Introduction
Glioma is an incurable primary malignant tumor with
a high recurrence rate. The life expectancy of patients with glioma
is ~1 year, and that of patients exhibiting recurrence is ~4 months
(1–3). Survey results have demonstrated that
the incidence of glioma has increased in recent years. The
principal treatment regimen used clinically is surgical resection
followed by chemotherapy (4);
however, this treatment regimen is not able to cure glioma as
gliocytes exhibit invasive growth with unclear boundaries from
normal brain tissue, resulting in incomplete surgical resection,
and may be resistant to chemotherapy (5,6).
Therefore, improved therapies are required to achieve improved
clinical outcomes and reduce the rate of recurrence in patients
with glioma.
Numerous studies have demonstrated that microRNAs
(miRNAs/miRs) are extensively involved in regulating mutations in
glioma-associated genes (7),
cellular invasion, migration, apoptosis (8) and blood vessel formation around the
residual tumor (9). Analysis of
the expression levels of miRNAs in normal brain tissue compared
with glioma has demonstrated that 22 miRNAs, including miR-15a,
miR-16, miR-21, the miR-17-92 cluster, miR-26a and miR-221/222, are
upregulated in glioma and promote the proliferation, migration and
invasion of cancer cells (10–13),
whereas 33 miRNAs, including miR-7, miR-137, miR-145, the miR-181
family and miR-195, are downregulated and inhibit apoptosis in
cancer cells (14–16). The oncogenic miRNA miR-21 has been
observed to be associated with malignant glioma, and its expression
level is directly associated with tumor grade; patients with low
miRNA-21 expression exhibit increased survival times and improved
clinical outcomes (17).
In long-term studies, researchers have demonstrated
that a single miRNA is able to regulate >100 proteins encoded by
target genes and that approximately one-third of proteins involved
in common biological activities are regulated by different miRNAs.
Therefore, various miRNAs exhibit synergistic activities and are
able to regulate identical target mRNAs to mediate the same
biological process (18,19). In a previous study, miR-1 and
miR-21 were demonstrated to function synergistically during
myocardial ischemia, as predicted by synergy score calculations and
validated by cellular experiments (20). Simultaneous modulation of two
miRNAs was observed to have apoptosis-inhibiting effects on cardiac
myocytes and to allow for a reduction in the concentration of
miRNAs in order to effectively avoid off-target effects (21).
Therefore, it was hypothesized that miRNAs may act
synergistically during the regulation of proliferation and
apoptosis in glioma. The present study aimed to identify
glioma-sensitive miRNA combinations with potential synergistic
regulatory effects based on synergy scores and to validate such
miRNA combinations at the cellular level, in order to provide a
rational, effective, novel noncoding RNA-based treatment strategy
for the prevention and treatment of glioma.
Materials and methods
Collection of patient tissues
The present study was performed with the ethical
approval of the Human Ethics Committee of The First Affiliated
Hospital of Harbin Medical University (Harbin, China) in accordance
with the Declaration of Helsinki, and written informed consent was
obtained from all enrolled patients. Noncancerous brain tissues
were collected from the temporal lobes of four patients with
epilepsy. All tissues were frozen in liquid nitrogen immediately
following surgical removal and stored at −80°C.
Cell culture and transfection
U87 cells, obtained from the Cell Resource Center of
the Shanghai Institute of Life Sciences (Chinese Academy of
Sciences, Shanghai, China), were cultured in high-glucose
Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum
(Invitrogen; Thermo Fisher Scientific, Inc.) and 100 U/ml
streptomycin/penicillin (Beyotime Institute of Biotechnology,
Haimen, China) at 37°C with 5% CO2. miRNA
mimics/inhibitors and negative control (NC) were synthesized by
Guangzhou RiboBio Co. Ltd (Guangzhou, China), and sequences are
presented in Table I. Following
starvation in serum-free medium for 12 h, U87 cells were
transfected with miR-7, miR-181 and miR-195 mimics, miR-15a,
miR-16, miR-20a, miR-21, miR-26a andmiR-222 inhibitors, or NC using
Xtreme GENE siRNA transfection reagent (Roche Diagnostics, Basel,
Switzerland). Transfection concentrations ranged between 10 and 50
nM.
 | Table I.Sequences of the miRNA mimics and
inhibitors. |
Table I.
Sequences of the miRNA mimics and
inhibitors.
| A, Sequences of miRNA
inhibitors |
|---|
|
|---|
| miRNA | Sequence |
|---|
| Hsa-miR-21 |
UAGCUUAUCAGACUGAUGUUGA |
| Hsa-miR-20a |
UAAAGUGCUUAUAGUGCAGGUAG |
| Hsa-miR-15a |
UAGCAGCACAUAAUGGUUUGUG |
| Hsa-miR-16 |
UAGCAGCACGUAAAUAUUGGCG |
| Hsa-miR-26 |
UUCAAGUAAUCCAGGAUAGGCU |
| Hsa-miR-222 |
CUCAGUAGCCAGUGUAGAUCCU |
|
| B, Sequences of miRNA
mimics |
|
| miRNA | Sequence |
|
| Hsa-miR-7 |
UGGAAGACUAGUGAUUUUGUUGU |
| Hsa-miR-181 |
AACAUUCAACGCUGUCGGUGAGU |
| Hsa-miR-195 |
UAGCAGCACAGAAAUAUUGGC |
|
| C, Sequence of
negative control |
|
| miRNA | Sequence |
|
| cel-miR-239b |
UUUGUACUACACAAAAGUACUG |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from normal brain tissue and
U87 cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. A total
of 0.5 µg RNA was reverse transcribed into first strand cDNA using
a ReverTraAce® qPCR RT kit (Toyobo Co., Ltd, Osaka, Japan) in a 10
µl reaction mixture containing the following: Total RNA (0.5 µg),
5X RT buffer (2 µl), RT Enzyme Mix (0.5 µl), Primer Mix (0.5 µl),
made up to 10 µl with Nuclease-free water. The RT reaction was
performed as follows: 25°C for 10 min, 37°C for 120 min, 85°C for 5
min and termination at 4°C for 5 min. The expression levels of
miR-15a, miR-16, miR-20a, miR-21, miR-26a, miR-222, miR-7, miR-181
and miR-195 were determined using SYBR Green Mix (Invitrogen;
Thermo Fisher Scientific, Inc.) on an ABI 7500 Fast Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
PCR conditions used to detect the miRNAs were as follows: 95°C for
10 min and 35 cycles at 95°C for 15 sec and 60°C for 1 min. The
miRNA expression relative to U6 was calculated using the
2−ΔΔCq method (22).
The primers for amplification of U6, miR-15a, miR-16, miR-21,
miR-20a, miR-26a, miR-7, miR-181, miR-222 and miR-195 are presented
in Table II.
 | Table II.Primers for reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primers for reverse
transcription-quantitative polymerase chain reaction.
|
| Primers |
|---|
|
|
|
|---|
| miR | Forward | Reverse |
|---|
| miR-21 |
GCGGCGGTAGCTTATCAGACTG |
TAGCTTATCAGACTGATGTTGA |
| miR-20a |
GCGGCGGTAAAGTGCTTATAGTG |
TAAAGTGCTTATAGTGCAGGTAG |
| miR-15a |
GCGGCGGTAGCAGCACATAATG |
TAGCAGCACATAATGGTTTGTG |
| miR-16 |
GCGGCGGTAGCAGCACGTAAAT |
TAGCAGCACGTAAATATTGGCG |
| miR-26a |
GCGGCGGTTCAAGTAATCCAGG |
TTCAAGTAATCCAGGATAGGCT |
| miR-222 |
GCGGCGGACCTGGCATACAATG |
ACCTGGCATACAATGTAGATTT |
| miR-7 |
GCGGCGGTGGAAGACTAGTG |
TGGAAGACTAGTGATTTTGTTGT |
| miR-181 |
GCGGCGGAACATTCAACGCTGTC |
AACATTCAACGCTGTCGGTGAGT |
| miR-195 |
GCGGCGGTAGCAGCACAGAAAT |
TAGCAGCACAGAAATATTGGC |
| U6 |
GCTTCGGCAGCACATATACTAAAAT |
CGCTTCACGAATTTGCGTGTCAT |
Cell viability assay
U87 cells were cultured in 96-well culture plates at
a density of 5×104 cells/well. Cell viability was assessed by
measuring mitochondrial dehydrogenase activity. Following miRNA
transfection, U87 cells were incubated with 10 µl MTT (0.5 mg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C for 4 h. The
purple formazan crystals were dissolved in 150 µl
dimethylsulfoxide. The absorbance was measured using a
spectrophotometer (Tecan Group, Ltd., Männedorf, Switzerland) at
490 nm.
Terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labeling
(TUNEL) staining assay
TUNEL staining was detected using an in situ
cell death detection kit (Roche Diagnostics) according to the
manufacturer's protocol. Following three washes with PBS, U87 cells
(2–5×106) were fixed with 4% paraformaldehyde at room temperature
and permeabilized in 0.1% Triton X-100 sodium citrate buffer on
ice. The kits were used to label apoptotic cells, and the nuclei
were stained with DAPI. The numbers of total cells and
TUNEL-positive cells were automatically counted using Image-Pro
Plus v6.0 software (Media Cybernetics, Inc., Rockville, MD, USA).
The apoptosis rate was defined as the ratio of apoptotic cells to
total cells.
Detection of apoptosis by flow
cytometry
Cell apoptosis was assayed using annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) using the
Annexin V-FITC Cell Apoptosis Detection kit (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. Cells
(2–3×105 cells/well) were cultured in 6-well plates. Following the
various indicated transfection treatments, the cells were digested
with trypsin and collected. Cells were washed twice with ice-cold
PBS and stained with annexin V-FITC for 12 min and PI for 5 min.
The results were analyzed with CellQuest software v5.1 (BD
Biosciences, Franklin Lakes, NJ, USA) and expressed as the
percentage of apoptotic cells at the early stage (annexin
V-FITC+/PI-; Q4) and the late stage (annexin V-FITC+/PI+; Q2).
Heat map analysis
MATLAB software (vR2012b; MathWorks, Inc., Natick,
MA, USA) was used to produce heat maps. Relative cell viability
data was inputted and calculated using the surf function to obtain
a three-dimensional colored surface model.
Statistical analyses
GraphPad Prism v6.0 software (GraphPad Software,
Inc. La Jolla, CA, USA) was used for statistical analysis. All data
are expressed as the mean ± standard error of the mean. Statistical
analysis was performed using one-way analysis of variance (ANOVA)
followed by Bonferroni's test. P<0.05 was considered to indicate
a statistically significant difference.
Results
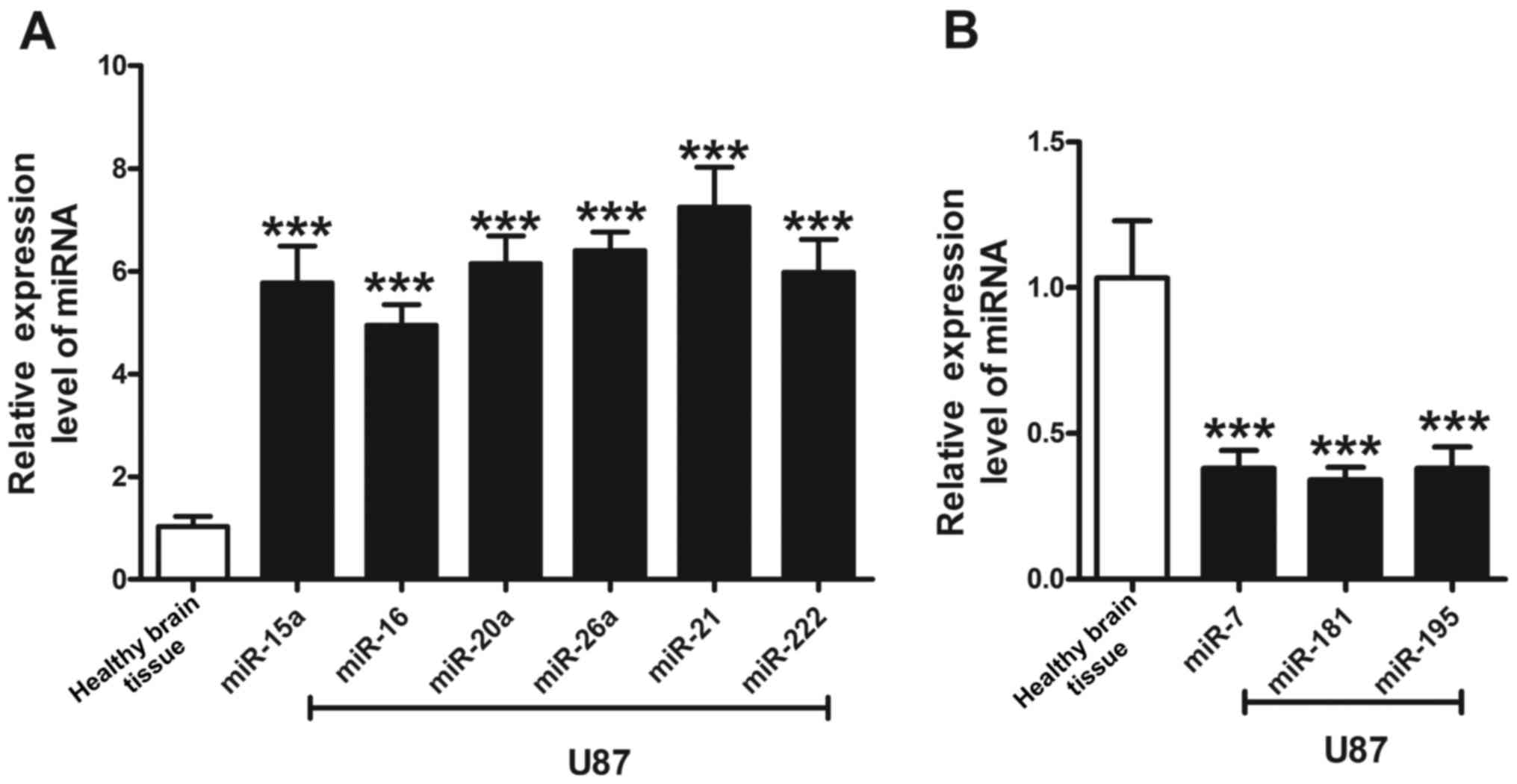
miRNAs are aberrantly expressed in U87
cells
Based on previous reports demonstrating aberrant
expression of miRNAs in glioma cells, the expression levels of nine
miRNAs in U87 cells and normal brain tissue were examined using
RT-qPCR. Compared with the expression levels of miRNAs in normal
tissue, miR-15a, miR-16, miR-20a, miR-21, miR-26a, and miR-222 were
upregulated in U87 cells by 5.8-, 5.0-, 6.2-, 6.4-, 7.3-, and
6.0-fold, respectively. By contrast, miR-7, miR-181, and miR-195
were downregulated. miR-21 exhibited the most marked alteration
among the six miRNAs, exhibiting increased expression in U87 cells.
The expression levels of the three downregulated miRNAs exhibited
similar rates of reduction, with an average relative expression
level of ~0.35 (Fig. 1).
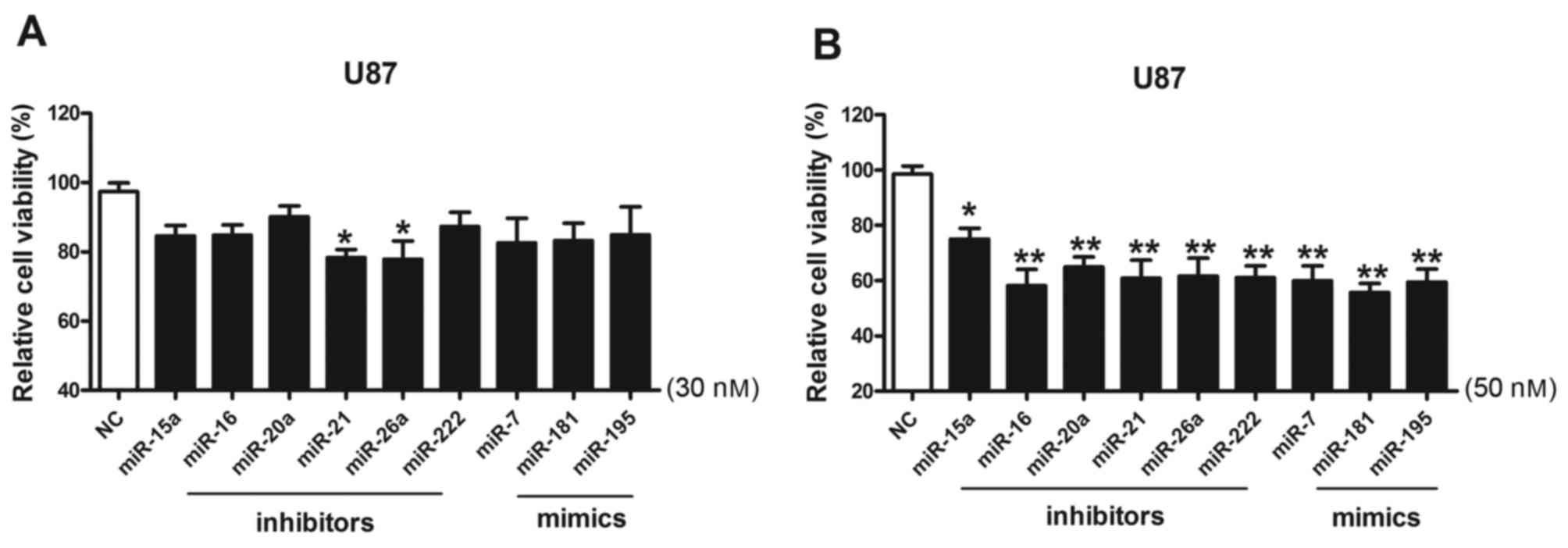
Effect of miRNAs on the cell viability
of U87 cells
U87 cells were transfected with miRNA inhibitors and
miRNA mimics at 30 and 50 nM, and their effects on the U87 cells
were investigated using cell viability assays (Fig. 2). The results of the present study
demonstrated that miR-21 and miR-26a inhibitors exhibited effects
on the viability of U87 cells when used at 30 nM; as the
transfection concentration increased to 50 nM, the inhibitory
effects of the miRNA inhibitors and mimics on cell viability were
markedly increased.
Synergistic effects of miRNAs on the
viability of U87 cells
In order to identify miRNA combinations with
synergistic effects, the above-described miRNAs were investigated
as miRNA-miRNA pairs according to calculated synergy scores.
According to the miRNA transfection concentration test, cells were
cotransfected with two miRNA inhibitors (1:1) at 30 nM each, in
order to avoid off-target effects induced by high concentrations of
miRNAs and to screen out miRNA combinations with potent synergistic
effects at low doses. A total of six upregulated miRNAs were
divided into miRNA pairs as follows: miR-20a/15a, miR-16/20a,
miR-21/16, miR-21/15a, miR-21/20a, miR-16/222, miR-21/222, and
miR-21/26, and cells were cotransfected with these miRNA pairs.
Compared with the NC group, U87 cells exhibited significantly
reduced cell viability following cotransfection with two low-dose
miRNAs. Additionally, following cotransfection with miR-20a/15a,
miR-16/20a, miR-21/16, miR-21/15a, and miR-21/20a inhibitors, the
combined inhibitory effect of each miRNA pair on the viability of
U87 cells was increased compared with the sum of the individual
inhibitory effects of each of the transfected miRNAs. Cell
viability was reduced to 77.2 and 88.4% following transfection with
miR-21 inhibitor and miR-20a inhibitor, respectively, whereas
viability was 48.5% following cotransfection with miR-21 and
miR-20a inhibitors together, indicating that miR-21 and miR-20a
exhibited the most potent synergistic inhibitory effect on the
viability of U87 cells (Fig.
3).
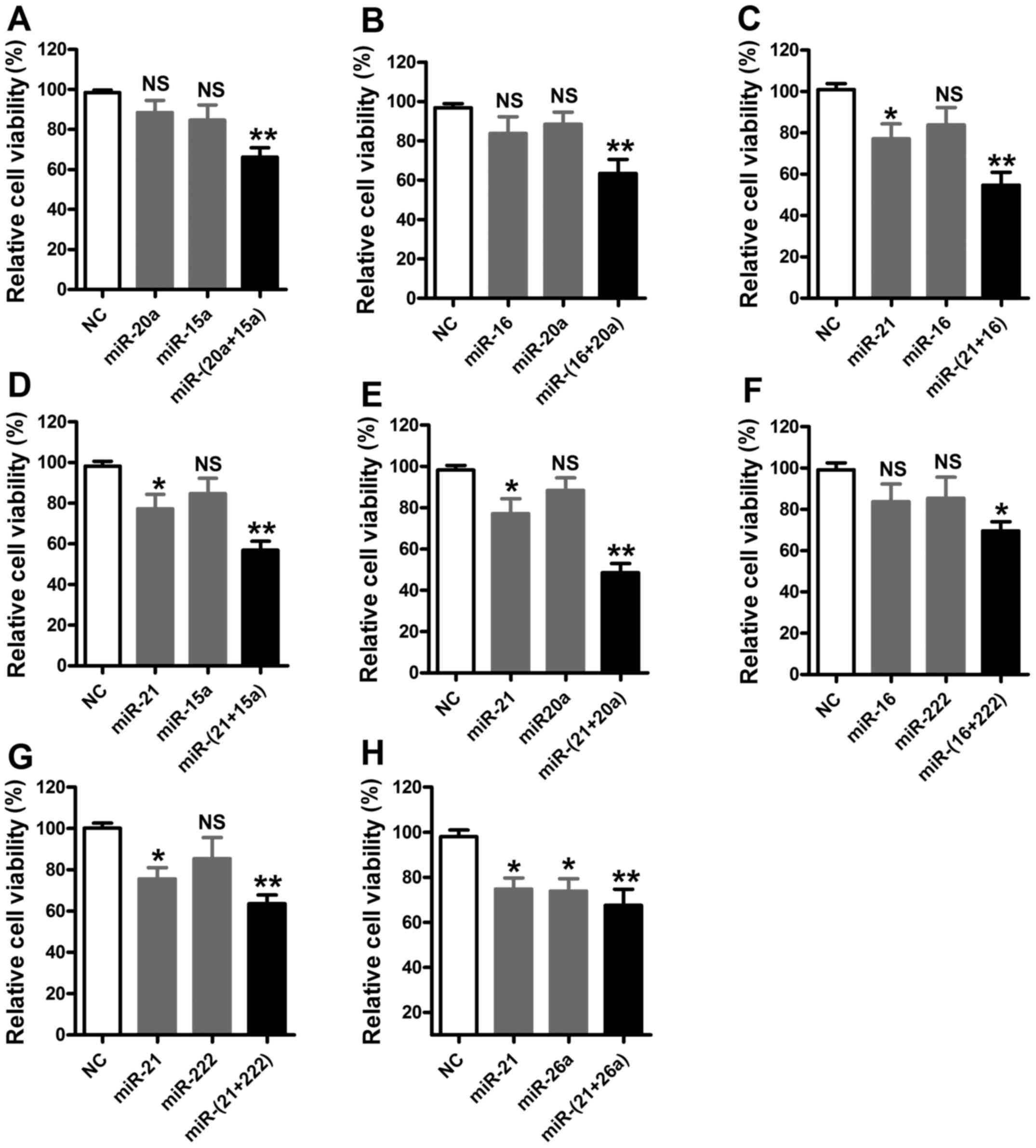 | Figure 3.Results of synergistic effects of
miRNA inhibitors (n=6). MTT assays of cells transfected with the
miRNA pairs (A) miR-20a:miR-15a, (B) miR-16:miR-20a, (C)
miR-21:miR-16, (D) miR-21:miR-15a, (E) miR-21:miR-20a, (F)
miR-16:miR-222, (G) miR-21:miR-222 and (H) miR-21:miR-26a. All
miRNAs were used at a concentration of 30 nM. *P<0.05,
**P<0.01 vs. NC. miRNA, microRNA; NC, negative control; NS, not
significant. |
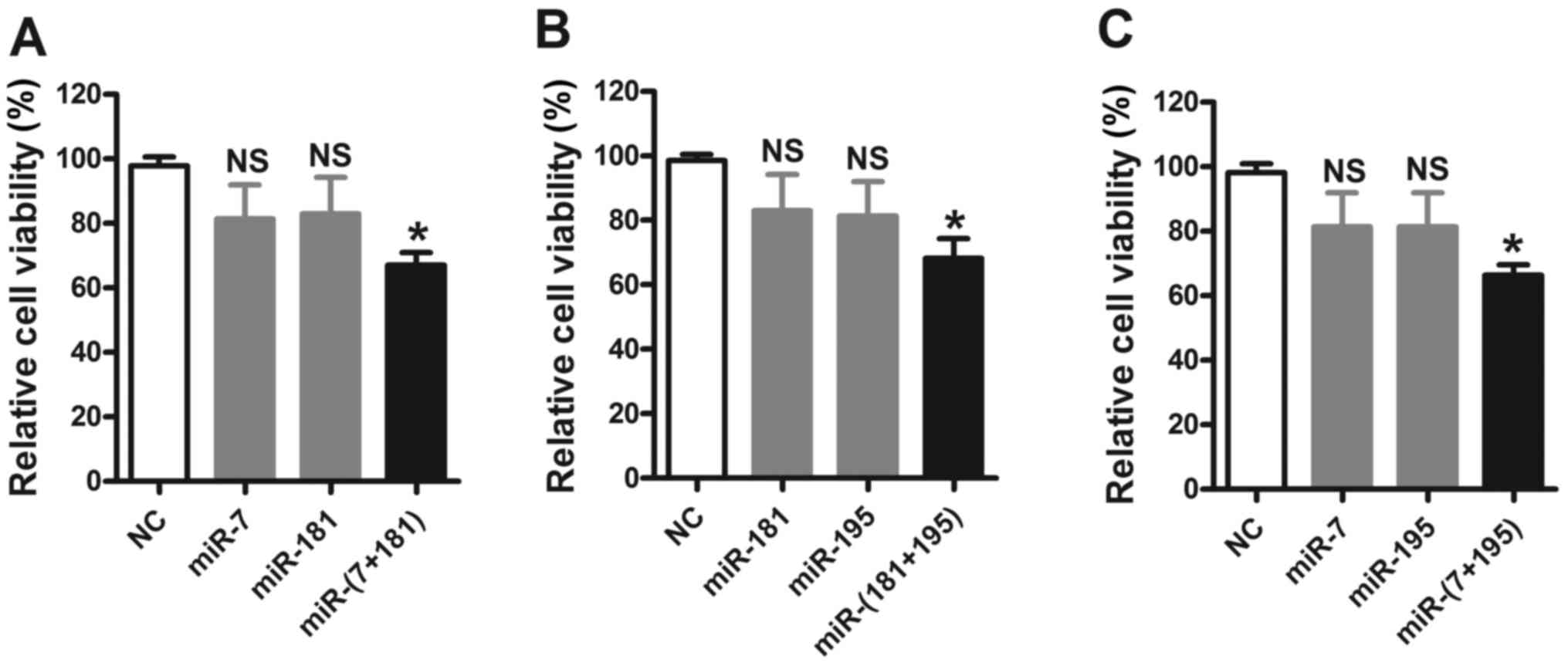
Following investigation of the synergism of
upregulated miRNAs, downregulated miRNAs, miR-7, miR-181 and
miR-195, were investigated as miRNA pairs. It was observed that,
although cotransfection with miR-7/181, miR-7/195, and miR-181/195
mimics resulted in marked reductions in cell viability compared
with that in the NC group, the effects were additive rather than
synergistic effects (Fig. 4).
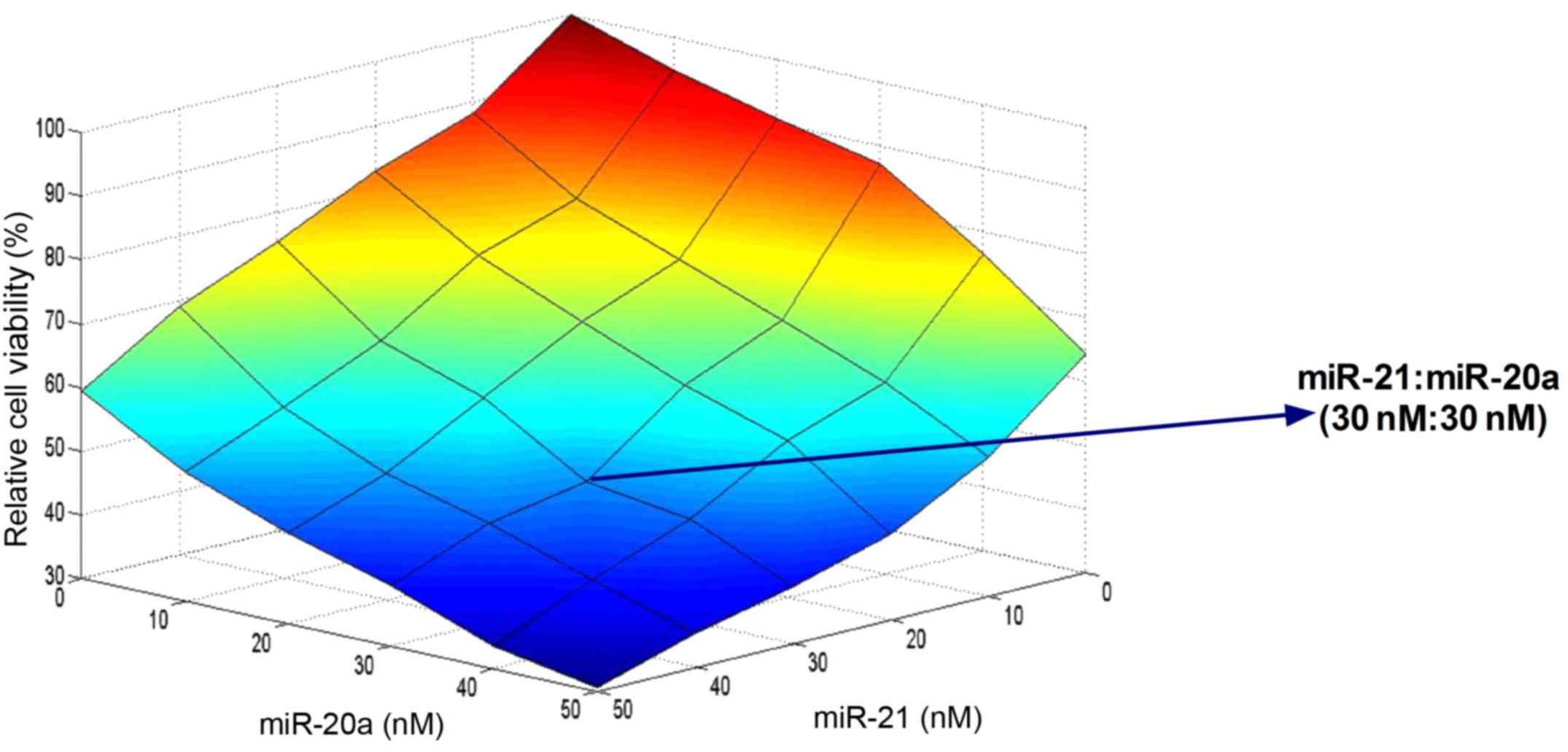
Screening of the potent synergistic
effects of miR-21 and miR-20a
Following identification of the potent synergistic
effects of miR-21 and miR-20a, the synergistic effects were further
investigated at different concentrations and concentration ratios
in the range of 10–50 nM. Cell viability assays and heat map
analyses demonstrated that with the same transfection
concentrations, miR-21 exerted increased effects on cells compared
with miR-20a. The miRNA pair of miR-21 (30 nM) and miR-20a (30 nM)
exhibited the most marked inhibitory effect on cell viability
(Fig. 5).
Synergistic effect of miR-21 and
miR-20a cotransfection on apoptosis in U87 cells
In order to further investigate the synergistic
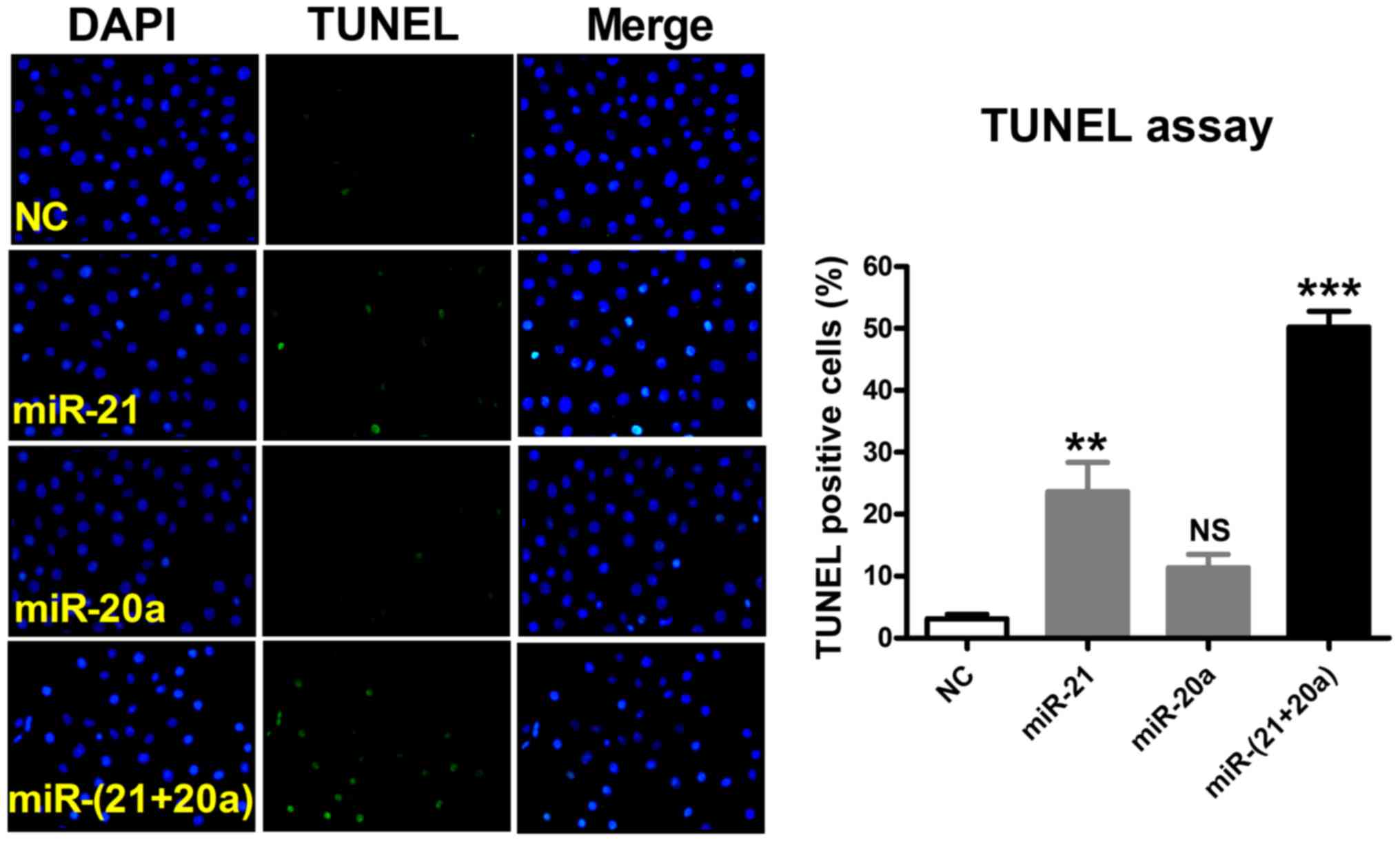
effects of miR-21 and miR-20a, TUNEL staining was used for
qualitative observation of apoptotic cells and the percentage of
apoptotic cells was quantified using Image-Pro Plus software.
Compared with the NC group, transfection with a miR-21 inhibitor
significantly promoted apoptosis in U87 cells, and transfection
with miR-20a promoted apoptosis to a limited degree. By contrast,
cotransfection with miR-21 and miR-20a markedly promoted apoptosis
with an evident synergistic effect (Fig. 6).
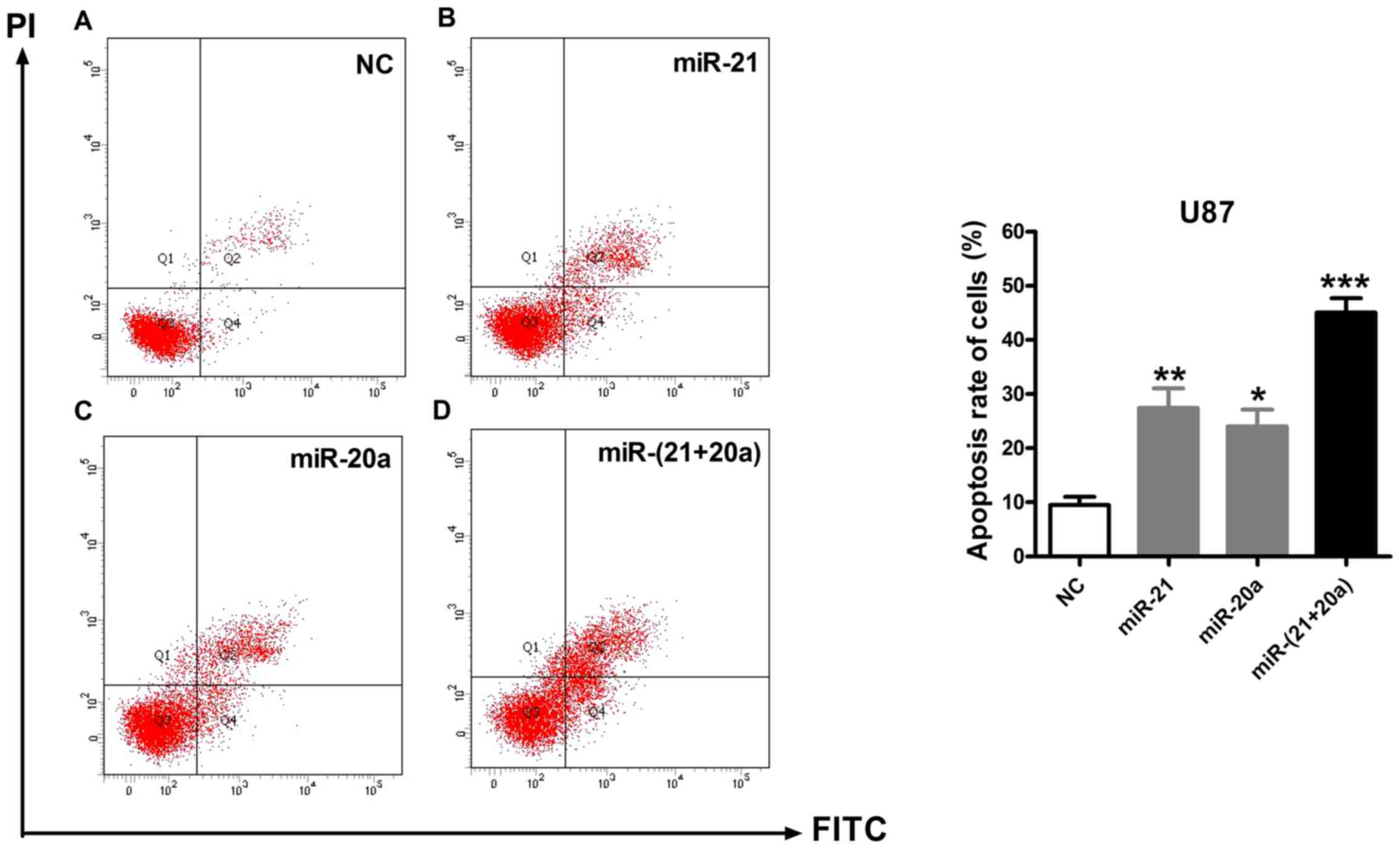
Flow cytometry was performed to quantitatively
analyze the types and proportions of apoptotic cells. As presented
in Fig. 7, compared with the NC
group, in which the apoptotic rate was 9.5% [quadrant (Q)2, 7.0%;
Q4, 2.5%], transfection with miR-21 or miR-20a alone markedly
promoted apoptosis in U87 cells, leading to an average apoptotic
rate of 27.4% (Q2, 21.7%; Q4, 5.7%) and 24.0% (Q2, 17.7%; Q4,
6.3%), respectively, whereas cotransfection with miR-21 and miR-20a
yielded an average apoptotic rate of 45.0% (Q2, 36.2%; Q4, 8.8%).
Flow cytometric analysis demonstrated that cotransfection with
miR-21 and miR-20a promoted apoptosis, particularly early
apoptosis, in U87 cells.
Discussion
In previous studies, researchers have confirmed the
important regulatory effects of miRNAs in glioma and identified
aberrantly expressed miRNAs through analysis and validation of
miRNAs in blood from tumor tissue and cerebrospinal fluid (23–26).
Based on the multi-target regulatory mechanism of miRNAs and
long-term study results, it is hypothesized that rational
intervention of aberrantly expressed miRNAs may become an important
approach to glioma treatment and prevention. Therefore, further
analysis of abnormally-expressed miRNAs in brain glioma is of
importance to the treatment of glioma.
In the present study, nine abnormal miRNAs in brain
glioma cells were selected, and their individual and synergistic
effects were examined and compared. The results of the present
study demonstrated that the individual regulatory effects of these
nine miRNAs on glioma cells are consistent with the results of
previous studies (13,27–30).
Additionally, the preliminary systems biology approach in the
present study indicated that combinations of miRNAs exhibited
certain synergistic regulatory effects. The synergistic regulatory
effects of miRNAs were experimentally confirmed at the cellular
level, and the results were consistent with the results of
bioinformatic predictions (20).
The present study demonstrated that miR-21 and miR-20a exhibited
the largest synergistic effects among all the tested miRNA pairs;
the combination of low-concentration (30 nM) miR-21 and miR-20a
inhibitors promoted apoptosis in U87 cells, increased the ratio of
early and late apoptotic cells, and exerted combined effects that
were increased compared with the sum of the individual effects.
The synergistic effects of miRNAs have been
demonstrated in the treatment of breast cancer and non-small cell
lung cancer (NSCLC). Kasinski et al (31) demonstrated that the combined
application of miR-34a and let-7miR precursor mimics was able to
improve the efficacy of treatment for NSCLC, and Stahlhut and Slack
(32) demonstrated that it was
able to further enhance the sensitivity of NSCLC to erlotinib
treatment. Devulapally et al (33) demonstrated that the combined use of
miR-21 and miR-10b inhibitors was able to increase the inhibitory
effects of the miRNAs on breast cancer cells and promote cancer
cell apoptosis. These previous experimental results are consistent
with the results of the present study. In addition to the
synergistic interference of miRNAs in tumor cells, synergistic
regulatory effects of miRNAs have been observed in the regulation
of cardiovascular diseases (20).
Compared with single drug administration targeting a
single miRNA, selecting two low-dose miRNAs for synergistic
interference has advantages. Synergistic interference is able to
increase the effects of the treatment and, additionally, reduce the
dose of any single miRNA in order to avoid the off-target effects
of miRNAs and achieve multi-target, high-efficiency regulatory
effects. However, there were limitations to the present study. The
analyses were only performed at the cellular level; detailed
studies and animal experiments are required in order to determine
the mechanisms of miRNA synergistic regulation. Additionally, the
miRNA delivery system used in the present study is associated with
certain side effects; therefore, in order to achieve targeted and
stable interference effects, drug delivery systems and materials
suitable for miRNAs need to be developed. In previous studies,
encapsulation of miRNAs with high-molecular-weight polymers,
including poly lactic-co-glycolic acid and polyethylene glycol, has
been observed to increase the in vivo stability and
targeting of miRNAs (33–35).
In conclusion, in the present study, it was observed
that there exist certain miRNA combinations, which are able to
exert synergistic tumor-inhibitory effects on the regulation of
apoptosis in brain glioma cells. Of the miRNAs examined, the
combined use of miR-21 and miR-20a inhibitors demonstrated the
clearest synergistic pro-apoptotic effects. The present study
supports the development of rational, effective and novel noncoding
RNA-based treatment strategies, for the prevention and treatment of
glioma.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81272788, 81472368
and 31301095), the Science Foundation of the Health Department of
Heilongjiang Province (grant no. 2014301), the Science Foundation
of the First Affiliated Hospital of Harbin University (grant no.
2015B014), and the Harbin medical university scientific research
innovation fund (No 2016LCZX68).
References
|
1
|
Gladson CL, Prayson RA and Liu WM: The
pathobiology of glioma tumors. Annu Rev Pathol. 5:33–50. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 15
Suppl 2:ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Morgan LL: The epidemiology of glioma in
adults: A ‘state of the science’ review. Neuro Oncol. 17:623–624.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hervey-Jumper SL and Berger MS: Maximizing
safe resection of low- and high-grade glioma. J Neuro Oncol.
130:pp269–282. 2016. View Article : Google Scholar
|
|
6
|
Stavrovskaya AA, Shushanov SS and
Rybalkina EY: Problems of glioblastoma multiforme drug resistance.
Biochemistry (Mosc). 81:91–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang C, Li C, Li J, Han J, Shang D, Zhang
Y, Zhang W, Yao Q, Han L, Xu Y, et al: Identification of
miRNA-mediated core gene module for glioma patient prediction by
integrating high-throughput miRNA, mRNA expression and pathway
structure. PLoS One. 9:e969082014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adlakha YK and Saini N: Brain microRNAs
and insights into biological functions and therapeutic potential of
brain enriched miRNA-128. Mol Cancer. 13:332014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Onishi M, Ichikawa T, Kurozumi K and Date
I: Angiogenesis and invasion in glioma. Brain Tumor Patho.
28:pp13–24. 2011. View Article : Google Scholar
|
|
10
|
Gaur AB, Holbeck SL, Colburn NH and Israel
MA: Downregulation of Pdcd4 by mir-21 facilitates glioblastoma
proliferation in vivo. Neuro Oncol. 13:580–590. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Zhao S, Zhen Y, Li Q, Teng L, Asai A
and Kawamoto K: A miR-21 inhibitor enhances apoptosis and reduces
G(2)-M accumulation induced by ionizing radiation in human
glioblastoma U251 cells. Brain Tumor Pathol. 28:209–214. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang C, Zhang J, Hao J, Shi Z, Wang Y,
Han L, Yu S, You Y, Jiang T, Wang J, et al: High level of
miR-221/222 confers increased cell invasion and poor prognosis in
glioma. J Transl Med. 10:e1192012. View Article : Google Scholar
|
|
13
|
Ernst A, Campos B, Meier J, Devens F,
Liesenberg F, Wolter M, Reifenberger G, Herold-Mende C, Lichter P
and Radlwimmer B: De-repression of CTGF via the miR-17-92 cluster
upon differentiation of human glioblastoma spheroid cultures.
Oncogene. 29:3411–3422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Costa PM, Cardoso AL, Mano M and de Lima
MC: MicroRNAs in glioblastoma: Role in pathogenesis and
opportunities for targeted therapies. CNS Neurol Disord Drug
Targets. 14:222–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu DG, Wang YY, Fan LG, Luo H, Han B, Sun
LH, Wang XF, Zhang JX, Cao L, Wang XR, et al: MicroRNA-7 regulates
glioblastoma cell invasion via targeting focal adhesion kinase
expression. Chin Med J (Engl). 124:2616–2621. 2011.PubMed/NCBI
|
|
16
|
Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P
and Hu W: MicroRNA-181a sensitizes human malignant glioma U87MG
cells to radiation by targeting Bcl-2. Oncol Rep. 23:997–1003.
2010.PubMed/NCBI
|
|
17
|
Gabriely G, Wurdinger T, Kesari S, Esau
CC, Burchard J, Linsley PS and Krichevsky AM: MicroRNA 21 promotes
glioma invasion by targeting matrix metalloproteinase regulators.
Mol Cell Biol. 28:5369–5380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu J, Li CX, Li YS, Lv JY, Ma Y, Shao TT,
Xu LD, Wang YY, Du L, Zhang YP, et al: MiRNA-miRNA synergistic
network: Construction via co-regulating functional modules and
disease miRNA topological features. Nucleic Acids Res. 39:825–836.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu S, Huang M, Nguyen PK, Gong Y, Li Z,
Jia F, Lan F, Liu J, Nag D, Robbins RC and Wu JC: Novel microRNA
prosurvival cocktail for improving engraftment and function of
cardiac progenitor cell transplantation. Circulation. 124 11
Suppl:S27–S34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu W, Zhao Y, Xu Y, Sun Y, Wang Z, Yuan W
and Du Z: Dissection of protein interactomics highlights microRNA
synergy. PLoS One. 8:e633422013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Singh S, Narang AS and Mahato RI:
Subcellular fate and off-target effects of siRNA, shRNA, and miRNA.
Pharm Res. 28:2996–3015. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Dutta A and Abounader R: The role
of microRNAs in glioma initiation and progression. Front Biosci
(Landmark Ed). 17:700–712. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Codo P, Weller M, Meister G, Szabo E,
Steinle A, Wolter M, Reifenberger G and Roth P: MicroRNA mediated
down-regulation of NKG2D ligands contributes to glioma immune
escape. Oncotarget. 5:7651–7662. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Visani M, de Biase D, Marucci G, Cerasoli
S, Nigrisoli E, Reggiani ML Bacchi, Albani F, Baruzzi A and Pession
A; PERNO study group, : Expression of 19 microRNAs in glioblastoma
and comparison with other brain neoplasia of grades I–III. Mol
Oncol. 8:417–430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feng SY, Dong CG, Wu K, Wang XG, Qiao J
and Shao JF: Lentiviral expression of anti-microRNAs targeting
miR-27a inhibits proliferation and invasiveness of U87 glioma
cells. Mol Med Rep. 6:275–281. 2012.PubMed/NCBI
|
|
27
|
Shi R, Wang PY, Li XY, Chen JX, Li Y,
Zhang XZ, Zhang CG, Jiang T, Li WB, Ding W and Cheng SJ: Exosomal
levels of miRNA-21 from cerebrospinal fluids associated with poor
prognosis and tumor recurrence of glioma patients. Oncotarget.
6:26971–26981. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gwak HS, Kim TH, Jo GH, Kim YJ, Kwak HJ,
Kim JH, Yin J, Yoo H, Lee SH and Park JB: Silencing of microRNA-21
confers radio-sensitivity through inhibition of the PI3K/AKT
pathway and enhancing autophagy in malignant glioma cell lines.
PLoS One. 7:e474492012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han L, Yue X, Zhou X, Lan FM, You G, Zhang
W, Zhang KL, Zhang CZ, Cheng JQ, Yu SZ, et al: MicroRNA-21
expression is regulated by β-catenin/STAT3 pathway and promotes
glioma cell invasion by direct targeting RECK. CNS Neurosci Ther.
18:573–583. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yao X, Li W, Wang Y, Ding Z, Hou W, Zeng
M, Deng G, Zhang J and Yang H: Expression of hsa-miR-20a in human
glioma tissues and its effect on the proliferation of human glioma
cells in vitro. Nan Fang Yi Ke Da Xue Xue Bao. 32:198–201. 2012.(In
Chinese). PubMed/NCBI
|
|
31
|
Kasinski AL, Kelnar K, Stahlhut C,
Orellana E, Zhao J, Shimer E, Dysart S, Chen X, Bader AG and Slack
FJ: A combinatorial microRNA therapeutics approach to suppressing
non-small cell lung cancer. Oncogene. 34:3547–3555. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stahlhut C and Slack FJ: Combinatorial
action of MicroRNAs let-7 and miR-34 Effectively synergizes with
erlotinib to suppress non-small cell lung cancer cell
proliferation. Cell Cycle. 14:2171–2180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Devulapally R, Sekar NM, Sekar TV, Foygel
K, Massoud TF, Willmann JK and Paulmurugan R: Polymer nanoparticles
mediated codelivery of antimiR-10b and antimiR-21 for achieving
triple negative breast cancer therapy. ACS Nano. 9:2290–2302. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jin H, Yu Y, Chrisler WB, Xiong Y, Hu D
and Lei C: Delivery of MicroRNA-10b with polylysine nanoparticles
for inhibition of breast cancer cell wound healing. Breast Cancer
(Auckl). 6:9–19. 2012.PubMed/NCBI
|
|
35
|
Fernandez-Fernandez A, Manchanda R and
McGoron AJ: Theranostic applications of nanomaterials in cancer:
Drug delivery, image-guided therapy, and multifunctional platforms.
Appl Biochem Biotechnol. 165:1628–1651. 2011. View Article : Google Scholar : PubMed/NCBI
|