Introduction
Colorectal cancer (CRC) is currently one of the most
commonly diagnosed types of cancer worldwide, with an estimated 1.4
million cases leading to 693,900 fatalities in 2012 (1). More than half of patients (50–60%)
diagnosed with CRC develop metastases (2,3).
Oxaliplatin is a third-generation platinum analog
that may disrupt DNA replication and transcription, leading to DNA
damage and cell apoptosis (4,5).
Oxaliplatin-based chemotherapy is a primary treatment for patients
with metastatic CRC; however, its efficacy is limited. The
combination of oxaliplatin-based chemotherapy and targeted therapy,
including bevacizumab, panitumumab and cetuximab, may prolong
progression-free survival by only 1–3 months (6–8).
Novel therapeutic agents are urgently required to improve survival
rates.
The cullin-ring ubiquitin ligases (CRL), a subset of
E3 ligases (9) and the predominant
multi-unit ubiquitin ligase family in cells, are involved in the
degradation of 20% of ubiquitinated cellular proteins to regulate a
wide variety of biologic processes (10,11).
MLN4924 is a first-in-class inhibitor of neural precursor cell
expressed, developmentally downregulated 8 (NEDD8)-activating
enzyme E1 that was discovered via high-throughput screening
(10,12), and which has entered various
phase-I/II clinical trials for cancer therapy due to its
significant anticancer efficacy in preclinical studies (13). Via the formation of an inactive
covalent NEDD8-MLN4924 adduct (14), MLN4924 blocks CRL neddylation,
leading to accumulation of a mass of CRL substrates (10,15,16),
and resulting in DNA re-replication, the DNA damage response (DDR),
abnormal cell cycle progression, apoptosis, autophagy and
senescence, thus inhibiting the growth of cancer cells (10,17–22).
MLN4924 and oxaliplatin are anticancer agents that
may induce the DDR. There is potentially a synergistic effect
between MLN4924 and oxaliplatin in inducing the DDR, leading to
cell cycle arrest and apoptosis, and resulting in improved
anticancer efficacy. The aim of the present study was to examine
the synergistic effect and specific underlying mechanisms of
MLN4924 and oxaliplatin combined treatment of CRC.
Materials and methods
Cell lines, culture and reagents
SW620 and HCT116 human colorectal carcinoma cell
lines were obtained from the American Type Culture Collection
(Manassas, VA, USA), and cultured in RPMI-1640 (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (Biochrom, Ltd., Cambridge, UK) and 1%
penicillin-streptomycin solution, at 37°C in 5% CO2.
MLN4924 (Takeda Pharmaceutical Co., Ltd., Tokyo, Japan) was
dissolved in dimethyl sulfoxide (DMSO) and stored at −20°C. MLN4924
solution was freshly made every week and stored in the dark at room
temperature prior to use. Oxaliplatin (Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany) was dissolved in sterile water and
stored at −20°C. SW620 and HCT116 were seeded into 24-well plates
at a density of 1×104 cells/well or 12-well at a density
of 2×105 cells/well, and treated with 0.1, 0.3 or 1.0
µmol/l MLN4924, 0.3 µmol/l oxaliplatin, or 0.3 µmol/l MLN4924
combined with 0.3 µmol/l oxaliplatin for 24, 48, 72 or 96 h at
37°C.
Western blotting
Cells were lysed with radioimmunoprecipitation assay
lysis buffer (Beyotime Institute of Biotechnology, Haimen, China)
containing phenylmethylsulfonyl fluoride (Beyotime Institute of
Biotechnology), and lysates were centrifuged at 13,500 × g for 5
min at 4°C. Protein concentrations were quantified using a
Bicinchoninic Acid kit (Thermo Fisher Scientific, Inc.). A total of
30 µg protein was separated using 10% SDS-PAGE gels, and
transferred to polyvinylidene difluoride membranes. The membranes
were blocked with 5% non-fat milk in TBS containing Tween-20 (TBST)
for 1 h at room temperature, and subsequently incubated with the
following primary antibodies overnight at 4°C: Mouse anti-cullin1
(1:1,000; catalog no. sc-17775; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), rabbit anti-p21 (1:1,000; catalog no. 3733-1;
Epitomics, Burlingame, CA, USA), rabbit anti-total checkpoint
kinase 2 (t-CHK2; 1:1,000; catalog no. 3428-1; Epitomics), rabbit
anti-total histone H2A (t-H2A; 1:1,000; catalog no. 3522-1;
Epitomics), rabbit anti-phosphorylated (p)-CHK2 (Thr68; 1:1,000;
catalog no. 2197; Cell Signaling Technology, Inc., Danvers, MA,
USA), rabbit anti-p-H2A (Ser139; 1:1,000; catalog no. 2577; Cell
Signaling Technology, Inc.), rabbit anti-p27 (1:1,000; catalog no.
3686; Cell Signaling Technology, Inc.), rabbit anti-p53 (1:1,000;
catalog no. 2527; Cell Signaling Technology, Inc.), rabbit
anti-cleaved poly adenosine diphosphate ribose polymerase (PARP;
1:1,000; catalog no. 9532; Cell Signaling Technology, Inc.) and
rabbit anti-β-actin (1:1,000; catalog no. ab8227; Abcam, Cambridge,
UK). Subsequently, membranes were washed twice with TBST and
incubated with a horseradish peroxidase (HRP)-conjugated goat
anti-rabbit secondary antibody (1:2,000; catalog no. SA00001-2;
Wuhan Sanying Biotechnology, Wuhan, China) or an HRP-conjugated
anti-mouse secondary antibody (1:2,000; catalog no. ab131368;
Abcam) for 1 h at room temperature. Protein bands were visualized
using an Immobilon Western Chemiluminescent HRP Substrate (EMD
Millipore, Billerica, MA, USA) and imaged using a Tanon 4200
Chemiluminescent Imaging system (Tanon Science and Technology Co.,
Ltd., Shanghai, China).
Cell counting and clonogenic
assay
For cell counting, cells were seeded into 24-well
plates at a density of 1×104 cells per well and treated
with 0.1 µmol/l MLN4924. Cells were trypsinized, resuspended and
counted using a Cellometer Auto T4 Cell Viability Counter (Nexcelom
Bioscience, Lawrence, MA, USA) at 0, 48, 72 and 96 h after MLN4924
treatment. For the clonogenic assay, cells were seeded into a 60-mm
dish at a density of 500 cells/well and cultured for 10 days after
0.1 µmol/l MLN4924 treatment. Colonies on the dish were fixed with
4% paraformaldehyde and stained with crystal violet. Colonies with
>50 cells were counted.
Flow cytometric analysis
Cells were seeded into 6-well plates at a density of
2×105 per well and treated with DMSO, MLN4924 or
oxaliplatin were harvested and fixed in 70% ethanol at −20°C
overnight, and stained with 36 mg/ml propidium iodide (PI;
Sigma-Aldrich; Merck Millipore) containing 10 mg/ml RNase
(Sigma-Aldrich; Merck Millipore) at 37°C for 15 min, following
which they were analyzed for apoptosis and cell cycle progression
using a CyAn™ ADP analyzer (Beckman Coulter, Inc., Brea, CA, USA).
Apoptosis was measured as the percentage of cells in the sub-G1
population. Data were analyzed using ModFit LT software version 4.0
(Verity Software House, Inc., Topsham, ME, USA).
Live-dead cell staining assay
A Live-Dead Cell Staining kit (Enzo Life Sciences,
Inc., Farmingdale, NY, USA) was utilized to stain cells to
discriminate between live and dead cells. Cells were seeded into
12-well plates at a density of 2×105 cells per well.
Solutions A (Live-Dye, a cell-permeable green fluorescent dye) and
B (PI, a cell non-permeable red fluorescent dye) were mixed in
solution buffer. Cells were washed once with PBS and stained with
mixed staining solution at 37°C for 15 min. Cells were immediately
observed under a fluorescence microscope using a band-pass filter.
Healthy cells were stained green and dead cells were stained
red.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Data were analyzed using GraphPad Prism software version 6
(GraphPad Software, Inc., La Jolla, CA, USA). An unpaired
two-tailed Student's t-test was used for the comparison of
parameters between groups. Multiple groups were compared using
analysis of variance followed by the Bonferroni post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
MLN4924 inhibits cullin neddylation
and growth of CRC cells
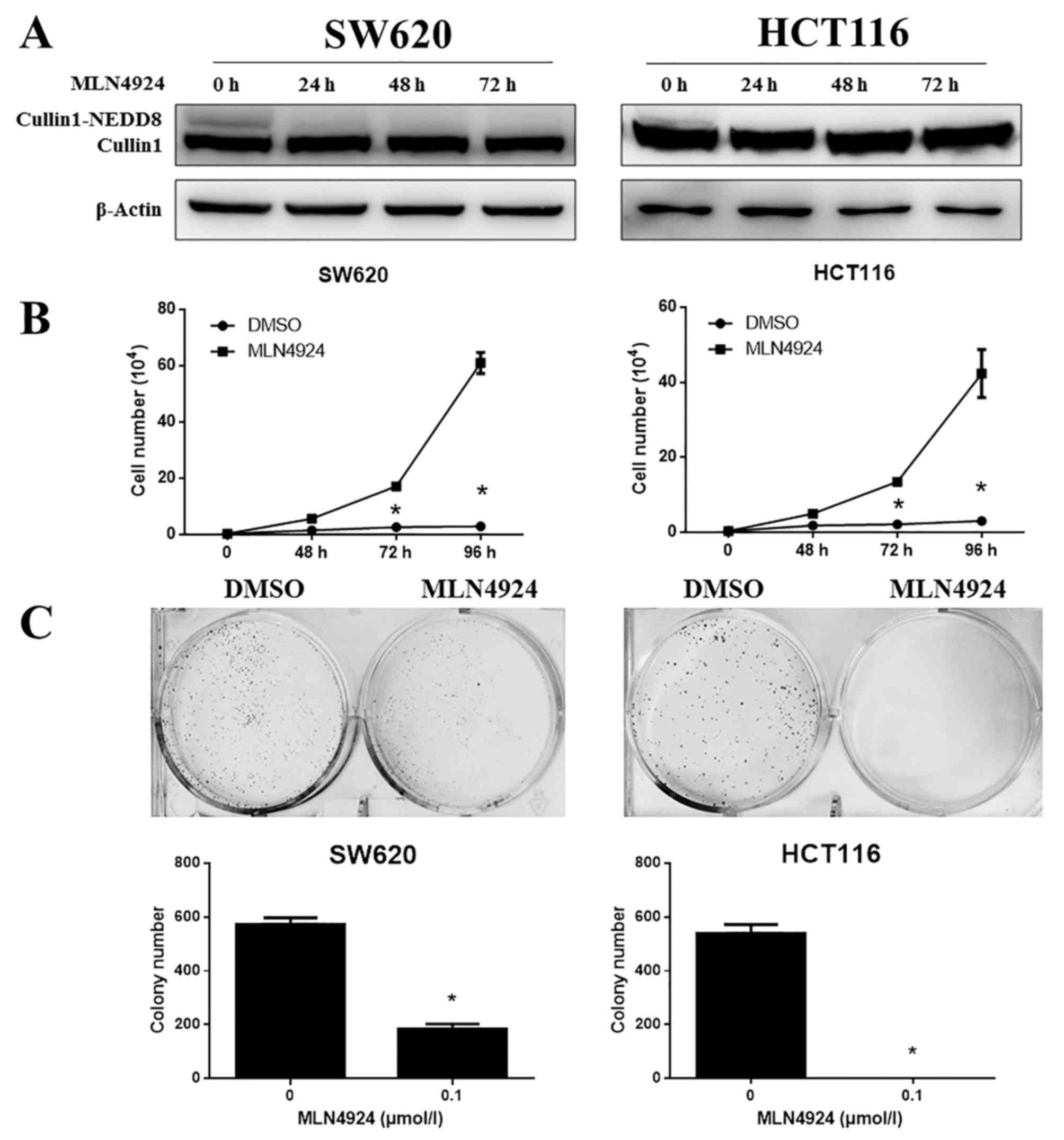
The efficacy of MLN4924 treatment on CRC cells was
examined. SW620 and HCT116 cells were treated with MLN4924 and
subjected to western blotting and cell growth analysis. MLN4924
completely inhibited cullin1-NEDD8 protein expression levels in
SW620 and HCT116 cells (Fig. 1A)
and significantly suppressed the proliferation of SW620 and HCT116
cells (Fig. 1B; P<0.001).
MLN4924 additionally markedly suppressed cell clonogenic survival
in these cells (Fig. 1C;
P<0.001). These results suggested that MLN4924 significantly
inhibited cullin1 neddylation and the growth of CRC cells.
MLN4924 induces the DDR, G2 cell cycle
arrest and apoptosis in CRC cells
Protein expression levels of p-H2A and p-CHK2,
well-known markers of the DDR, were increased as MLN4924
concentrations increased in SW620 and HCT116 cells. Additionally,
p53, p21 and p27 protein expression levels increased as MLN4924
concentrations increased in the two cell lines. p21 and p27 are
classical CRL substrates and p21 and p53 are involved in G2 cell
cycle arrest. Protein expression levels of cleaved-PARP, an
indicator of cell apoptosis, increased in a dose-dependent manner
(Fig. 2A). Cell cycle analysis
demonstrated that MLN4924 treatment triggered sub G1 (an indicator
of apoptosis) and G2 cell cycle arrest in CRC cells (Fig. 2B and C; P<0.001). These findings
suggested that MLN4924 treatment induced the DDR by inactivating
CRL, leading to cell cycle disturbance and apoptosis, which was
consistent with previous studies (20–23).
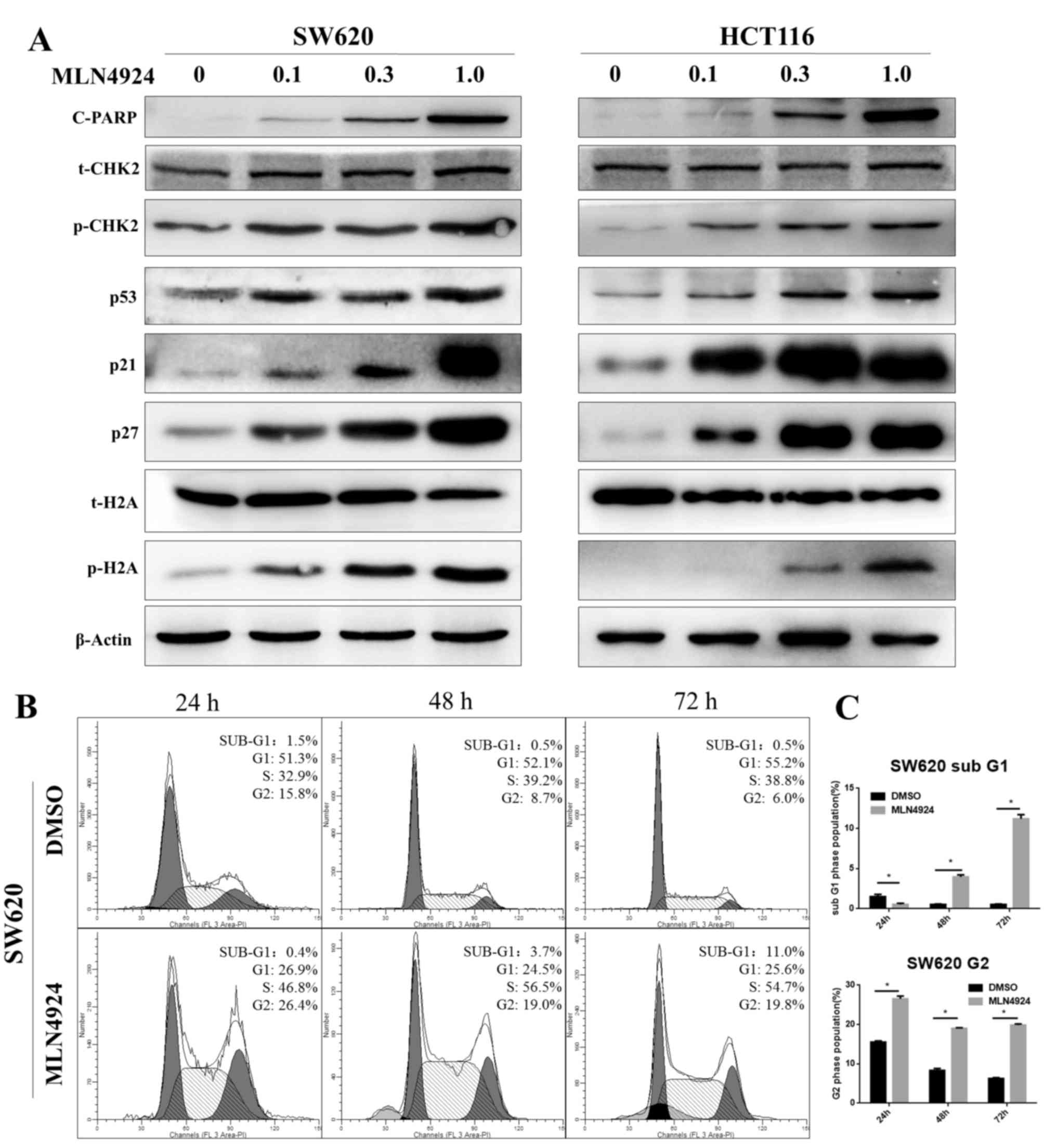 | Figure 2.MLN4924 treatment induces G2 cell
cycle arrest and apoptosis in colorectal cancer cells. (A) The
protein expression levels of c-PARP, p-CHK2, p53, p21, p27 and
p-H2A were assessed in MLN4924-treated SW620 and HCT116 cells by
western blot analysis; β-actin served as a loading control. (B)
MLN4924 induced cell-cycle arrest and apoptosis, as examined by
propidium iodide staining and flow cytometric analysis. (C) The
percentage of cells at the G2 and sub-G1 phases. Data are expressed
as the mean ± standard deviation. *P<0.001. c-PARP, cleaved poly
adenosine diphosphate ribose polymerase; CHK2, checkpoint kinase 2;
p, phosphorylated; H2A, histone H2A; DMSO, dimethyl sulfoxide. |
MLN4924 increases the
oxaliplatin-induced DDR and G2 cell cycle arrest
It has previously been reported that oxaliplatin is
involved in DNA damage by inducing DNA cross-links and activating
the DDR (4,5). The present study demonstrated that
MLN4924 and oxaliplatin treatment induced increased protein
expression levels of p-H2A and p-CHK2 compared with single agent
treatment in CRC cells. However, MLN4924 treatment in combination
with oxaliplatin did not increase protein expression levels of p21
and p53 compared with single agent treatment (Fig. 3A). These results suggested that
MLN4924 treatment increased the oxaliplatin-induced DDR.
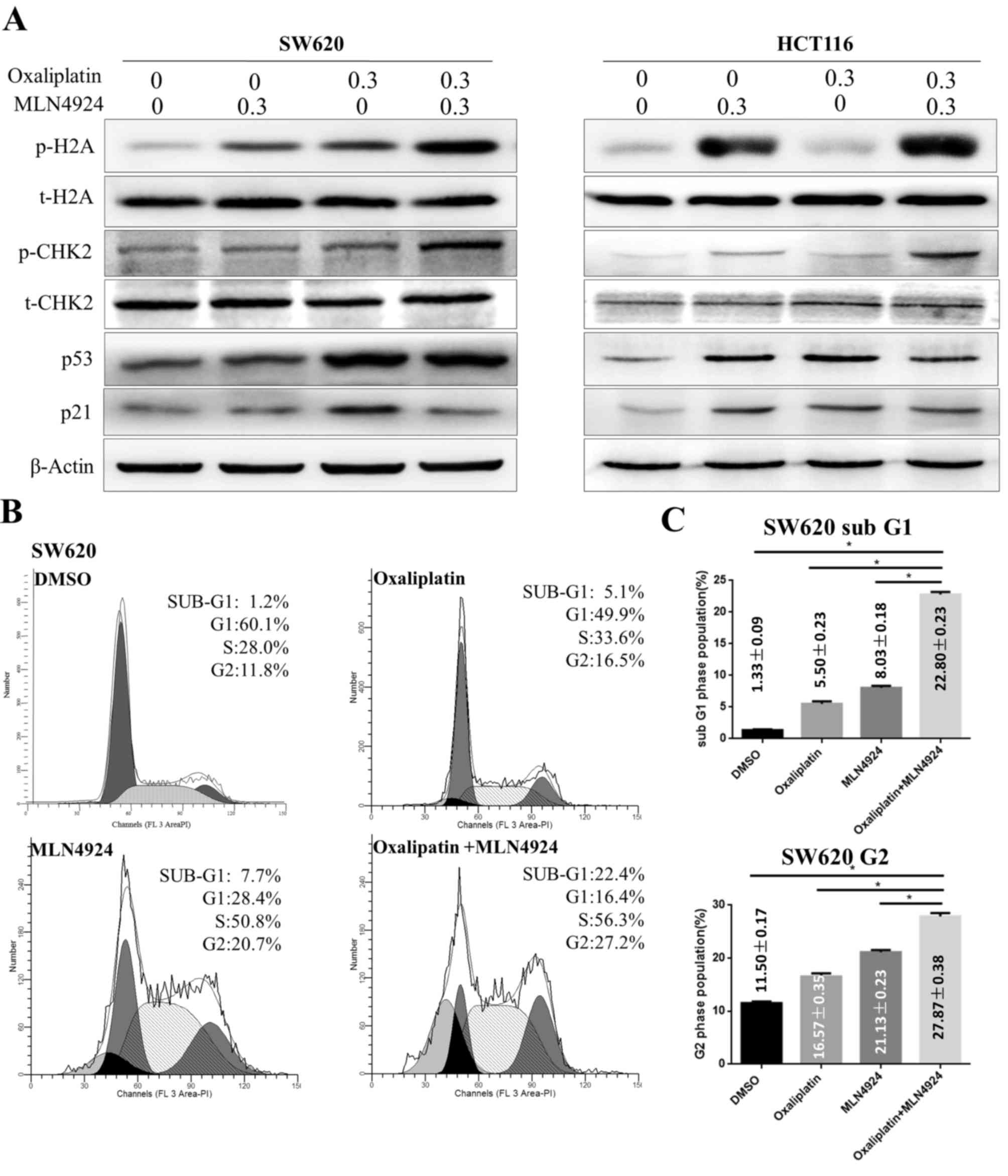 | Figure 3.MLN4924 increases the
oxaliplatin-induced DNA damage response, G2 cell cycle arrest and
apoptosis. (A) Western blot analysis of protein expression levels
of p-H2A, t-H2A, p-CHK2, t-CHK2, p53 and p21 in SW620 and HTC116
cells following single or combined MLN4924 and oxaliplatin
treatment; β-actin served as a loading control. (B) Flow cytometric
analysis of the cell cycle of SW620 cells following single or
combined oxaliplatin and MLN4924 treatment. (C) Quantification of
the percentage of cells in G2 and sub-G1 phases. Data are expressed
as the mean ± standard deviation. *P<0.001. H2A, histone H2A;
CHK2, checkpoint kinase 2; p, phosphorylated; t, total; DMSO,
dimethyl sulfoxide. |
Cell cycle analysis indicated that oxaliplatin
treatment induced G2 cell cycle arrest in CRC cells (Fig. 3B). Cells treated with MLN4924 and
oxaliplatin had an increased proportion of cells in G2 phase
compared with cells treated with a single agent (Fig. 3B). These findings demonstrated that
MLN4924 treatment increased oxaliplatin-induced G2 cell cycle
arrest, accompanied by an additional increase in the sub-G1
population (Fig. 3C; P<0.001);
however, p21 and p53 were not active in this process.
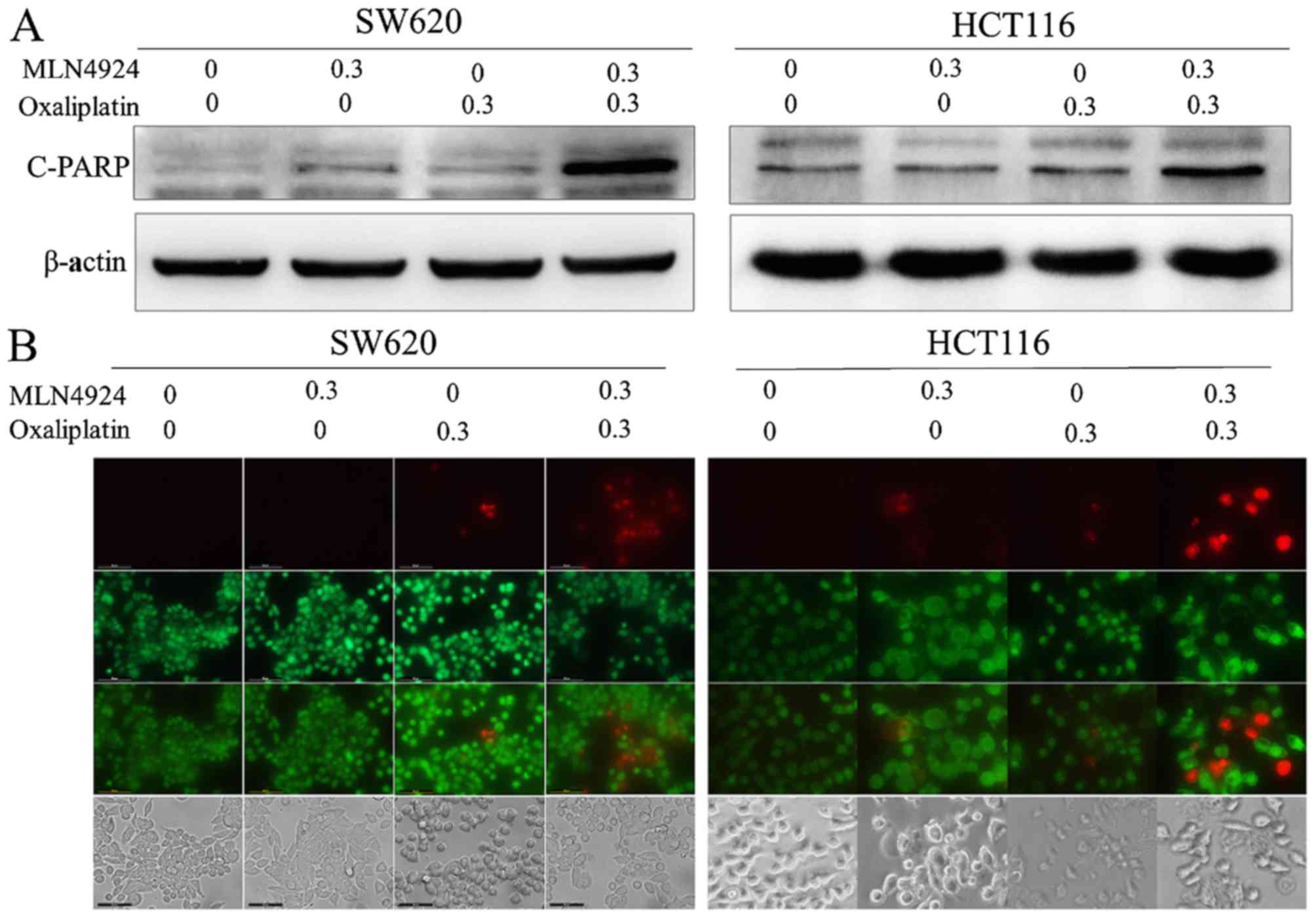
MLN4924 increases oxaliplatin-induced
apoptosis
MLN4924 and oxaliplatin treatment increased the
protein expression levels of cleaved-PARP compared with single
agent treatment (Fig. 4A). Cell
apoptosis was analyzed using the Live-Dead Cell Staining kit. CRC
cells treated with combined MLN4924 and oxaliplatin exhibited
increased apoptotic cells (stained red) compared with MLN4924 or
oxaliplatin only treatment (Fig.
4B). These results indicated that MLN4924 increased
oxaliplatin-induced apoptosis.
Discussion
Over half of patients (50–60%) diagnosed with CRC
develop metastases (2,3). Oxaliplatin-based chemotherapy is the
first line of treatment for patients with metastatic CRC; however,
its efficacy is limited. The combination of oxaliplatin-based
chemotherapy with anti-vascular endothelial growth factor or
anti-epidermal growth factor receptor agents prolongs
progression-free survival by only 1–3 months (6–8).
Novel therapeutic agents are urgently required to improve the
anticancer efficacy of oxaliplatin.
Our previous study demonstrated that MLN4924 may
induce the DDR in liver cancer cells in vivo and in
vitro (23). Other previous
studies additionally reported that MLN4924 may induce the DDR and
apoptosis in multiple cancer cell lines (17,18).
The present study hypothesized that MLN4924 treatment may induce
the DDR in CRC cells, which may sensitize these cells to
oxaliplatin. MLN4924 treatment was demonstrated to enhance the
anticancer efficacy of oxaliplatin in CRC cells by increasing
oxaliplatin-induced DDR, G2 cell cycle arrest and apoptosis.
Oxaliplatin forms inter- and intra-strand
cross-links with DNA, leading to the DDR (24). MLN4924 additionally induces the DDR
by accumulation of chromatin licensing and DNA replication factor 1
(21), inhibition of the nuclear
factor-κB signaling pathway (19)
and increase of reactive oxygen species (ROS) generation (25). p-H2A expression levels are
increased according to the DNA damage level, and are thus used as a
typical maker of the DDR (26).
CHK2 is phosphorylated in response to DNA damage, is involved in
the DNA damage signaling pathway, and is additionally used as a
maker of the DDR (27). The
present study demonstrated that combined MLN4924 and oxaliplatin
treatment induced increased protein expression levels of p-H2A and
p-CHK2 compared with single treatment. These results indicated that
MLN4924 may significantly increase oxaliplatin-induced DDR.
Furthermore, MLN4924 treatment was revealed to
increase oxaliplatin-induced G2 cell cycle arrest. p21 and p53 are
two well-known functional proteins of this cell cycle disturbance
process (28,29). The present study demonstrated that
MLN4924 or oxaliplatin treatment increased the protein expression
levels of p21 and p53 in CRC cells; however, combined treatment did
not increase protein expression levels of p21 and p53 further.
These results suggested that p21 and p53 were not active in the
process by which MLN4924 increased oxaliplatin-induced G2 cell
cycle arrest. DNA damage may activate checkpoints, and
phosphorylation of CHK2 is widely reported to lead to G2 cell cycle
arrest (27,30). The present study demonstrated that
p-CHK2 protein expression levels were increased following combined
treatment, indicating that it may have mediated the G2 cell cycle
arrest process.
Previous studies have reported that MLN4924
treatment increases the cisplatin-induced DDR and apoptosis in
multiple cell lines, via inhibiting monoubiquitination of fanconi
anemia group D2 (31) and
increasing ROS generation (25).
Oxaliplatin is a third-generation platinum analog, and is more
effective in the treatment of CRC compared with cisplatin (32). The specific mechanisms underlying
the increase of the oxaliplatin-induced DDR and G2 cell cycle
arrest in CRC cells by MLN4924 require further investigation.
In conclusion, the present study revealed that the
NEDD8-activating enzyme inhibitor MLN4924 sensitizes CRC cells to
oxaliplatin by inducing the DDR and increasing protein expression
levels of p-CHK2, leading to G2 cell cycle arrest and apoptosis.
These findings provide evidence for the potential efficacy of
combined MLN4924- and oxaliplatin-based chemotherapy for the
treatment of CRC.
Acknowledgements
The present study was supported by the Shanghai
Municipal Commission of Health and Family Planning (grant no.
20144Y0094) and the National Natural Science Foundation of China
(grant nos. 81500503 and 81420108005).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee WS, Yun SH, Chun HK, Lee WY, Yun HR,
Kim J, Kim K and Shim YM: Pulmonary resection for metastases from
colorectal cancer: Prognostic factors and survival. Int J
Colorectal Dis. 22:699–704. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yoo PS, Lopez-Soler RI, Longo WE and Cha
CH: Liver resection for metastatic colorectal cancer in the age of
neoadjuvant chemotherapy and bevacizumab. Clin Colorectal Cancer.
6:202–207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mowaka S, Ziehe M, Mohamed D, Hochkirch U,
Thomale J and Linscheid MW: Structures of
oxaliplatin-oligonucleotide adducts from DNA. J Mass Spectrom.
47:1282–1293. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Virag P, Perde-Schrepler M, Fischer-Fodor
E, Tatomir C, Dorneanu SA, Cernea VI and Irimie A: Superior
cytotoxicity and DNA cross-link induction by oxaliplatin versus
cisplatin at lower cellular uptake in colorectal cancer cell lines.
Anticancer Drugs. 23:1032–1038. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saltz LB, Clarke S, Diaz-Rubio E,
Scheithauer W, Figer A, Wong R, Koski S, Lichinitser M, Yang TS,
Rivera F, et al: Bevacizumab in combination with oxaliplatin-based
chemotherapy as first-line therapy in metastatic colorectal cancer:
A randomized phase III study. J Clin Oncol. 26:2013–2019. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Douillard JY, Siena S, Cassidy J,
Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham
D, Jassem J, et al: Randomized, phase III trial of panitumumab with
infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4)
versus FOLFOX4 alone as first-line treatment in patients with
previously untreated metastatic colorectal cancer: The PRIME study.
J Clin Oncol. 28:4697–4705. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bokemeyer C, Bondarenko I, Makhson A,
Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G,
Stroh C, et al: Fluorouracil, leucovorin, and oxaliplatin with and
without cetuximab in the first-line treatment of metastatic
colorectal cancer. J Clin Oncol. 27:663–671. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng N, Schulman BA, Song L, Miller JJ,
Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al:
Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase
complex. Nature. 416:703–709. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Soucy TA, Smith PG, Milhollen MA, Berger
AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP,
Critchley S, et al: An inhibitor of NEDD8-activating enzyme as a
new approach to treat cancer. Nature. 458:732–736. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Petroski MD and Deshaies RJ: Function and
regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol.
6:9–20. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Petroski MD: Mechanism-based neddylation
inhibitor. Chem Biol. 17:6–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Soucy TA, Dick LR, Smith PG, Milhollen MA
and Brownell JE: The NEDD8 conjugation pathway and its relevance in
cancer biology and therapy. Genes Cancer. 1:708–716. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brownell JE, Sintchak MD, Gavin JM, Liao
H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt
AL, et al: Substrate-assisted inhibition of ubiquitin-like
protein-activating enzymes: The NEDD8 E1 inhibitor MLN4924 forms a
NEDD8-AMP mimetic in situ. Mol Cell. 37:102–111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liao H, Liu XJ, Blank JL, Bouck DC,
Bernard H, Garcia K and Lightcap ES: Quantitative proteomic
analysis of cellular protein modulation upon inhibition of the
NEDD8-activating enzyme by MLN4924. Mol Cell Proteomics.
10:M111.0091832011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bennett EJ, Rush J, Gygi SP and Harper JW:
Dynamics of cullin-RING ubiquitin ligase network revealed by
systematic quantitative proteomics. Cell. 143:951–965. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Milhollen MA, Traore T, Adams-Duffy J,
Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E,
Langston SP, et al: MLN4924, a NEDD8-activating enzyme inhibitor,
is active in diffuse large B-cell lymphoma models: Rationale for
treatment of NF-{kappa}B-dependent lymphoma. Blood. 116:1515–1523.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jia L, Li H and Sun Y: Induction of
p21-dependent senescence by an NAE inhibitor, MLN4924, as a
mechanism of growth suppression. Neoplasia. 13:561–569. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Swords RT, Kelly KR, Smith PG, Garnsey JJ,
Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M,
Nawrocki ST, et al: Inhibition of NEDD8-activating enzyme: A novel
approach for the treatment of acute myeloid leukemia. Blood.
115:3796–3800. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin JJ, Milhollen MA, Smith PG, Narayanan
U and Dutta A: NEDD8-targeting drug MLN4924 elicits DNA
rereplication by stabilizing Cdt1 in S phase, triggering checkpoint
activation, apoptosis, and senescence in cancer cells. Cancer Res.
70:10310–10320. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Milhollen MA, Narayanan U, Soucy TA, Veiby
PO, Smith PG and Amidon B: Inhibition of NEDD8-activating enzyme
induces rereplication and apoptosis in human tumor cells consistent
with deregulating CDT1 turnover. Cancer Res. 71:3042–3051. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei D, Li H, Yu J, Sebolt JT, Zhao L,
Lawrence TS, Smith PG, Morgan MA and Sun Y: Radiosensitization of
human pancreatic cancer cells by MLN4924, an investigational
NEDD8-activating enzyme inhibitor. Cancer Res. 72:282–293. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D,
Pan Y, Ding C, Qian J, Wu L, et al: The Nedd8-activating enzyme
inhibitor MLN4924 induces autophagy and apoptosis to suppress liver
cancer cell growth. Cancer Res. 72:3360–3371. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Woynarowski JM, Faivre S, Herzig MC,
Arnett B, Chapman WG, Trevino AV, Raymond E, Chaney SG, Vaisman A,
Varchenko M and Juniewicz PE: Oxaliplatin-induced damage of
cellular DNA. Mol Pharmacol. 58:920–927. 2000.PubMed/NCBI
|
|
25
|
Nawrocki ST, Kelly KR, Smith PG, Espitia
CM, Possemato A, Beausoleil SA, Milhollen M, Blakemore S, Thomas M,
Berger A and Carew JS: Disrupting protein NEDDylation with MLN4924
is a novel strategy to target cisplatin resistance in ovarian
cancer. Clin Cancer Res. 19:3577–3590. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rogakou EP, Nieves-Neira W, Boon C,
Pommier Y and Bonner WM: Initiation of DNA fragmentation during
apoptosis induces phosphorylation of H2AX histone at serine 139. J
Biol Chem. 275:9390–9395. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsuoka S, Rotman G, Ogawa A, Shiloh Y,
Tamai K and Elledge SJ: Ataxia telangiectasia-mutated
phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA.
97:10389–10394. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bunz F, Dutriaux A, Lengauer C, Waldman T,
Zhou S, Brown JP, Sedivy JM, Kinzler KW and Vogelstein B:
Requirement for p53 and p21 to sustain G2 arrest after DNA damage.
Science. 282:1497–1501. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hirao A, Kong YY, Matsuoka S, Wakeham A,
Ruland J, Yoshida H, Liu D, Elledge SJ and Mak TW: DNA
damage-induced activation of p53 by the checkpoint kinase Chk2.
Science. 287:1824–1827. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rainey MD, Black EJ, Zachos G and
Gillespie DA: Chk2 is required for optimal mitotic delay in
response to irradiation-induced DNA damage incurred in G2 phase.
Oncogene. 27:896–906. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kee Y, Huang M, Chang S, Moreau LA, Park
E, Smith PG and D'Andrea AD: Inhibition of the Nedd8 system
sensitizes cells to DNA interstrand cross-linking agents. Mol
Cancer Res. 10:369–377. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fink D, Nebel S, Aebi S, Zheng H, Cenni B,
Nehmé A, Christen RD and Howell SB: The role of DNA mismatch repair
in platinum drug resistance. Cancer Res. 56:4881–4886.
1996.PubMed/NCBI
|