Krüger M, Hein L and Braun T: Acquisition of the contractile
phenotype by murine arterial smooth muscle cells depends on the
Mir143/145 gene cluster. J Clin Invest. 119:2634–2647. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cordes KR, Sheehy NT, White MP, Berry EC,
Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN and Srivastava D:
miR-145 and miR-143 regulate smooth muscle cell fate and
plasticity. Nature. 460:705–710. 2009.PubMed/NCBI
|
|
7
|
Xin M, Small EM, Sutherland LB, Qi X,
McAnally J, Plato CF, Richardson JA, Bassel-Duby R and Olson EN:
MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and
responsiveness of smooth muscle cells to injury. Genes Dev.
23:2166–2178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deacon DC, Nevis KR, Cashman TJ, Zhou Y,
Zhao L, Washko D, Guner-Ataman B, Burns CG and Burns CE: The
miR-143-adducin3 pathway is essential for cardiac chamber
morphogenesis. Development. 137:1887–1896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Quintavalle M, Elia L, Condorelli G and
Courtneidge SA: MicroRNA control of podosome formation in vascular
smooth muscle cells in vivo and in vitro. J Cell Biol. 189:13–22.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Davis BN, Hilyard AC, Nguyen PH, Lagna G
and Hata A: Induction of microRNA-221 by platelet-derived growth
factor signaling is critical for modulation of vascular smooth
muscle phenotype. J Biol Chem. 284:3728–3738. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Cheng Y, Zhang S, Lin Y, Yang J and
Zhang C: A necessary role of miR-221 and miR-222 in vascular smooth
muscle cell proliferation and neointimal hyperplasia. Circ Res.
104:476–487. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen
H, Dean DB and Zhang C: MicroRNA expression signature and
antisense-mediated depletion reveal an essential role of MicroRNA
in vascular neointimal lesion formation. Circ Res. 100:1579–1588.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chan MC, Hilyard AC, Wu C, Davis BN, Hill
NS, Lal A, Lieberman J, Lagna G and Hata A: Molecular basis for
antagonism between PDGF and the TGFbeta family of signalling
pathways by control of miR-24 expression. EMBO J. 29:559–573. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Elia L, Quintavalle M, Zhang J, Contu R,
Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J,
et al: The knockout of miR-143 and −145 alters smooth muscle cell
maintenance and vascular homeostasis in mice: Correlates with human
disease. Cell Death Differ. 16:1590–1598. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang C: MicroRNA-145 in vascular smooth
muscle cell biology: A new therapeutic target for vascular disease.
Cell Cycle. 8:3469–3473. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu ML, Wang JF, Wang GK, You XH, Zhao XX,
Jing Q and Qin YW: Vascular smooth muscle cell proliferation is
influenced by let-7d microRNA and its interaction with KRAS. Circ
J. 75:703–709. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Louis WJ and Howes LG: Genealogy of the
spontaneously hypertensive rat and Wistar-Kyoto rat strains:
Implications for studies of inherited hypertension. J Cardiovasc
Pharmacol. 16:(Suppl 7). S1–S5. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leeper NJ, Raiesdana A, Kojima Y, Chun HJ,
Azuma J, Maegdefessel L, Kundu RK, Quertermous T, Tsao PS and Spin
JM: MicroRNA-26a is a novel regulator of vascular smooth muscle
cell function. J Cell Physiol. 226:1035–1043. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie C, Huang H, Sun X, Guo Y, Hamblin M,
Ritchie RP, Garcia-Barrio MT, Zhang J and Chen YE: MicroRNA-1
regulates smooth muscle cell differentiation by repressing
Kruppel-like factor 4. Stem Cells Dev. 20:205–210. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Villeneuve LM, Kato M, Reddy MA, Wang M,
Lanting L and Natarajan R: Enhanced levels of microRNA-125b in
vascular smooth muscle cells of diabetic db/db mice lead to
increased inflammatory gene expression by targeting the histone
methyltransferase Suv39h1. Diabetes. 59:2904–2915. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Small EM, Sutherland LB, Rajagopalan KN,
Wang S and Olson EN: MicroRNA-218 regulates vascular patterning by
modulation of Slit-Robo signaling. Circ Res. 107:1336–1344. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fichtlscherer S, De Rosa S, Fox H,
Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Röxe T,
Müller-Ardogan M, et al: Circulating microRNAs in patients with
coronary artery disease. Circ Res. 107:677–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hermeking H: The miR-34 family in cancer
and apoptosis. Cell Death Differ. 17:193–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu
P, Zhong Y, Kim SY, Bennett MJ, Chen C, et al: miR-34 miRNAs
provide a barrier for somatic cell reprogramming. Nat Cell Biol.
13:1353–1360. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bouhallier F, Allioli N, Lavial F, Chalmel
F, Perrard MH, Durand P, Samarut J, Pain B and Rouault JP: Role of
miR-34c microRNA in the late steps of spermatogenesis. RNA.
16:720–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu WM, Pang RT, Chiu PC, Wong BP, Lao K,
Lee KF and Yeung WS: Sperm-borne microRNA-34c is required for the
first cleavage division in mouse. Proc Natl Acad Sci USA.
109:490–494. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Haramati S, Navon I, Issler O, Ezra-Nevo
G, Gil S, Zwang R, Hornstein E and Chen A: MicroRNA as repressors
of stress-induced anxiety: The case of amygdalar miR-34. J
Neurosci. 31:14191–14203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Aranha MM, Santos DM, Solá S, Steer CJ and
Rodrigues CM: miR-34a regulates mouse neural stem cell
differentiation. PLoS One. 6:e213962011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guan H, Zhu L, Fu M, Yang D, Tian S, Guo
Y, Cui C, Wang L and Jiang H: 3,3′Diindolylmethane suppresses
vascular smooth muscle cell phenotypic modulation and inhibits
neointima formation after carotid injury. PLoS One. 7:e349572012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hotchkiss A, Robinson J, MacLean J,
Feridooni T, Wafa K and Pasumarthi KB: Role of D-type cyclins in
heart development and disease. Can J Physiol Pharmacol.
90:1197–1207. 2012. View Article : Google Scholar : PubMed/NCBI
|