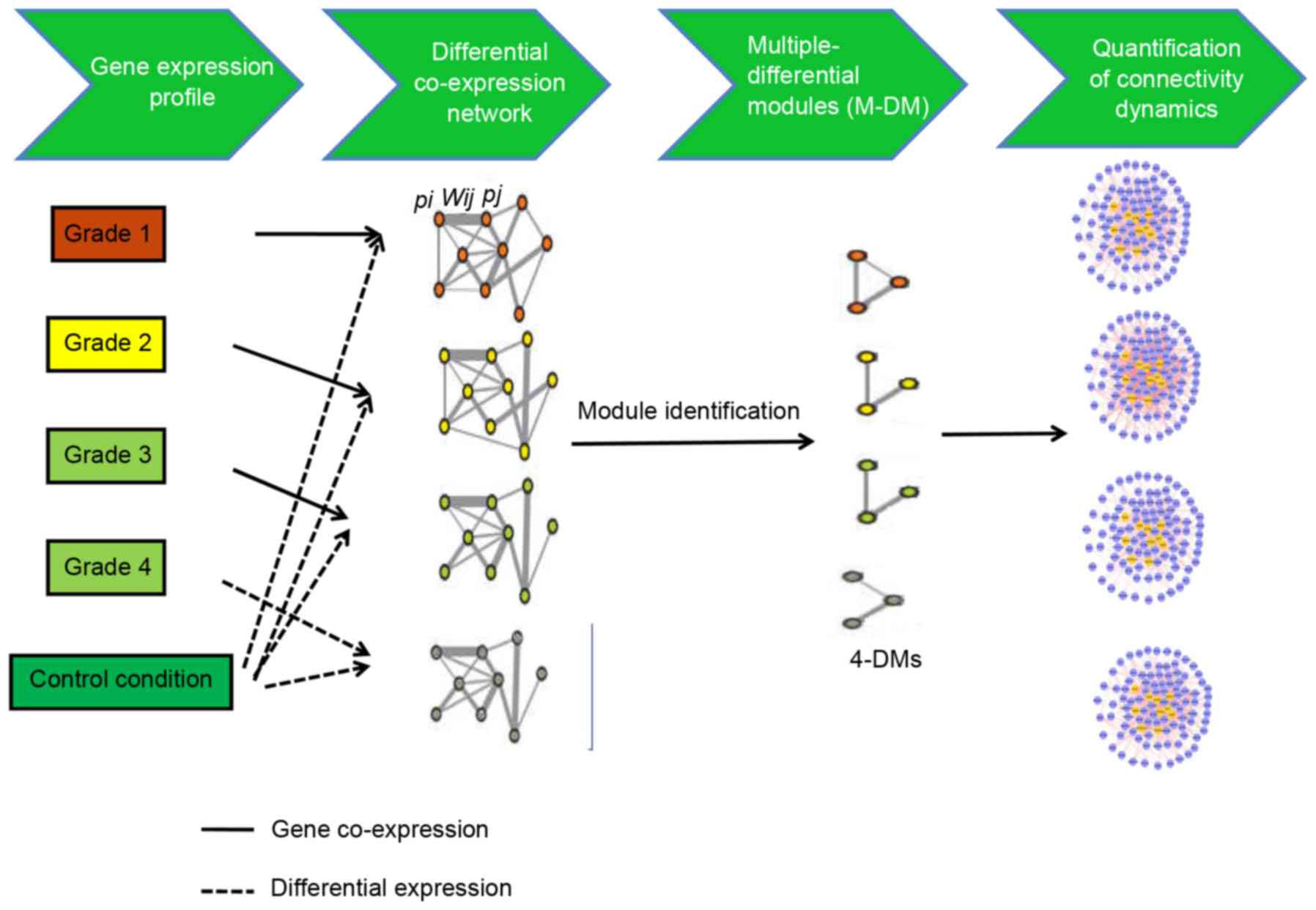
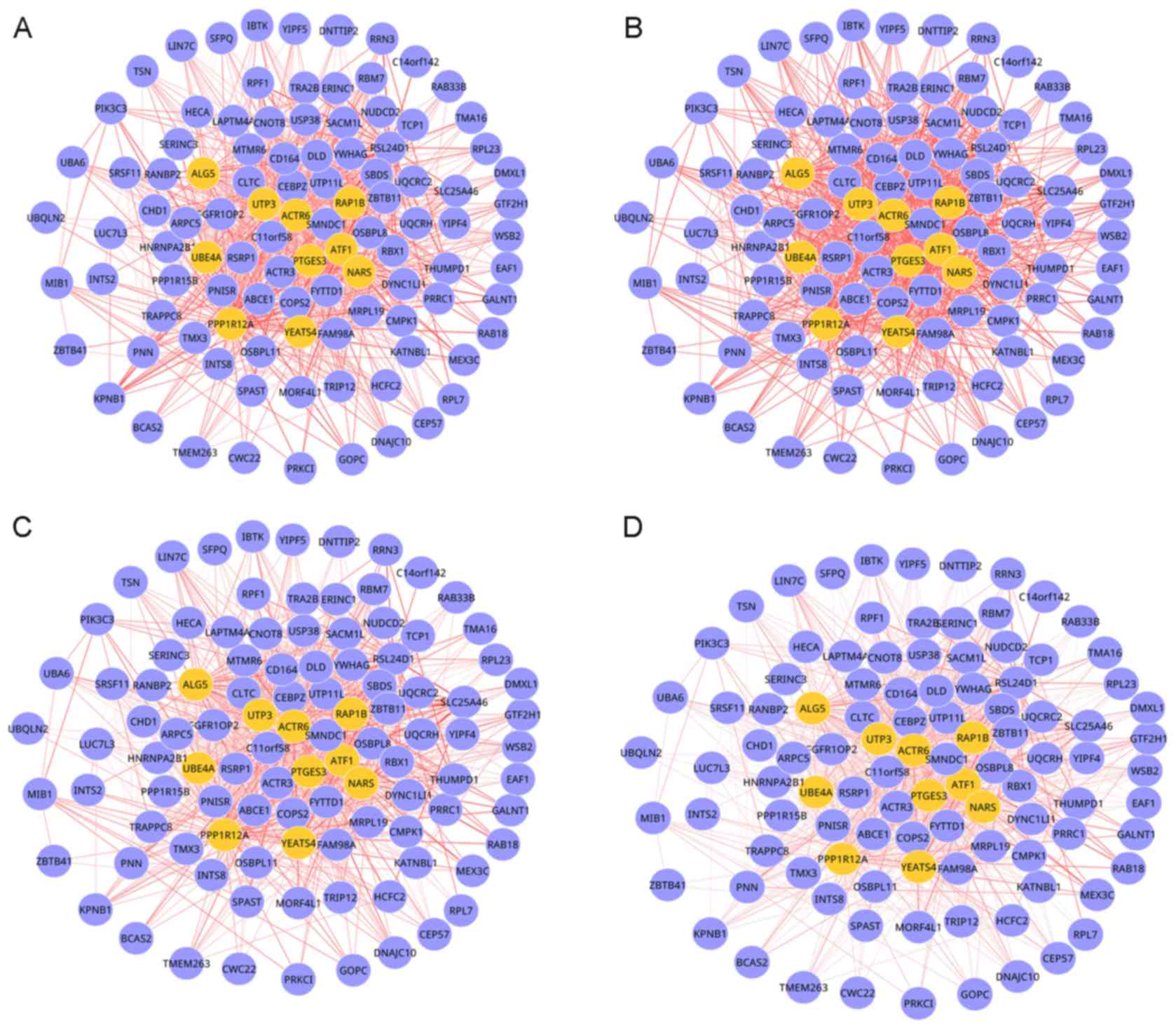
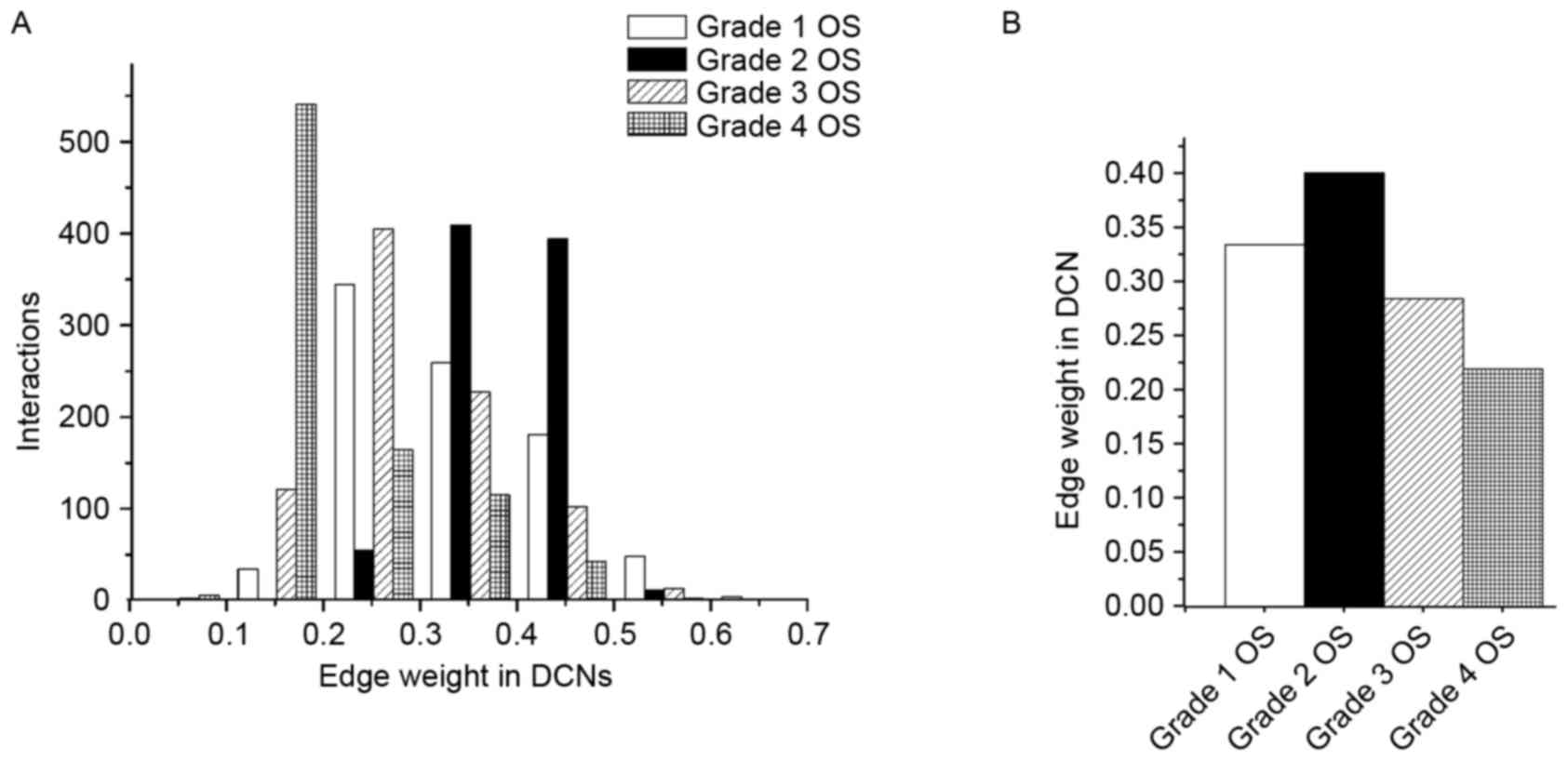
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcomaPediatric and adolescent osteosarcoma. Springer; pp.
3–13. 2010
|
|
2
|
Walkley CR, Qudsi R, Sankaran VG, Perry
JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, et
al: Conditional mouse osteosarcoma, dependent on p53 loss and
potentiated by loss of Rb, mimics the human disease. Genes Dev.
22:1662–1676. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Buddingh EP, Anninga JK, Versteegh MI,
Taminiau AH, Egeler RM, van Rijswijk CS, Hogendoorn PC, Lankester
AC and Gelderblom H: Prognostic factors in pulmonary metastasized
high-grade osteosarcoma. Pediatr Blood Cancer. 54:216–221.
2010.PubMed/NCBI
|
|
4
|
Rettew AN, Getty PJ and Greenfield EM:
Receptor tyrosine kinases in osteosarcoma: Not just the usual
suspectsCurrent Advances in Osteosarcoma. Springer; pp. 47–66.
2014
|
|
5
|
Hameed M and Dorfman H: Primary malignant
bone tumors-recent developmentsSeminars in diagnostic pathology.
Elsevier; pp. 86–101. 2011, View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Polager S and Ginsberg D: p53 and E2f:
Partners in life and death. Nat Rev Cancer. 9:738–748. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu X, Tang WH, Zhao XM and Chen L: A
network approach to predict pathogenic genes for Fusarium
graminearum. PLoS One. 5:pii: e130212010. View Article : Google Scholar
|
|
8
|
Chen L, Wang RS and Zhang XS: Biomolecular
networks: Methods and applications in systems biology. John Wiley
& Sons; 2009, View Article : Google Scholar
|
|
9
|
Ellis JD, Barrios-Rodiles M, Colak R,
Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O'Hanlon D, Kim PM, et
al: Tissue-specific alternative splicing remodels protein-protein
interaction networks. Mol Cell. 46:884–892. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Harbison CT, Gordon DB, Lee TI, Rinaldi
NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo
J, et al: Transcriptional regulatory code of a eukaryotic genome.
Nature. 431:99–104. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang B, Tian Y, Jin L, Li H, Shih IeM,
Madhavan S, Clarke R, Hoffman EP, Xuan J, Hilakivi-Clarke L and
Wang Y: DDN: A caBIG® analytical tool for differential network
analysis. Bioinformatics. 27:1036–1038. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma X, Gao L, Karamanlidis G, Gao P, Lee
CF, Garcia-Menendez L, Tian R and Tan K: Revealing pathway dynamics
in heart diseases by analyzing multiple differential networks. PLoS
Comput Biol. 11:e10043322015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuijjer ML, Rydbeck H, Kresse SH, Buddingh
EP, Lid AB, Roelofs H, Bürger H, Myklebost O, Hogendoorn PC,
Meza-Zepeda LA and Cleton-Jansen AM: Identification of osteosarcoma
driver genes by integrative analysis of copy number and gene
expression data. Genes Chromosomes Cancer. 51:696–706. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma X, Gao L and Tan K: Modeling disease
progression using dynamics of pathway connectivity. Bioinformatics.
30:2343–2350. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Watson-Haigh NS, Kadarmideen HN and
Reverter A: PCIT: An R package for weighted gene co-expression
networks based on partial correlation and information theory
approaches. Bioinformatics. 26:411–413. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sarawagi S and Kirpal A: Efficient set
joins on similarity predicates. Proceedings of the 2004 ACM SIGMOD
international conference on Management of data ACM. 743–754. 2004.
View Article : Google Scholar
|
|
17
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J R Stat Soc. Series B (Methodological).
57:289–300. 1995.
|
|
18
|
Taylor IW, Linding R, Warde-Farley D, Liu
Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q and Wrana JL:
Dynamic modularity in protein interaction networks predicts breast
cancer outcome. Nat Biotechnol. 27:199–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bisson N, James DA, Ivosev G, Tate SA,
Bonner R, Taylor L and Pawson T: Selected reaction monitoring mass
spectrometry reveals the dynamics of signaling through the GRB2
adaptor. Nat Biotechnol. 29:653–658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
21
|
da W Huang, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ciechanover A, Orian A and Schwartz AL:
Ubiquitin-mediated proteolysis: Biological regulation via
destruction. Bioessays. 22:442–451. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo G, Gui Y, Gao S, Tang A, Hu X, Huang
Y, Jia W, Li Z, He M, Sun L, et al: Frequent mutations of genes
encoding ubiquitin-mediated proteolysis pathway components in clear
cell renal cell carcinoma. Nat Genet. 44:17–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rivard A, Berthou-Soulie L, Principe N,
Kearney M, Curry C, Branellec D, Semenza GL and Isner JM:
Age-dependent defect in vascular endothelial growth factor
expression is associated with reduced hypoxia-inducible factor 1
activity. J Biol Chem. 275:29643–29647. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo M, Cai C, Zhao G, Qiu X, Zhao H, Ma Q,
Tian L, Li X, Hu Y, Liao B, et al: Hypoxia promotes migration and
induces CXCR4 expression via HIF-1α activation in human
osteosarcoma. PLoS One. 9:e905182014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nishioka T, Shohag MH, Amano M and
Kaibuchi K: Developing novel methods to search for substrates of
protein kinases such as Rho-kinase. Biochim Biophys Acta.
1854:1663–1666. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chiyoda T, Sugiyama N, Shimizu T, Naoe H,
Kobayashi Y, Ishizawa J, Arima Y, Tsuda H, Ito M, Kaibuchi K, et
al: LATS1/WARTS phosphorylates MYPT1 to counteract PLK1 and
regulate mammalian mitotic progression. J Cell Biol. 197:625–641.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li J, Liu X, Liao J, Tian J, Wang J, Wang
X, Zhang J and Xu X: MYPT1 sustains centromeric cohesion and the
spindle-assembly checkpoint. J Genet Genomics. 40:575–578. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Grassie ME, Moffat LD, Walsh MP and
MacDonald JA: The myosin phosphatase targeting protein (MYPT)
family: A regulated mechanism for achieving substrate specificity
of the catalytic subunit of protein phosphatase type 1δ. Arch
Biochem Biophys. 510:147–159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lafleur EA, Koshkina NV, Stewart J, Jia
SF, Worth LL, Duan X and Kleinerman ES: Increased Fas expression
reduces the metastatic potential of human osteosarcoma cells. Clin
Cancer Res. 10:8114–8119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Horowitz JM, Park SH, Bogenmann E, Cheng
JC, Yandell DW, Kaye FJ, Minna JD, Dryja TP and Weinberg RA:
Frequent inactivation of the retinoblastoma anti-oncogene is
restricted to a subset of human tumor cells. Proc Natl Acad Sci
USA. 87:2775–2779. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tanaka M, Setoguchi T, Hirotsu M, Gao H,
Sasaki H, Matsunoshita Y and Komiya S: Inhibition of Notch pathway
prevents osteosarcoma growth by cell cycle regulation. Br J Cancer.
100:1957–1965. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ellenbroek SI, Iden S and Collard JG: Cell
polarity proteins and cancer. Seminars in cancer biology Elsevier.
208–215. 2012. View Article : Google Scholar
|
|
34
|
Vasiliev JM: Cytoskeletal mechanisms
responsible for invasive migration of neoplastic cells. Int J Dev
Biol. 48:425–440. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wodarz A and Näthke I: Cell polarity in
development and cancer. Nat Cell Biol. 9:1016–1024. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jaffee EA, Jhaveri DT and Anders R:
Diagnostic biomarkers and therapeutic targets for pancreatic cancer
US Patent 20,150,316,554. Filed December 2, 2013; issued November
5. 2015
|
|
37
|
Phipps KR, Charette J and Baserga SJ: The
small subunit processome in ribosome biogenesis-progress and
prospects. Wiley Interdiscip Rev RNA. 2:1–21. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ruggero D and Pandolfi PP: Does the
ribosome translate cancer? Nat Rev Cancer. 3:179–192. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Montanaro L, Treré D and Derenzini M:
Nucleolus, ribosomes, and cancer. Am J Pathol. 173:301–310. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kondoh N, Shuda M, Tanaka K, Wakatsuki T,
Hada A and Yamamoto M: Enhanced expression of S8, L12, L23a, L27
and L30 ribosomal protein mRNAs in human hepatocellular carcinoma.
Anticancer Res. 21:2429–2433. 2001.PubMed/NCBI
|
|
41
|
Jorgensen P, Nishikawa JL, Breitkreutz BJ
and Tyers M: Systematic identification of pathways that couple cell
growth and division in yeast. Science. 297:395–400. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bernstein KA and Baserga SJ: The small
subunit processome is required for cell cycle progression at G1.
Mol Biol Cell. 15:5038–5046. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Colotta F, Allavena P, Sica A, Garlanda C
and Mantovani A: Cancer-related inflammation, the seventh hallmark
of cancer: Links to genetic instability. Carcinogenesis.
30:1073–1081. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cebola I, Custodio J, Muñoz M,
Díez-Villanueva A, Paré L, Prieto P, Aussó S, Coll-Mulet L, Boscá
L, Moreno V and Peinado MA: Epigenetics override pro-inflammatory
PTGS transcriptomic signature towards selective hyperactivation of
PGE2 in colorectal cancer. Clin Epigenetics. 7:742015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liang Y, Liu M, Wang P, Ding X and Cao Y:
Analysis of 20 genes at chromosome band 12q13: RACGAP1 and MCRS1
overexpression in nonsmall-cell lung cancer. Genes Chromosomes
Cancer. 52:305–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lomnytska MI, Becker S, Gemoll T, Lundgren
C, Habermann J, Olsson A, Bodin I, Engström U, Hellman U, Hellman
K, et al: Impact of genomic stability on protein expression in
endometrioid endometrial cancer. Br J Cancer. 106:1297–1305. 2012.
View Article : Google Scholar : PubMed/NCBI
|