Introduction
Microvascular related diseases are the most frequent
complications of diabetes (1);
especially diabetic foot, which afflicts ~10% of people with
diabetes (2). The vascular
endothelium is considered to serve an essential role in
diabetes-associated microvascular dysfunction, especially diabetic
foot. The endothelium serve a significant role in the regulation of
microvascular function and development of physiological and
pathophysiological inflammation. Endothelial cell (EC) injury is a
critical element of diabetic foot (2). Previous studies have reported that
high blood glucose induces EC apoptosis, and causes cellular
dysfunction and cell death (1).
However, the vascular function pathogenesis is complicated and
there are a number of signaling pathways involved, including the
Notch pathway.
The Notch1 signaling pathway is significant in cell
differentiation, primarily determining and regulating cell survival
(3). In mammals, four receptors
(Notch1-Notch4) and five ligands, Jagged1, Jagged2, Delta-like
(Dl)1, Dll3 and Dll4, have been discovered (4,5).
Dysregulation or loss of Notch signaling underlies a wide range of
human disorders (6). A previous
study indicates that Notch signals inhibit the development of
erythroid/megakaryocytic cells by suppressing GATA-1 activity
through the induction of Hes1 (7),
Mutations in Notch1 cause aortic valve disease (8).
The Notch/Hes1 pathway has been identified to play a
key role in mediating ECs function (8,9),
participating in regulating angiogenesis, development and vascular
inflammation reaction related pathophysiology process. DLL4-Notch
signaling has a key role in regulation of tumor angiogenesis
(4,10), and serve an important role in
placental angiogenesis (11).
Previous studies demonstrated that vaccarin attenuates the human
EA.hy926 ECs oxidative stress injury through inhibition of Notch
signaling (12). Another report
indicated that Dll4 signaling through Notch1 regulates formation of
tip cells during angiogenesis (4).
Hyperglycemia can cause inflammation and oxidative
stress, leading to EC damage. It has also been demonstrated to
mediate NF-κB activation, increase Nox4 expression, and increase
inflammatory cytokine activation in vascular smooth muscle
(13,14). Furthermore, hyperglycemia also
causes the Notch signal pathway to be aberrantly activated
(15).
MicroRNAs (miRNAs) are endogenous conservative
noncoding small RNAs of 21–25 nucleotides and through binding to
3′-untranslated regions (3′UTRs) of the target mRNA (16), usually resulting in translational
repression or target mRNA degradation and gene silencing. miRNAs
regulate many cellular biological activities ranging from cell
growth and differentiation, apoptosis, metabolism and angiogenesis.
Studies demonstrated that hyperglycemia may cause abnormal high
expression of clusters of miRNAs affecting a series of
pathophysiology process (17). In
addition, series of miRNAs serve an important role in the
hyperglycemia-related pathophysiology process (18).
miR-200c is upregulated by oxidative stress and
induces EC apoptosis and senescence via zinc finger E-box-binding
homeobox (ZEB)1 inhibition (19).
miR-200c may regulate vascular endothelial growth factor A, ZEBs,
fms related tyrosine kinase 1, I kappa B kinase β, Krüppel-like
factor 9, fibulin-5 (8) or ZEB1
(20) to participate in regulating
EC function. The present study predicted that miR-200c and Notch1
had complementary pairing in promoter regions by using the computer
software TargetScan version 7.1. Hyperglycemia-induced vascular
lesions are the leading cause of diabetic lower limb ischemia.
Previous studies have identified that miR-200c was significantly
increased expression in the serum of type 2 diabetes patients with
critical limb ischemia, which may suggest that miR-200c is possibly
related to regulating mechanism of diabetic vascular endothelial
injury. However, how miR-200c is involved in the diabetic vascular
injury has never been reported.
The miR-200 family includes miR-200a, miR-200b,
miR-200c, miR-429 and miR-141; they have emerged as noticeable
markers for predicting cancer prognosis and tumors progression.
miR-200c is a tumor suppressor in various cancers, such as
pancreatic, gastric and breast cancer through the inhibition of
Kirsten rat sarcoma viral oncogene homolog (KRAS) (21,22),
and the mediation of Leydig tumor cells and murine adrenocortical
tumor cells by vimentin. Another study using luciferase reporter
plasmids observed that miR-200c inhibited the AKT and ERK pathways
by directly targeting KRAS. It is suggested that miR-200c may be
related to normal cells survival and function, rather than
malignant cells. A previous study indicated that miR-200c served an
important role in EC differentiation and vasculogenesis by
targeting the transcription repressor ZEB1 (18).
The purpose of the present study was to investigate
how high glucose mediated miR-200c and Notch1, and whether miR-200c
can regulate EC differentiation and function via the Notch1/Hes1
signal pathway in vitro, in addition to the role of miR-200c
in H5V apoptosis under high glucose conditions.
Materials and methods
Cell lines and cell culture
H5V cells, which are murine heart endothelial
immortalized cells (23) (kindly
provided by Nanjing Medical University (Nanjing, China), were
cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, USA)
containing fetal calf serum 10% (v/v), 100 U/ml penicillin and 100
µg/ml streptomycin, in a humidified incubator at 37°C with 5%
CO2.
miRNA overexpression
An miRNA mimic (a synthetic RNA oligonucleotide
duplex mimicking miRNA precursor) purchased from Suzhou GenePharma
Co., Ltd, (Suzhou, China) was used for overexpression of miRNA. The
synthetic RNA molecules were purchased from Shanghai GenePharma
Co., Ltd. (Shanghai, China) including pre-miR-200c and
pre-miR-control (scrambled negative control RNA). H5V cells were
seeded in six-well plates (2×105) in complete medium
without antibiotics at least 24 h prior to transfection, when cell
confluence was ~70%, a final concentration of 50 nM
miRNA-200c-mimics, miRNA-200c-inhibitor and NC RNA were transfected
into the cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
instructions. A total of 6 h later, the culture medium was replaced
with DMEM supplemented with 2% fetal bovine serum, the cells
continued to be cultured for 24 or 48 h; they were then harvested
and processed for further analysis.
Western blot analysis
H5V cells were lysed in RIPA lysis buffer (Beyotime
Institute of Biotechnology, Shanghai, China), total protein was
separated by 10% SDS-PAGE and blotted onto polyvinylidene
difluoride membranes. The membrane was blocked with 5% skimmed milk
and incubated with primary antibodies (anti-Notch1; ab52627;
1:2,000; anti-Hes1; ab108937; 1:1,000, anti-GAPDH; ab181602;
1:5,000; all from Abcam, Cambridge, MA, USA) overnight at 4°C. It
was then incubated with horseradish peroxidase-conjugated
anti-rabbit or anti mouse secondary immunoglobulins (B0014K052600;
1:2,000; BioSharp, Anhui, China) for 2 h at room temperature. The
signal was then detected by enhanced chemiluminescence (Pierce;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. The data were quantified by ImageJ version 1.43 (National
Institutes of Health, Bethesda, MD, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted with the TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. cDNA was reverse transcribed using
RevertAid First Strand cDNA Synthesis Kit (Invitrogen; Thermo
Fisher Scientific, Inc.). Briefly, cDNA was synthesized using a
miR-200c specific primer (Sangon Biotech Co., Ltd., Shanghai,
China) in a reverse transcription system. The reaction conditions
were as follows: 16°C for 30 min, 42°C for 42 min, 85°C for 5 min.
Quantitative detection of reverse transcription products was
performed using specific sense and antisense primers of miR-200c
and SYBR Green I dye (Molecular Probes; Thermo Fisher Scientific,
Inc.). The PCR reaction condition was as follows: 95°C for 5 min,
94°C for 20 sec, 55°C for 20 sec, and 72°C for 20 sec, for 40
cycles, to obtain fluorescence intensity. U6 was used as an
internal control. The sequence of specific primer for miR-200c was:
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATGCACTGGATACGACTCCATC-3′;
miR-200c sense primer, 5′-TAATACTGCCGGGTAAT-3′; miR-200c antisense
primer, 5′-GTGCAGGGTCCGAGGT-3′. The Cq value was analyzed using the
RFQ-PCR 7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.)
analysis program version 2.0.6). Relative mRNA expression levels
were determined by the 2−ΔΔCq method (24) in comparison with control cells.
Cell survival assay
The cell suspension was inoculated in 96-well plates
(100 µl/well). The culture plate was maintained in the incubator
(37°C and 5% CO2). A total of 10 µl CCK (Shanghai Qcbio
Science & Technologies Co., Ltd., Shanghai, China) was added to
each well, and the culture plate was incubated in incubator for 1–4
h. Absorbance was measured at 450 nm with a microplate reader
(BioTex, Houston, TX, USA).
Immunofluorescence staining for Notch1
and Hes1
Cells were fixed with PBS containing 4%
paraformaldehyde (Sigma-Aldrich; Merck KGaA) for 20 min,
permeabilized and blocked with blocking buffer (Abcam) for 30 min.
Cells were then incubated with rat anti-mouse Notch1 primary
antibody (ab52627; 1:2,000; Abcam) at 4°C overnight, followed by
incubation with Alexa fluoro 555 conjugated secondary antibodies
(A32727; 1:1,000; Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C for 30 min. Nuclei were stained with DAPI (Sigma-Aldrich;
Merck KGaA). Images were acquired using a 50i Nikon fluorescence
microscope (Nikon Corporation, Tokyo, Japan). Images were processed
with Adobe Photoshop CS4 software version 11.0 (Adobe Systems,
Inc., San Jose, CA, USA). The proliferation rate referred to the
number of Notch/Hes1-positive cells divided by the number of
DAPI-stained cells.
Luciferase reporter assay
Using NCBI GeneBank database (https://www.ncbi.nlm.nih.gov/gene/), the authors
synthesized 3′ untranslated region (3′-UTR) sequences of target
genes at 100–120 nt in length containing the seed sequence. The
full-length of 3′-UTR of the Notch1 gene was amplified by PCR from
H5V genomic DNA and inserted into pGL3 control vector (Promega
Corporation, Madison, WI, USA). Using the Qiagen XL-site directed
Mutagenesis Kit (Qiagen, Valencia, CA, USA), the authors
constructed several inserts by deletions of 4 bp from the
complementarity site of the Notch1 gene. Lipofectamine 2000 was
used to transfect H5V cells when cell confluence reached 80% in a
24-well plate with a 0.5 µg firefly luciferase reporter vector
(Sangon Biotech Co., Ltd.) and 0.5 µg control vector containing
Renilla luciferase (Sangon Biotech Co., Ltd.), phRL-TK (Promega
Corporation) by Amaxa Nucleofector (Lonza Group, Basel,
Switzerland). Each Nucleofector used 50 nM of the miR-200c or a
scrambled oligonucleotide. At 48 h, the relative luciferase
activities of Firefly/Renilla were consecutively measured through
the dual luciferase assay (Promega Corporation). All data are
presented as mean ± standard deviation.
Statistical analysis
All experiments were performed at least three times.
Comparisons between two observations were analyzed by Student's
paired t-test, and multiple comparisons with a one- or two-way
analysis of variance. Multiple testing bias was assessed with the
Bonferroni test. Data are expressed as mean ± standard error of the
mean. P<0.05 was considered to indicate a statistically
significant difference.
Results
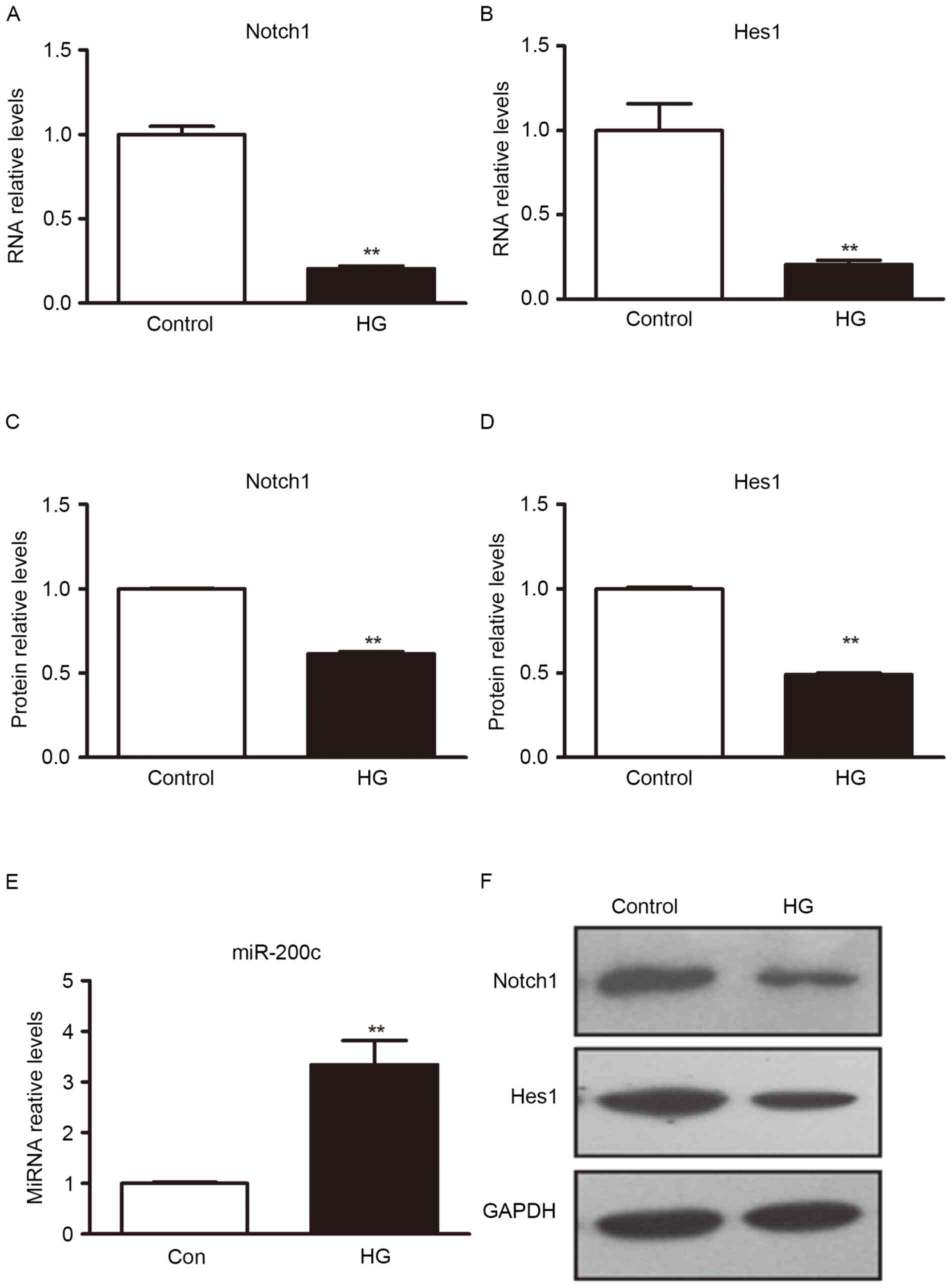
High glucose induced miR-200c
expression
Exposing ECs to high glucose is thought to mimic the
hyperglycemia of diabetic patients. miR-200c is upregulated by
oxidative stress and induces EC apoptosis and senescence via Notch1
inhibition (19), hyperglycemia is
the principal cause of induced oxidative stress (13). To investigate whether high glucose
was able to induce expression of miR-200c in H5V cells, the H5V
cells were cultured in different glucose concentration for
comparison. Cells were cultured under normal glucose (5 mM) or high
glucose (25 mM). Taking normal glucose as a control, the H5V cells
were cultured for 8 h and miRNAs expression level was determined
through RT-qPCR. When compared with cells in normal culture medium,
miR-200c was upregulated under high glucose conditions (Fig. 1).
Notch1 and Hes1 gene was repressed by
high glucose
The Notch1/Hes1 signal pathway serves an important
role in EC development and its function (6,8). To
determine how hyperglycemic stress affects Notch1 and Hes1, H5V
cells were exposed to high glucose conditions, with normal culture
medium as the control. Western blot analysis was used to detect
Notch1 and Hes1 protein expression. RT-qPCR was used to detect
Notch1 and Hes1 mRNA expression. The result indicated that, under
the condition of high sugar medium, Notch1 and Hes1 protein and
mRNA level expression were significantly decreased, compared with
normal culture medium (P<0.01; Fig.
1).
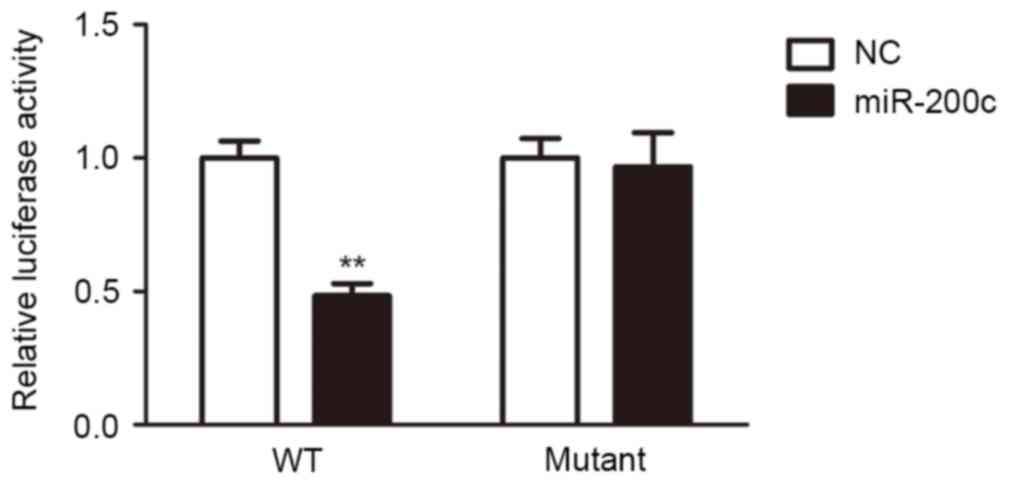
miR-200c inhibited the Notch1/Hes
pathway by targeting Notch1
To explore the function of miR-200c, computational
algorithms (TargetScan and miRanda) were used to identify the
potential target gene of miR-200c. It demonstrated that Notch1 was
a target gene of miR-200c (Table
I). Luciferase reporter assays were performed to clarify this
finding. The full-length Notch1 was cloned downstream of the
firefly luciferase gene and co-transfected with miR-200c mimics or
scrambled oligonucleotides controls. Luciferase activity was
measured 48 h following transfection. Luciferase expression was
decreased in H5V cells that were co-transfected with miR-200c and
the wild-type Notch 3′-UTR, compared to the control. However, no
decrease can be observed in cells co-transfected with miR-200c and
the mutant Notch1 3′-UTR in comparison with controls (Fig. 2). These results suggested that
miR-200c can directly target Notch1.
 | Table I.The algorithm predicted that Notch1
was a target gene of miR-200c. |
Table I.
The algorithm predicted that Notch1
was a target gene of miR-200c.
| Name of target region
and miRNA | Predicted
consequential pairing of target region (top) and miRNA
(bottom) |
|---|
| Position 736–743 of
Notch1 3′UTR | 5′
…UCUUUGUUUCAGGUUCAGUAUUA… |
| hsa-miR-200c-3p | 3′
AGGUAGUAAUGGGCCGUCAUAAU |
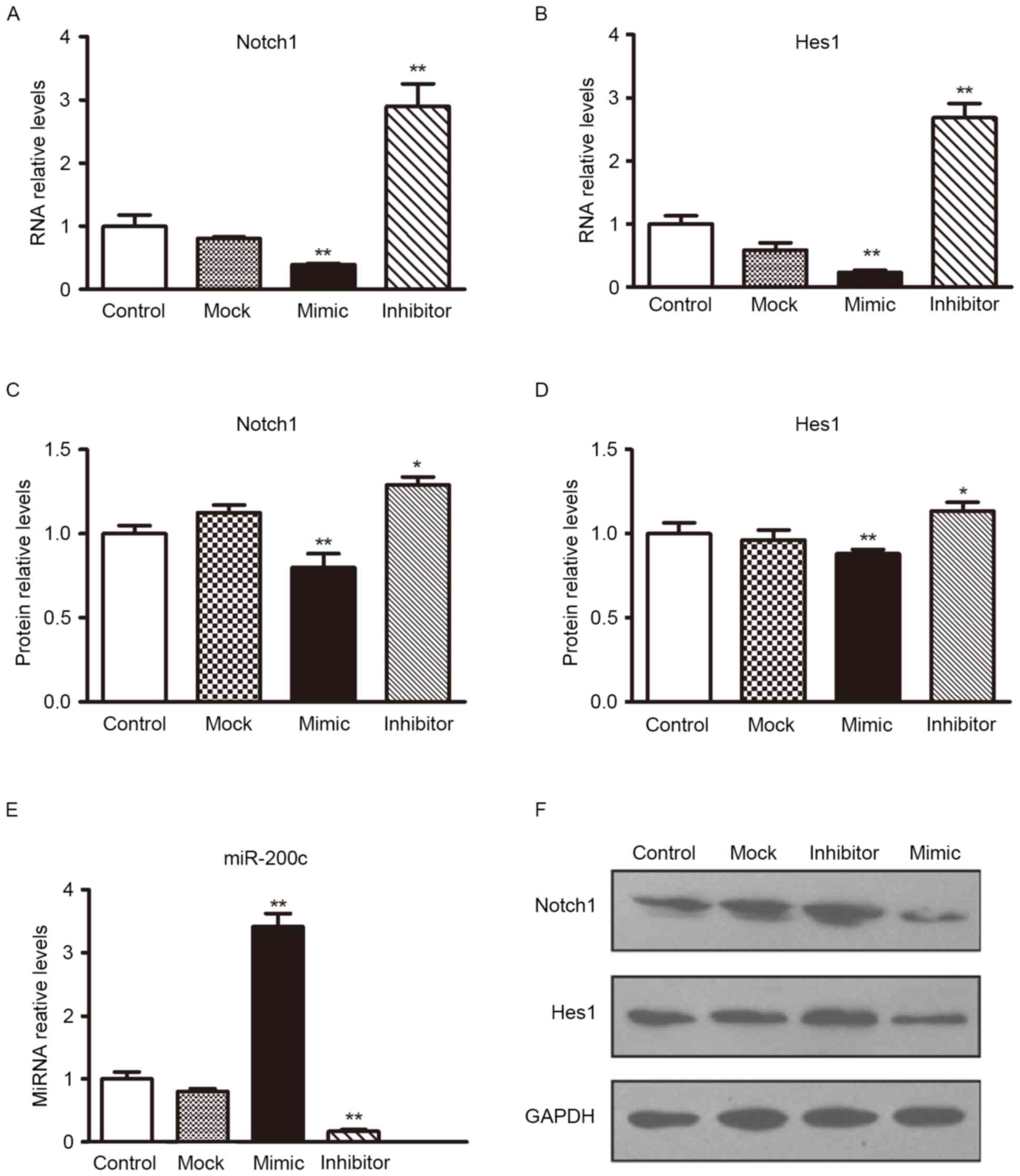
To determine the level at which miR-200c influences
Notch1 expression, the expression of Notch1 mRNA was analyzed
following transfection with miR-200c mimics or scramble
oligonucleotides controls in H5V cells. Following transfection with
miR-200c mimics, transfection efficiency could be observed in
Fig. 3E; the expression of
Notch1/Hes1 mRNA in the H5V cell line was lower than in the
controls (Figs. 3A and B). The
effect of miR-200c on Notch1/Hes1 expression was also verified by
western blotting analyses (Figs. 3C, D
and F). The overexpression of miR-200c reduced Notch1/Hes1
protein levels significantly. These results provide evidence that
miR-200c directly recognizes the 3′-UTR of Notch 1 mRNA and
inhibits Notch 1 translation.
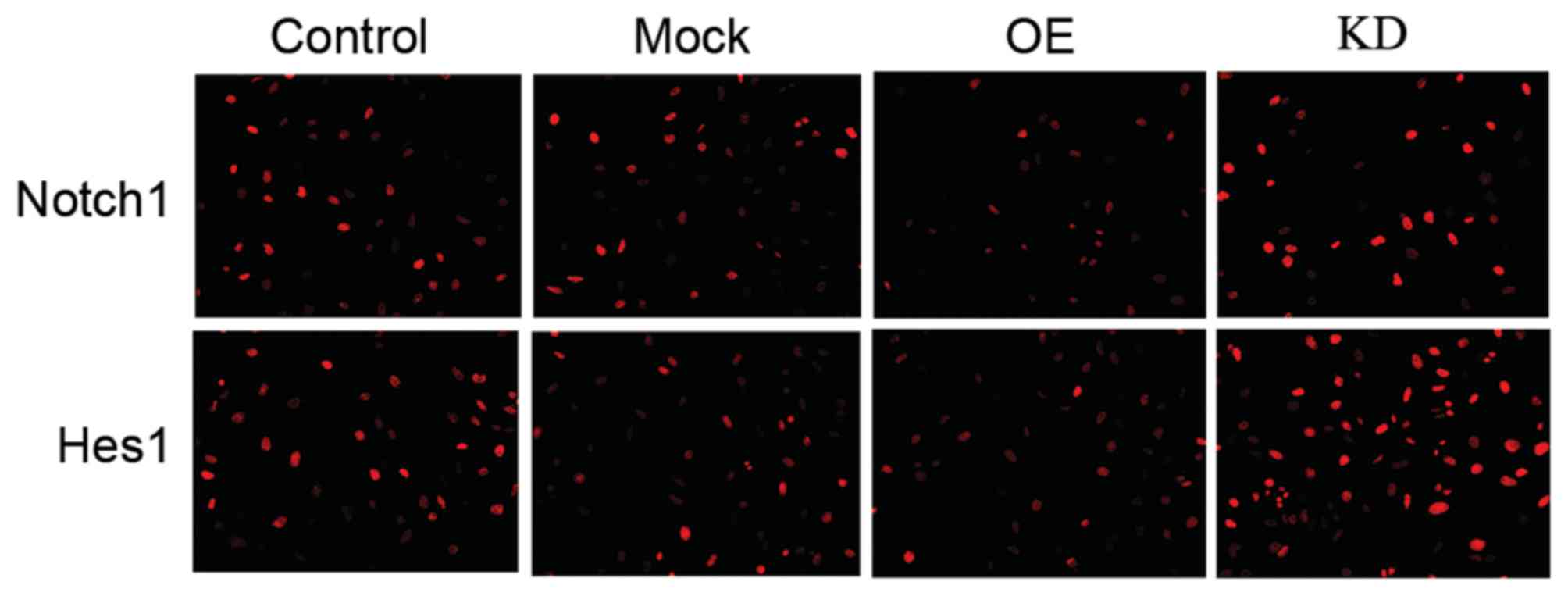
Although Notch 1 serves a wide-ranging role in
controlling cell fate, differentiation and development (25), in ECs, the activation of Notch1 can
trigger the Notch1/Hes1 signal pathway (7), which serves an important role in ECs'
development and function. Therefore, it was presumed that miR-200c
may regulate that pathway by targeting Notch1. Therefore, the
authors upregulated miR-200c levels via miR-200c mimics in H5V
cells. As verified by similar results from protein quantification
and an immunocytochemistry assay (Fig.
4), the western blotting results demonstrated that miR-200c
decreased the expression levels of Notch 1 and Hes1 (Figs. 5C, D and F).
Subsequently, H5V cells were treated with miR-200c
or a scramble control. At 24 h, they were transfected with
Notch1-encoding vector (without an endogenous 3′-UTR) or mock
vector (Fig. 3E presented that the
transfection was successful in H5V cells). The miR-200c mimic
inhibited the expression of Notch1, a similar alteration was
observed in the expression levels of Hes1. However, the expression
level of Notch1 and Hes1 was increased by enhancing miR-200c
inhibition in H5V cells (Fig. 3).
These results suggested that miR-200c inhibits the Notch1 and Hes1
pathway by targeting Notch1.
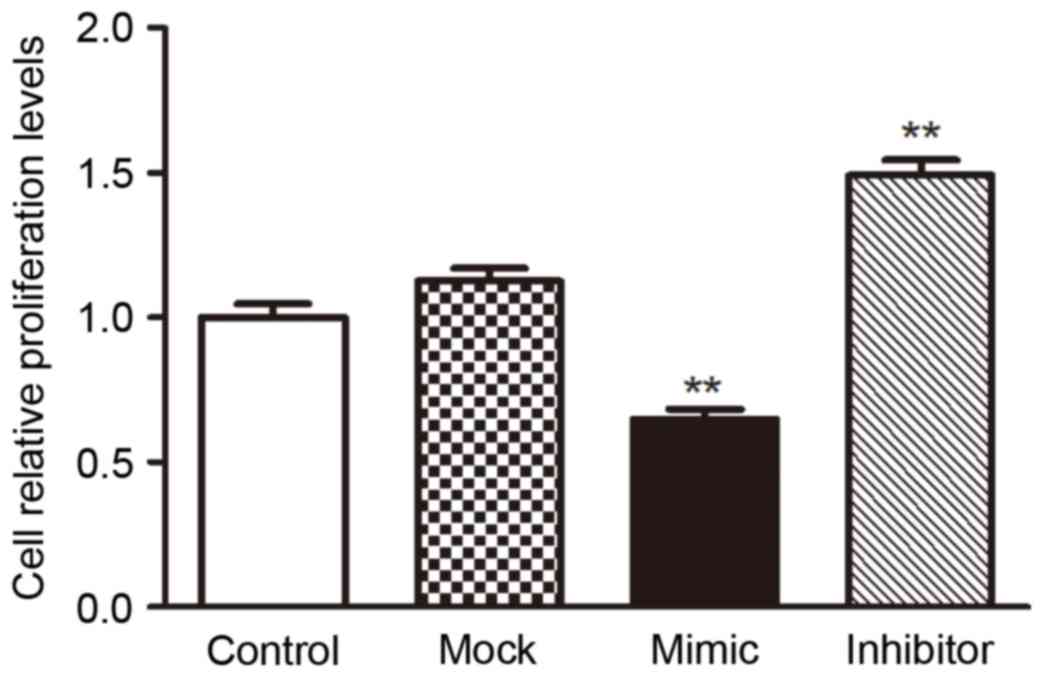
miR-200c inhibited the proliferation
of VEC H5V by targeting Notch1
To investigate the biological consequences of the
miR-200c-driven repression of Notch1 in H5V cells, the authors
investigated whether increasing or decreasing the expression of
miR-200c would have an impact on cell proliferation in H5V cells.
H5V cells were transfected with miR-200c mimics, miR-200c
inhibitors or their scrambled oligonucleotides controls,
respectively, then were counted after 24 h by CCK8 assay. The
result showed that the increased expression of miR-200c in H5V
cells inhibited the cell proliferation compared with the control
cells, while the decreased expression of miR-200c promoted the
proliferation (Fig. 5).
The above results prompted us to validate that
miR-200c could inhibit the proliferation of H5V cell by repressing
Notch1 expression, it was involved in the development of
vasculopathy induced by hyperglycemia. For this purpose, H5V cells
were transfected with mock vector, scrambled oligonucleotides
controls, Notch1-encoding vector + miR-200c or mock vector +
scrambled oligonucleotides controls, respectively. The results
indicated that increased expression of miR-200c inhibited
proliferation by repressing Notch1 in high glucose condition
(Fig. 5).
Discussion
Exposure of the vascular endothelial tissue to high
blood sugar causes vascular lesions of diabetic lower limbs,
thereby causing ischemia, which serves a critical role in the
pathogenesis of diabetic vascular complications. Previous studies
indicated that high blood sugar dysregulated multiple miRNAs,
including miR-200c (1,17,18),
which results in cellular dysfunction, cell apoptosis and cell
death through targeting their downstream proteins. Notch signaling
has been widely connected in epithelial-mesenchymal transition, EC
proliferation and apoptosis regulation (15), it is not surprising that aberrant
gain or loss of Notch signaling components has been directly linked
to multiple human disorders (6).
The present study has indicated that miR-200c served
a key role in mice EC H5V apoptosis induced by a high glucose
environment, the results demonstrated that miR-200c can
downregulate Notch1 and Hes1 by directly binding 3′UTR of Notch1′s
promoter. This is similar to previous reports, which indicated that
the Notch1/Hes1 signal pathway is vital to EC function.
Diabetic foot results in limited joint mobility of
the ankle and foot, impairs muscular performance and reduces gait
speed, increasing risk factors for ulceration (26). Diabetic feet are difficult to heal
and are a significant risk factor for non-traumatic foot
amputation. At present, popular diabetic foot treatments clinically
include low-level light therapy and hyperbaric oxygen therapy, as
well as various drugs and therapies such as antibiotics,
neuropathic drugs, wound dressings, skin substitutes, growth
factors and inflammatory modulators. The majority of these
therapies target the treatment of diabetic foot ulcer to address
the altered biochemical composition of the diabetic wound (2). However, no single treatment can be
definitively recommended for the treatment of diabetic foot ulcers.
These are clinical treatments according to disease symptoms, which
cannot solve the key problem. In pathology, diabetic foot is caused
by vascular lesions caused by insufficient blood supply of ischemic
necrosis of lower limbs. Obviously, ECs are a key regulator of
blood vessel function. This study aimed to explore the
pathophysiology of diabetic foot, and offer some possible basis for
molecular therapy.
From the current experiments, it was demonstrated
that miR-200c was significantly upregulated in H5V cells stimulated
by high glucose conditions. Overexpression of miR-200c may reduce
H5V cells proliferation by targeting the Notch1/Hes1 pathway. These
findings suggested that miR-200c mediating Notch1/Hes1 may involve
in the process of vascular damage caused by hyperglycemia, the
method of inhibiting miR-200c may provide a novel therapeutic
approach for patients with diabetic foot.
The deficiencies of the present study are that it is
only a cell study, the vasculogenesis process could not be
presented, the process of vascular impairment in hyperglycemia
could not be observed, and the regulation of miR-200c on the
morphology and function of damaged vascular in high sugar could not
be confirmed; therefore, more animal studies are required to
further verify the function of miR-200c in ECs of diabetics.
Acknowledgements
The present study was financially supported by Xing
Jin (Shandong University) and Qiang Guan (Shanxi Province People's
Hospital).
References
|
1
|
Fiorentino TV, Prioletta A, Zuo P and
Folli F: Hyperglycemia-induced oxidative stress and its role in
diabetes mellitus related cardiovascular diseases. Curr Pharm Des.
19:5695–5703. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jeffcoate WJ and Harding KG: Diabetic foot
ulcers. Lancet. 361:1545–1551. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chiaramonte R, Basile A, Tassi E,
Calzavara E, Cecchinato V, Rossi V, Biondi A and Comi P: A wide
role for NOTCH1 signaling in acute leukemia. Cancer Lett.
219:113–120. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hellström M, Phng LK, Hofmann JJ, Wallgard
E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N,
et al: Dll4 signalling through Notch1 regulates formation of tip
cells during angiogenesis. Nature. 445:776–780. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Williams CK, Li JL, Murga M, Harris AL and
Tosato G: Up-regulation of the Notch ligand Delta-like 4 inhibits
VEGF-induced endothelial cell function. Blood. 107:931–939. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kopan R and Ilagan MX: The canonical Notch
signaling pathway: Unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishiko E, Matsumura I, Ezoe S, Gale K,
Ishiko J, Satoh Y, Tanaka H, Shibayama H, Mizuki M, Era T, et al:
Notch signals inhibit the development of erythroid/megakaryocytic
cells by suppressing GATA-1 activity through the induction of HES1.
J Biol Chem. 280:4929–4939. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garg V, Muth AN, Ransom JF, Schluterman
MK, Barnes R, King IN, Grossfeld PD and Srivastava D: Mutations in
NOTCH1 cause aortic valve disease. Nature. 437:270–274. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iso T, Kedes L and Hamamori Y: HES and
HERP families: Multiple effectors of the Notch signaling pathway. J
Cell Physiol. 194:237–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liao WR, Hsieh RH, Hsu KW, Wu MZ, Tseng
MJ, Mai RT, Wu Lee YH and Yeh TS: The CBF1-independent Notch1
signal pathway activates human c-myc expression partially via
transcription factor YY1. Carcinogenesis. 28:1867–1876. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jaiswal MK, Agrawal V, Pamarthy S, Katara
GK, Kulshrestha A, Gilman-Sachs A, Beaman KD and Hirsch E: Notch
signaling in inflammation-induced preterm labor. Sci Rep.
5:152212015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie F, Cai W, Liu Y, Li Y, Du B, Feng L
and Qiu L: Vaccarin attenuates the human EA.hy926 endothelial cell
oxidative stress injury through inhibition of Notch signaling. Int
J Mol Med. 35:135–142. 2015.PubMed/NCBI
|
|
13
|
Xi G, Shen X, Wai C, Vilas CK and Clemmons
DR: Hyperglycemia stimulates p62/PKCζ interaction, which mediates
NF-κB activation, increased Nox4 expression, and inflammatory
cytokine activation in vascular smooth muscle. FASEB J.
29:4772–4782. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang Q, Wang J, Wang L, Zhang Y, Yin H, Li
Y, Tong C, Liang G and Zheng C: Attenuation of inflammatory
response by a novel chalcone protects kidney and heart from
hyperglycemia-induced injuries in type 1 diabetic mice. Toxicol
Appl Pharmacol. 288:179–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ferrer-Lorente R, Bejar MT and Badimon L:
Notch signaling pathway activation in normal and hyperglycemic rats
differs in the stem cells of visceral and subcutaneous adipose
tissue. Stem Cells Dev. 23:3034–3048. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou Y, Gu P, Shi W, Li J, Hao Q, Cao X,
Lu Q and Zeng Y: MicroRNA-29a induces insulin resistance by
targeting PPARδ in skeletal muscle cells. Int J Mol Med.
37:931–938. 2016.PubMed/NCBI
|
|
17
|
Silambarasan M, Tan JR, Karolina DS,
Armugam A, Kaur C and Jeyaseelan K: MicroRNAs in hyperglycemia
induced endothelial cell dysfunction. Int J Mol Sci. 17:5182016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ocłoń E, Latacz A, Zubel-Łojek J and
Pierzchała-Koziec K: Hyperglycemia-induced changes in miRNA
expression patterns in epicardial adipose tissue of piglets. J
Endocrinol. 229:259–266. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Magenta A, Cencioni C, Fasanaro P,
Zaccagnini G, Greco S, Sarra-Ferraris G, Antonini A, Martelli F and
Capogrossi MC: miR-200c is upregulated by oxidative stress and
induces endothelial cell apoptosis and senescence via ZEB1
inhibition. Cell Death Differ. 18:1628–1639. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo Z, Wen G, Wang G, Pu X, Ye S, Xu Q,
Wang W and Xiao Q: MicroRNA-200C and −150 play an important role in
endothelial cell differentiation and vasculogenesis by targeting
transcription repressor ZEB1. Stem Cells. 31:1749–1762. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song C, Liu LZ, Pei XQ, Liu X, Yang L, Ye
F and Xie X, Chen J, Tang H and Xie X: miR-200c inhibits breast
cancer proliferation by targeting KRAS. Oncotarget. 6:34968–34978.
2015.PubMed/NCBI
|
|
22
|
Kumar K, Chow CR, Ebine K, Arslan AD, Kwok
B, Bentrem DJ, Eckerdt FD, Platanias LC and Munshi HG: Differential
Regulation of ZEB1 and EMT by MAPK-interacting protein kinases
(MNKs) and eIF4E in pancreatic cancer. Mol Cancer Res. 14:216–227.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guilini C, Urayama K, Turkeri G, Dedeoglu
DB, Kurose H, Messaddeq N and Nebigil CG: Divergent roles of
prokineticin receptors in the endothelial cells: Angiogenesis and
fenestration. Am J Physiol Heart Circ Physiol. 298:H844–H852. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu S, Ma X, Ai Q, Huang Q, Shi T, Zhu M,
Wang B and Zhang X: NOTCH1 functions as an oncogene by regulating
the PTEN/PI3K/AKT pathway in clear cell renal cell carcinoma. Urol
Oncol. 31:938–948. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Y, Li JS, Zhang X, Wu YJ, Huang K and
Zheng L: Ursolic acid inhibits early lesions of diabetic
nephropathy. Int J Mol Med. 26:565–570. 2010.PubMed/NCBI
|