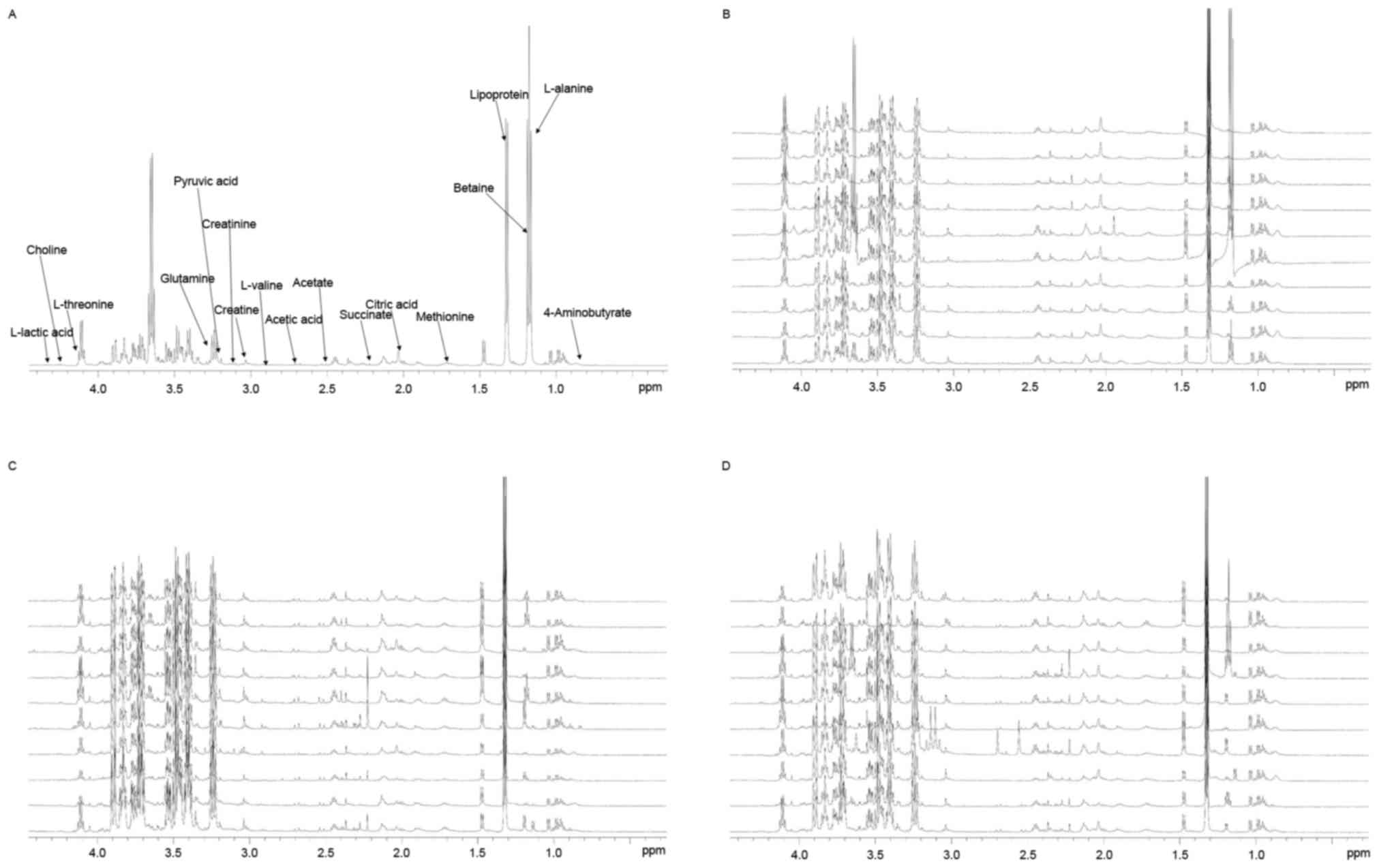
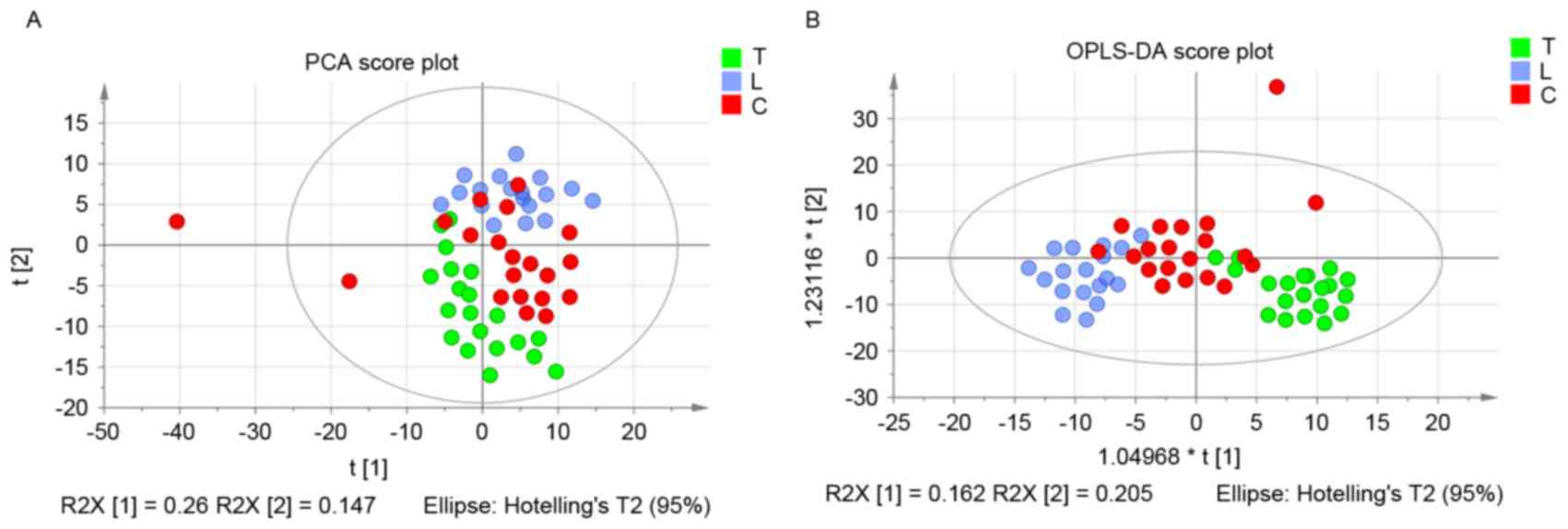
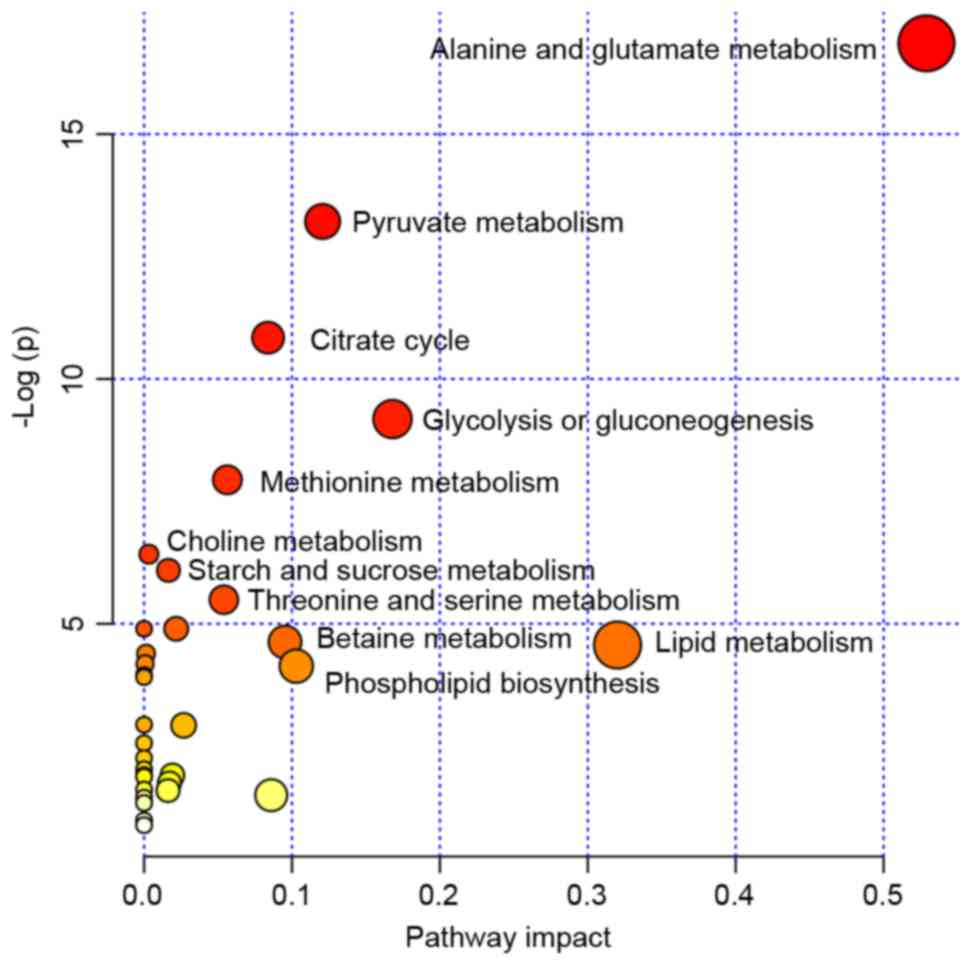
|
1
|
Maskell N: British Thoracic Society
Pleural Disease Guideline Group: British thoracic society pleural
disease guidelines-2010 update. Thorax. 65:667–669. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Light RW: Clinical practice: Pleural
effusion. N Engl J Med. 346:1971–1977. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Daniil ZD, Zintzaras E, Kiropoulos T,
Papaioannou AI, Koutsokera A, Kastanis A and Gourgoulianis KI:
Discrimination of exudative pleural effusions based on multiple
biological parameters. Eur Respir J. 30:957–964. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
World Health Organization, . Global
Tuberculosis report. Geneva, Switzerland: 2015
|
|
5
|
Light RW: Update on tuberculous pleural
effusion. Respirology. 15:451–458. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dodd PJ, Gardiner E, Coghlan R and Seddon
JA: Burden of childhood tuberculosis in 22 high-burden countries: A
mathematical modelling study. Lancet Glob Health. 2:e453–e459.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nicholson JK, Lindon JC and Holmes E:
‘Metabonomics’: Understanding the metabolic responses of living
systems to pathophysiological stimuli via multivariate statistical
analysis of biological NMR spectroscopic data. Xenobiotica.
29:1181–1189. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmidt CW: Metabolomics: What's happening
downstream of DNA. Environ Health Perspect. 112:A410–A415. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Illig T, Gieger C, Zhai G, Römisch-Margl
W, Wang-Sattler R, Prehn C, Altmaier E, Kastenmüller G, Kato BS,
Mewes HW, et al: A genome-wide perspective of genetic variation
inhuman metabolism. Nat Genet. 42:137–141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang J, Xu G, Zheng Y, Kong H, Pang T, Lv
S and Yang Q: Diagnosis of liver cancer using HPLC-based
metabonomics avoiding false-positive result from hepatitis and
hepatocirrhosis diseases. J Chromatogr B Analyt Technol Biomed Life
Sci. 813:59–65. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen J, Zhang X, Cao R, Lu X, Zhao S,
Fekete A, Huang Q, Schmitt-Kopplin P, Wang Y, Xu Z, et al: Serum
27-nor-5β-cholestane-3,7,12,24,25 pentol glucuronide discovered by
metabolomics as potential diagnostic biomarker for epithelium
ovarian cancer. J Proteome Res. 10:2625–2632. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hodavance MS, Ralston SL and Pelczer I:
Beyond blood sugar: The potential of NMR-based metabonomics for
type 2 human diabetes, and the horse as a possible model. Anal
Bioanal Chem. 387:533–537. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mäkinen VP, Soininen P, Forsblom C,
Parkkonen M, Ingman P, Kaski K, Groop PH and Ala-Korpela M:
FinnDiane Study Group: Diagnosing diabetic nephropathy by 1H NMR
metabonomics of serum. Magma. 19:281–296. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Messana I, Forni F, Ferrari F, Rossi C,
Giardina B and Zuppi C: Proton nuclear magnetic resonance spectral
profiles of urine in type II diabetic patients. Clin Chem.
44:1529–1534. 1998.PubMed/NCBI
|
|
15
|
Brindle JT, Nicholson JK, Schofield PM,
Grainger DJ and Holmes E: Application of chemometrics to 1H NMR
spectroscopic data to investigate a relationship between human
serum metabolic profiles and hypertension. Analyst. 128:32–36.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brindle JT, Antti H, Holmes E, Tranter G,
Nicholson JK, Bethell HW, Clarke S, Schofield PM, McKilligin E,
Mosedale DE and Grainger DJ: Rapid and noninvasive diagnosis of the
presence and severity of coronary heart disease using 1H-NMR-based
metabonomics. Nat Med. 8:1439–1444. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sabatine MS, Liu E, Morrow DA, Heller E,
Mc Carroll R, Wiegand R, Berriz GF, Roth FP and Gerszten RE:
Metabolonmic identification of novel biomarkers of myocardial
ischemia. Circulation. 112:3868–3875. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barba I, de León G, Martín E, Cuevas A,
Aguade S, Candell-Riera J, Barrabés JA and Garcia-Dorado D: Nuclear
magnetic resonance-based metabolomics predicts exercise-induced
ischemia in patients with suspected coronary artery disease. Magn
Reson Med. 60:27–32. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun L, Hu W, Liu Q, Hao Q, Sun B, Zhang Q,
Mao S, Qiao J and Yan X: Metabonomics reveals plasma metabolic
changes and inflammatory marker in polycystic ovary syndrome
patients. J Proteome Res. 11:2937–2946. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng P, Gao HC, Li Q, Shao WH, Zhang ML,
Cheng K, Yang DY, Fan SH, Chen L, Fang L and Xie P: Plasma
metabonomics as novel diagnostic approach for major depressive
disorder. J Proteome Res. 11:1741–1748. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia J, Mandal R, Sinelnikov IV, Broadhurst
D and Wishart DS: MetaboAnalyst 2.0-a comprehensive server for
metabolomic data analysis. Nucleic Acids Res. 40:W127–W133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xia J and Wishart DS: Metabolomic data
processing, analysis, and interpretation using MetaboAnalyst. Curr
Protoc Bioinformatics Chapter. 14:Unit 14.102011.
|
|
23
|
Yang WB, Liang QL, Ye ZJ, Niu CM, Ma WL,
Xiong XZ, Du RH, Zhou Q, Zhang JC and Shi HZ: Cell origins and
diagnostic accuracy of interleukin 27 in pleural effusions. PLoS
One. 7:e404502012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong X and Yang J: High IL-35 pleural
expression in patients with tuberculous pleural effusion. Med Sci
Monit. 21:1261–1268. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garrido V Villena, Viedma E Cases,
Fernández Villar A, de Pablo Gafas A, Pérez Rodríguez E, Pérez JM
Porcel, Rodríguez Panadero F, Martínez C Ruiz, Velázquez A
Salvatierra and Valdés Cuadrado L: Recommendations of diagnosis and
treatment of pleural effusion. Update. Arch Bronconeumol.
50:235–249. 2014.(In English, Spanish). View Article : Google Scholar
|
|
26
|
Frediani JK, Jones DP, Tukvadze N, Uppal
K, Sanikidze E, Kipiani M, Tran VT, Hebbar G, Walker DI, Kempker
RR, et al: Plasma metabolomics in human pulmonary tuberculosis
disease: A pilot study. PLoS One. 9:e1088542014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deja S, Porebska I, Kowal A, Zabek A, Barg
W, Pawelczyk K, Stanimirova I, Daszykowski M, Korzeniewska A,
Jankowska R and Mlynarz P: Metabolomics provide new insights on
lung cancer staging and discrimination from chronic obstructive
pulmonary disease. J Pharm Biomed Anal. 100:369–380. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nandakumar M, Nathan C and Rhee KY:
Isocitrate lyase mediates broad antibiotic tolerance in
Mycobacterium tuberculosis. Nat Commun. 5:43062014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eoh H and Rhee KY: Multifunctional
essentiality of succinate metabolism in adaptation to hypoxia in
Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 110:6554–6559.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Micklinghoff JC, Breitinger KJ, Schmidt M,
Geffers R, Eikmanns BJ and Bange FC: Role of the transcriptional
regulator RamB (Rv0465c) in the control of the glyoxylate cycle in
Mycobacterium tuberculosis. J Bacteriol. 191:7260–7269. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kusao I, Troelstrup D and Shiramizu B:
Possible mitochondria-associated enzymatic role in Non-Hodgkin
lymphoma residual disease. Cancer Growth Metastasis. 1:3–8.
2008.PubMed/NCBI
|
|
32
|
Schlichtholz B, Turyn J, Goyke E,
Biernacki M, Jaskiewicz K, Sledzinski Z and Swierczynski J:
Enhanced citrate synthase activity in human pancreatic cancer.
Pancreas. 30:99–104. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qiu Y, Cai G, Su M, Chen T, Liu Y, Xu Y,
Ni Y, Zhao A, Cai S, Xu LX and Jia W: Urinary metabonomic study on
colorectal cancer. J Proteome Res Mar. 9:1627–1634. 2010.
View Article : Google Scholar
|
|
34
|
Chen L, Liu T, Zhou J, Wang Y, Wang X, Di
W and Zhang S: Citrate synthase expression affects tumor phenotype
and drug resistance in human ovarian carcinoma. PLoS One.
9:e1157082014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chajès V, Cambot M, Moreau K, Lenoir GM
and Joulin V: Acetyl-CoA carboxylase alpha is essential to breast
cancer cell survival. Cancer Res. 66:5287–5294. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brusselmans K, de Schrijver E, Verhoeven G
and Swinnen JV: RNA interference-mediated silencing of the
acetyl-CoA-carboxylase-α gene induces growth inhibition and
apoptosis of prostate cancer cells. Cancer Res. 65:6719–6725. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kayser G, Kassem A, Sienel W,
Schulte-Uentrop L, Mattern D, Aumann K, Stickeler E, Werner M,
Passlick B and zur Hausen A: Lactate-dehydrogenase 5 is
overexpressed in non-small cell lung cancer and correlates with the
expression of the transketolase-like protein 1. Diagn Pathol.
5:222010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen JQ and Russo J: Dysregulation of
glucose transport, glycolysis, TCA cycle and glutaminolysis by
oncogenes and tumor suppressors in cancer cells. Biochim Biophys
Acta. 1826:370–384. 2012.PubMed/NCBI
|
|
40
|
Gilmore SA, Schelle MW, Holsclaw CM, Leigh
CD, Jain M, Cox JS, Leary JA and Bertozzi CR: Sulfolipid-1
biosynthesis restricts Mycobacterium tuberculosis growth in human
macrophages. ACS Chem Biol. 7:863–870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Brown AK, Bhatt A, Singh A, Saparia E,
Evans AF and Besra GS: Identification of the dehydratase component
of the mycobacterial mycolic acid-synthesizing fatty acid
synthase-II complex. Microbiology. 153:4166–4173. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Klink M, Brzezinska M, Szulc I, Brzostek
A, Kielbik M, Sulowska Z and Dziadek J: Cholesterol oxidase is
indispensable in the pathogenesis of Mycobacterium tuberculosis.
PLoS One. 8:e733332013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ouellet H, Johnston JB and de Montellano
PR: Cholesterol catabolism as a therapeutic target in Mycobacterium
tuberculosis. Trends Microbiol. 19:530–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Szatmari I and Nagy L: Nuclear receptor
signalling in dendritic cells connects lipids, the genome and
immune function. EMBO J. 27:2353–2362. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Almeida PE, Silva AR, Maya-Monteiro CM,
Töröcsik D, D'Avila H, Dezsö B, Magalhães KG, Castro-Faria-Neto HC,
Nagy L and Bozza PT: Mycobacterium bovis bacillus Calmette-Guérin
infection induces TLR2-dependent peroxisome proliferator-activated
receptor gamma expression and activation: Functions in
inflammation, lipid metabolism, and pathogenesis. J Immunol.
183:1337–1345. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen W, Zu Y, Huang Q, Chen F, Wang G, Lan
W, Bai C, Lu S, Yue Y and Deng F: Study on metabonomic
characteristics of human lung cancer using high resolution
magic-angle spinning 1H NMR spectroscopy and multivariate data
analysis. Magn Reson Med. 66:1531–1540. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Collins MD, Goodfellow M and Minnikin DE:
Fatty acid composition of some mycolic acid-containing coryneform
bacteria. J Gen Microbiol. 128:2503–2509. 1982.PubMed/NCBI
|
|
48
|
Baracos VE and Mackenzie ML:
Investigations of branched-chain amino acids and their metabolites
in animal models of cancer. J Nutr. 136 Suppl 1:S237–S242.
2006.
|
|
49
|
Choudry HA, Pan M, Karinch AM and Souba
WW: Branched-chain amino acid-enriched nutritional support in
surgical and cancer patients. J Nutr. 136 Suppl 1:S314–S318.
2006.
|
|
50
|
Liu B, Cui C, Duan W, Zhao M, Peng S, Wang
L, Liu H and Cui G: Synthesis and evaluation of anti-tumor
activities of N4 fatty acyl amino acid derivatives of
1-beta-arabinofuranosylcytosine. Eur Med Chem. 44:3596–3600. 2009.
View Article : Google Scholar
|