Introduction
Autophagy is a highly conserved, physiological,
catabolic process that engulfs organelles and cytoplasmic contents,
including macromolecules such as proteins and lipids (1,2).
These are broken down to their basic components to sustain cellular
metabolism. In addition to providing a basic catabolic function,
autophagy is believed to be essential for the maintenance of
cellular homeostasis via coping with stressful conditions to
improve survival (3). Unlike the
ubiquitin-proteasome system which selectively degrades proteins
attached by ubiquitin (4),
autophagy nonselectively degrades cytoplasmic proteins and
dysfunctional organelles (5). In
mammalian cells, there are predominantly three autophagic pathways
that have been identified, including macroautophagy, microautophagy
and chaperone-mediated autophagy (6,7).
Emerging evidence suggests that autophagy plays a
context-dependent role in cancer (8–10),
autophagy suppresses tissue injury and tumor initiation by
elimination of damaged cellular components on one side, however, in
an established tumor, autophagy promotes cancer progression by
providing substrates for metabolism and fostering survival
(11,12). The survival of organisms is
dependent upon their ability to efficiently generate energy through
the process of mitochondrial oxidative phosphorylation and when
cells subjected to prolonged hypoxia, autophagy is an adaptive
metabolic response to let cells go through energy deficiency and
this process requires the hypoxia-inducible factor 1 to maintain
oxygen homestasis (13–15).
HIF-1 composed of a constitutively expressed HIF-1β
subunit and an O2-regulated HIF-1α subunit is a
heterodimer and plays a key role in the regulation of oxygen
homestasis (16,17). Under aerobic conditions, HIF-1α
subunit is rapidly degraded but stabilized when the O2
dependent prolyl hydroxylases (PHDs) are inhibited under hypoxia
(16). HIF-1 regulates the
transcription of hundreds of genes in response to hypoxia whose
products restore blood supply and nutrients (18,19).
So far, there is growing evidence suggested autophagy is linked to
hypoxia, however an understanding of the precise role of HIF-1 in
the course of autophagy remains dismal.
In this present study, we examined the role of
HIF-1α/p27 in the regulation of autophagy. Our data indicate that
HIF-1α induces autophagy by promoting p27 activity, and p27 silence
inhibits HIF-1α induced autophagy. HIF-1α could also promote
esophageal carcinoma cells proliferation and tumorigenesis in
xenograft.
Materials and methods
Cell lines, cell culture
Human esophageal cancer EC109 and IMR90 human
diploid fibroblasts cells were purchased from National Institute of
Biological Products, Beijing, China. Young IMR90 cells are defined
as having completed <30 PD, while replicative senescent IMR90
cells are defined as having completed >50 PD. EC109 cell and
IMR90 cell were cultured in RPMI-1640 and DMEM media, respectively,
supplemented with 10% fetal bovine serum at 37°C. In the
experiments 2.4 g/l HEPES was added into the medium to inhibit cell
apoptosis caused by acidosis under hypoxia. Cultures at 90%
confluence were digested with 0.25% trypsin after washing with a
PBS solution and then split at a ratio of 1:2. For hypoxia culture,
cells were placed in a hypoxic (1% O2, 5%
CO2, 94% N2, 37°C) incubator (New Brunswick
Scientific Co., Ltd., Enfield, CT, USA) for indicated time. Control
cells were incubated for equivalent periods under normoxic
conditions (21% O2, 5% CO2, 37°C).
Plasmids, antibodies and regent
HIF-1α wild type (HIF-1α WT) plasmid and HIF-1α
constitutively active form of HIF-1α (HIF-1α ∆CA) plasmid were
kindly gift from Dr Makio Hayakawa. As HIF-1α can be degraded
through the ubiquitin-proteasome pathway upon normoxia by von
Hippel-Lindau (VHL) protein and VHL protein binds to HIF-1α by
recognizing two highly conserved proline residues (Pro-402 and
Pro-564) for polyubitylation, so let alanine to substitute the
conserved proline will keep HIF-1α constitutively active.
Transfection into EC109 cells was performed using Lipofectamine
Plus (Invitrogen Life Technologies, Carlsbad, CA, USA) according to
the manufacturer's instructions. The antibodies used in this study
were antibodies against HIF-1α (BD Transduction Laboratories,
Lexington, KY, USA), p16 (Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA), p27 (Santa Cruz Biotechnology, Inc.), PTEN (Santa
Cruz Biotechnology), E2F1 (Cell Signaling Technology, Inc.,
Danvers, MA, USA), LC3 (Cell Signaling Technology, Inc.), Bcl-2
(Cell Signaling Technology, Inc.), PARP (Santa Cruz Biotechnology,
Inc.), actin (Santa Cruz Biotechnology, Inc.). Besides,
CoCl2 (Sigma, St. Louis, MO, USA) was dissolved in
sterile water.
Western blot analysis
Cells were scraped from the plate and lysed in RIPA
buffer (10 mm Tris-HCl, 150 mm NaCl, 1% Triton X-100, 5 mm EDTA, 1%
sodium deoxycholate, 0.1% SDS, 1.2% aprotinin, 5 µm leupeptin, 4 µm
antipain, 1 mm phenylmethylsulphonyl fluoride, and 0.1 mm
Na3VO4) on ice for 1 h containing a protease
inhibitor mixture. Cell lysates were then centrifuged at 13,000 × g
for 15 min at 4°C, and the insoluble debris were discarded. Protein
concentration of each sample was determined by BCA Protein Assay
Reagent (Pierce Biotechnology, Inc., Rockford, IL, US). Then total
proteins were subjected to SDS-polyacrylamide gel electrophoresis
(SDS/PAGE) and transferred to nitrocellulose membranes (Millipore
Corp., Billerica, MA, USA). After blocking in 5% non-fat dry milk
in TBST (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.05% Tween-20), the
membranes were incubated with primary antibodies overnight at 4°C.
The membranes were then washed four times with TBST and then
incubated with HRP-conjugated secondary antibodies for 1 h at room
temperature. Proteins were visualized using chemiluminescent
substrate (Millipore Corp.).
Determination of apoptosis
Apoptotic cells were stained with Annexin V-FITC
using an Annexin V-FITC Apoptosis Detection kit I (BD Biosciences,
Franklin Lakes, NJ, USA) according to the manufacturer's
instructions. Stained cells were then analyzed by flow cytometry
with a FACScan cytometer.
RNA interference
The p27 siRNA (si-p27) sequence and the scrambled
siRNA control (si-ctrl) sequence were 5′-GCAACCGACGAUUCUUCUATT-3′
and 5′-TTCTCCGAACGTGTCACGTTT-3′, respectively. The cells were
transfected with siRNA duplexes for 48 h using Oligofectamine
(Invitrogen Life Technologies) following the manufacturer's
recommendations.
Tumorigenicity assay
In the experiment, NOD/SCID mice at age of 6 weeks
were injected with 1×106 cells in 100 µl PBS into left and right
flanks, respectively. Tumor sizes were measured every few days, and
tumor volumes were calculated as volume = length × width2 × (1/2).
Animals were maintained of regular food and water and after several
weeks, and then were killed and tumors were harvested. All
procedures were approved by the Ethics Committee of The First
Affiliated Hospital of Zhengzhou University.
Statistical analysis
The statistical significance of differences was
evaluated by unpaired t-test, and P<0.05 was considered to
indicate a statistically significant difference.
Results
Young IMR90 cells present an increase
of autophagy responses than senescence cells when exposed to
hypoxia
The existence of evidences that HIF-1α could induce
autophagy and RB-E2F1 pathway triggers autophagy led us to explore
whether HIF-1α could regulate autophagy through RB-E2F1 pathway
(14,20,21).
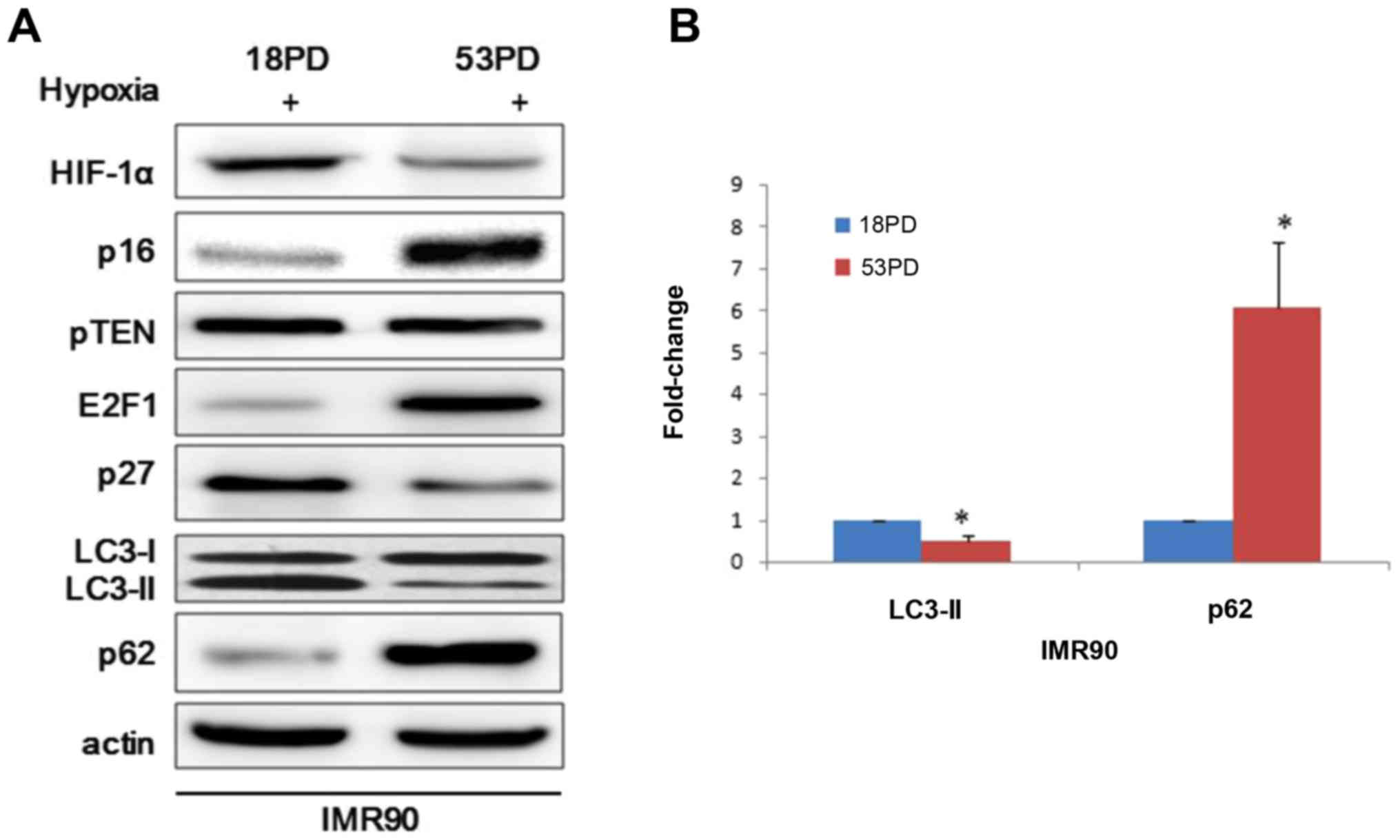
To investigate this hypothesis, young and sensescent IMR90 cells
were exposed to hypoxia and then collected and analyzed. Consistent
with previous findings, hypoxia induces leads to prolyl hydroxylase
inhibition and stabilization of HIF-1α and the conversion of the
microtubule-associated protein LC3-I to LC3-II, a biochemical
marker of autophagy that is correlated with the formation of
autophagosomes (Fig. 1A). To
further support the observation, we performed western blot analyses
of p62, which is an autophagic substrate. In response to hypoxia,
the degradation of p62 was much faster in young IMR90 cells than
senescent cells (Fig. 1A).
Immunoblotting analyses revealed an accumulation of E2F1 in
senescent IMR90 cells compared with young IMR90 cells and p27
presented an opposite expression profile to E2F1 (Fig. 1A). However, that p16INK4a showed
upregulation as cell senescence didn't accord with the previous
report that the RB activators p16INK4a and p27 induce autophagy
through modulating E2F1 (21).
Here ImageJ software was used to analyze the ratio of LC3-II bands
compared with reference bands of actin, which demonstrated that
LC3-II decreased in senescent IMR90 cells relative to young IMR90
cells (Fig. 1B). Taking all the
previous findings into consideration, we found that young IMR90
could produce more autophagosomes and this may indicate that young
IMR90 cells fit better than senescent cell when face hypoxia.
Hypoxia induces autophagy responses in
EC109 cells
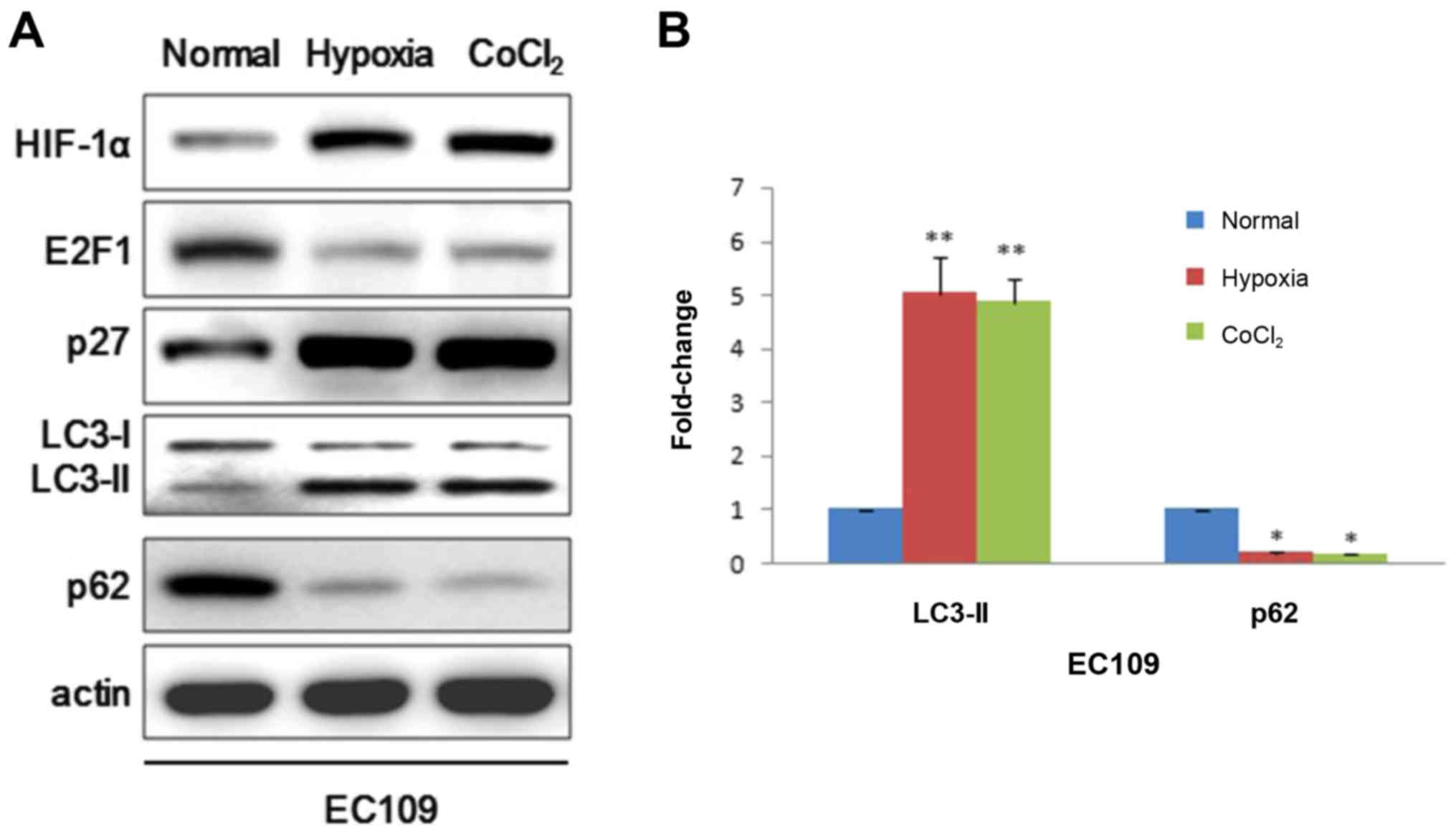
We further extended this study to EC109 cells to
verify whether HIF-1α could also play an important role in cancer
cell. EC109 cells were exposed to 20 or 1% for 24 h. Similarly,
western blot demonstrated that HIF-1α could induce autophagy by
positively modulating p27 which represses E2F1 activity just in
line with results in IMR90 cells (Fig.
2A). Given that hypoxia can alter a variety of metabolic
parameters that could modulate autophagy activity, here we used
CoCl2 which is a chemical hypoxia mimetic agent to
further strengthen the argument that hypoxia could induce
autophagy. Consistently, CoCl2 resulted in a decrease of
E2F1, p62 and increases of p27 and LC3-II (Fig. 2A). LC3-II bands were also analyzed
using ImageJ software (Fig. 2B).
Taken together, these results support that hypoxia not only induces
autophagosomes in IMR90 cells but also in EC109 cells.
HIF-1α induces autophagy response in
EC109 cells
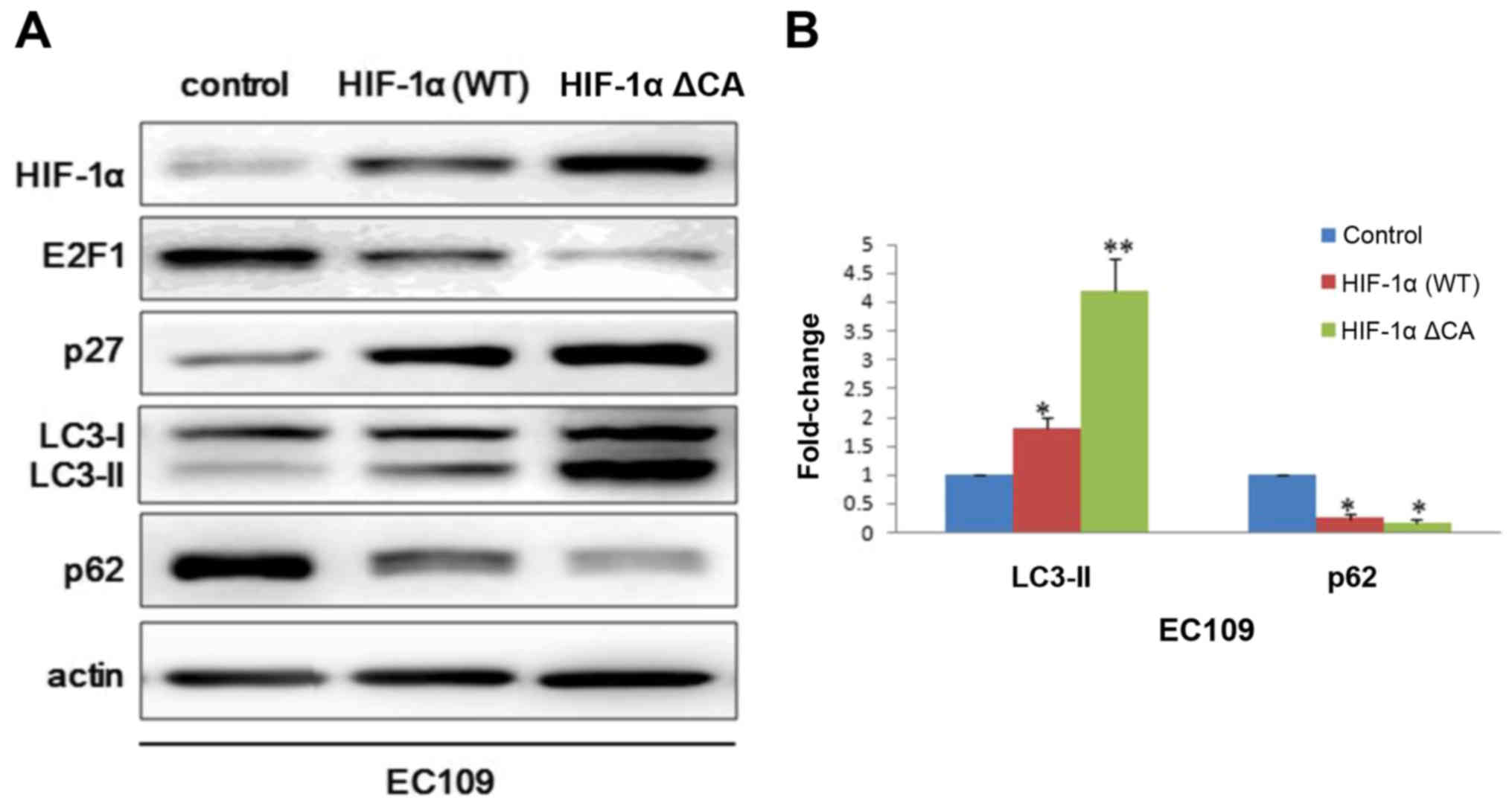
In order to further explore the role of HIF-1α in
autophagy. Vector control plasmid, HIF-1α wild type plasmid and
HIF-1α constitutive active plasmid were transfected into EC109
cells. Consistent with above results, we observed that p27
expression was significant higher in HIF-1α ΔCA group than HIF-1α
wild type group and blank control group. While E2F1 and p62
presented an opposite expression profile with p27 (Fig. 3A). To further assess autophagosome
maturation, we performed degree scanning of LC3-II which is an
autophagic marker (Fig. 3B).
P27 silence inhibits hypoxia induced
autophagy
Because E2F1 activity is negatively regulated by
CDKIs, such as p16 and p27 (22),
and previous results demonstrated that p16 and p27 could induce
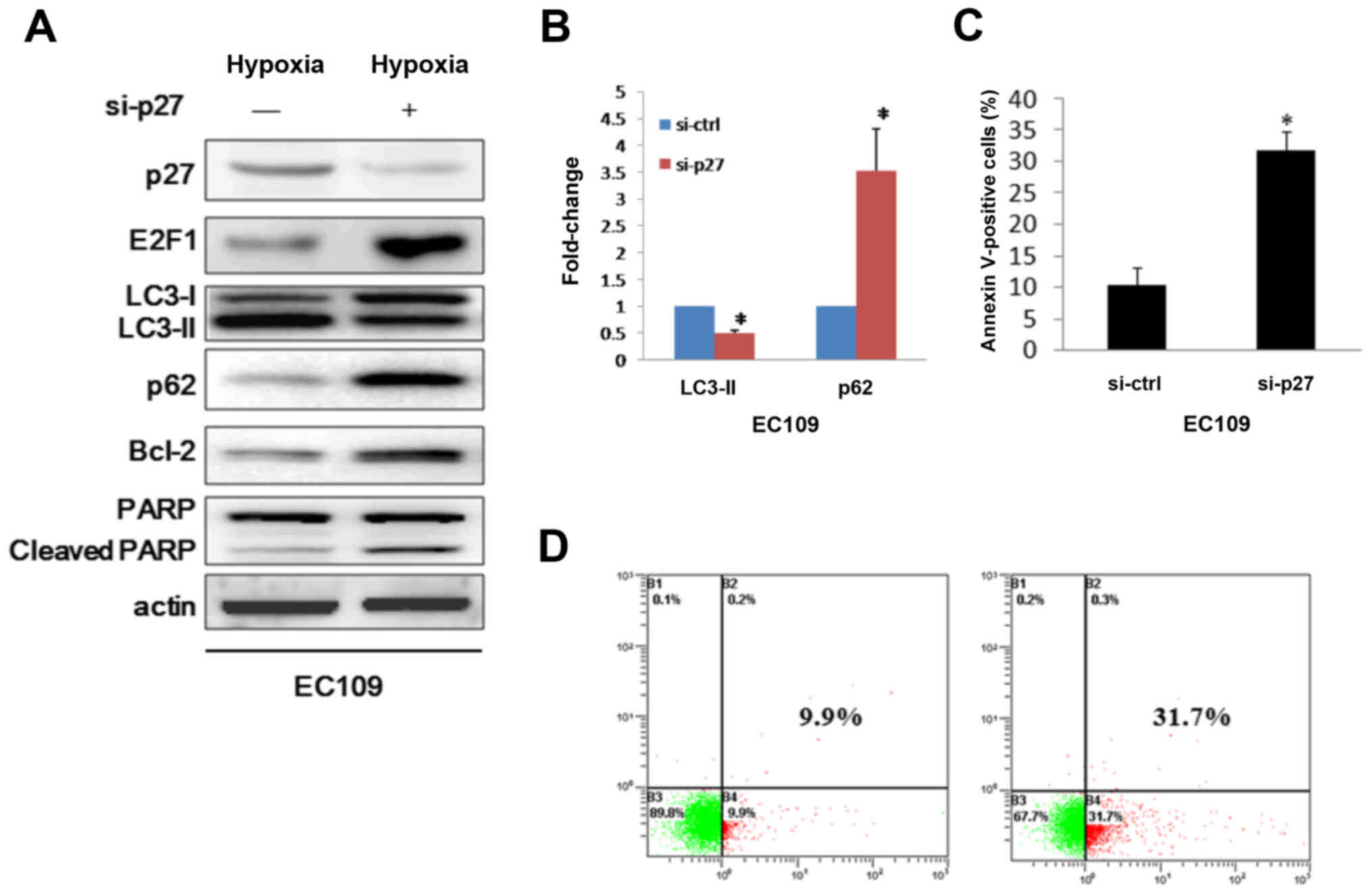
autophagy in some cell lines. To this end, we presumed that HIF-1α
may modulate autophagy via p16 and p27 pathways. Whereas p16 has a
different expression profile with p27, so we discreetly think p27
is the downstream target in HIF-1α induced autophagy. We then
challenged the autophagy mechanism by down-modulating p27 when in
hypoxia condition. As expected, silencing p27 in EC109 cells
decreased HIF-1α mediated processing of formation of LC3-II and
increased p62 expression (Fig. 4A and
B). Consistently, immunoassay showed E2F1 appeared high
expression when p27 silenced (Fig.
4A). Because expression of E2F1 induces apoptosis (23), we asked whether the cells that were
transduced with siRNA against p27 were undergoing cell death
through apoptosis. Therefore, Bcl-2 which is an autophagy inhibitor
and upregulated by E2F1 was examined (24,25).
Here we showed that repression of p27 lead to up-modulating of
Bcl-2 and cleavage of PARP (Fig.
4A), contributing to the triggering of apoptosis. To further
determine whether the cause of the cell death was apoptosis, we
used flow cytometry to examine the ability of the cells binding to
Annexin V. We found that p27 silence caused about 32% of the cells
to undergo apoptosis, much higher than the control group (Fig. 4C and D). Thus, we conclude that
HIF-1α plays a role in the control of autophagy via regulating
p27-E2F1 pathway.
HIF-1α promotes tumor cell
proliferation and tumorigenesis in vivo
Given several lines of evidence indicate that HIF-1α
function contributes to tumor growth, we next examined the effect
of HIF-1α on cell proliferation and tumor growth. To determine
whether HIF-1α rendered growth advantage to tumor cell in
vivo, we performed tumor xenograft studies. EC109 cell lines
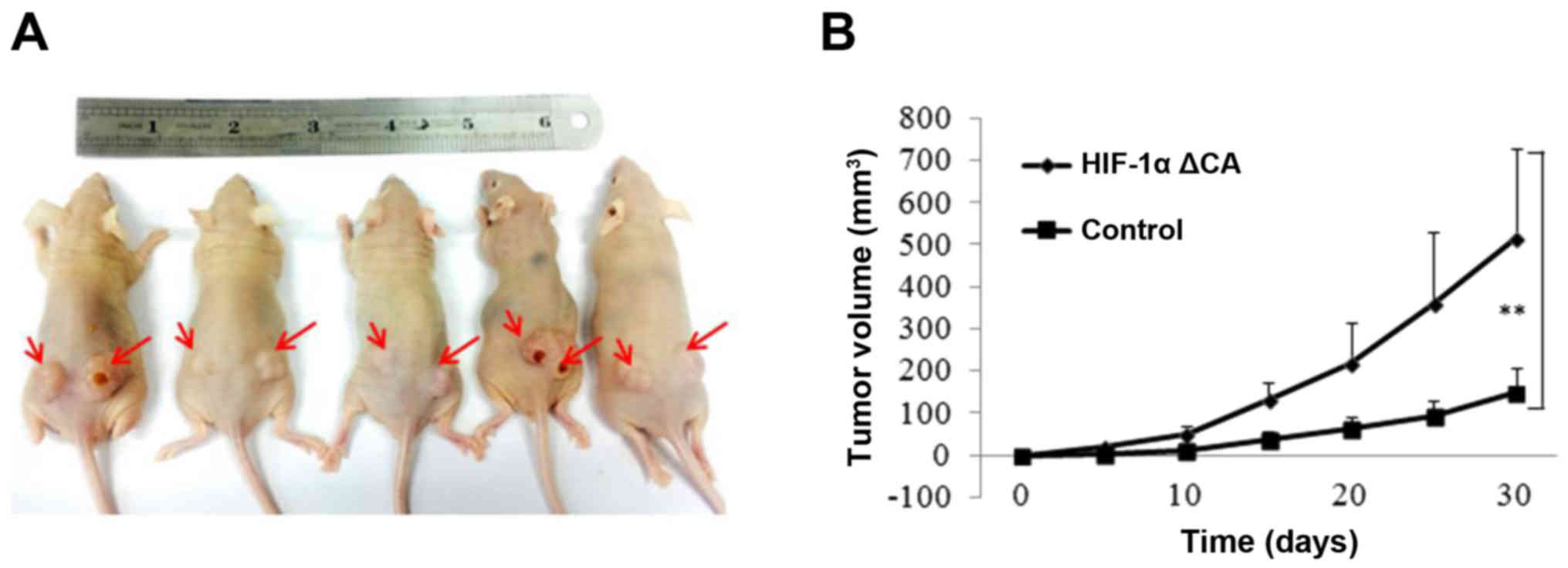
with stable expression of vector control and HIF-1α ΔCA were
injected into the left and right flanks of mude mice and tumor cell
growth was monitored over a period of 30 days. We found that cells
expressing HIF-1α% ΔCA grew at a faster rate than that the cells
expressing vector control. Representative kinetics of tumor growth
are shown in Fig. 5B. Therefore,
these results demonstrate that HIF-1α promotes cell proliferation
and tumor growth by inducing autophagy in hypoxia environment.
Discussion
As we all known autophagy is an evolutionarily
conserved process that occurs as a physiological process in normal
cells at a basal level to assure cellular homeostasis, or as a
strategic survival mechanism under hypoxia, stress and nutrient
deprivation conditions (3,26). Though various kinds of research
have focused on the mechanisms of autophagy and several gene have
been identified that could take part in the autophagy process.
However, the autophagy mechanisms that are involved in cancer
remain largely obscure. As we know that IMR90 cells have served as
a non tumor control cell line in a wide variety of studies and
esophageal cancer is one of the most malignant cancers in China
(27,28). So in this study, we used IMR90
cells and esophageal carcinoma cell line EC109 cells to investigate
the role of autophagy in normal human cell and tumor cell.
Thereafter, we identified HIF-1α regulates autophagy via modulating
p27-E2F1 pathway and we uncover a previously unidentified
connection between HIF-1α and autophagy for the first time.
HIF-1α has been found to be the main reason for
malignant tumor survival in hypoxia (29,30).
As a transcriptional activator, HIF-1α can induce the expression of
kinds of target genes, which promote angiogenesis, enhance hypoxia
tolerance (31). Here, we found
HIF-1α can induce autophagy through p27-E2F1 pathway under hypoxia
which could degrade cell constituents and supply energy for cell
survival. Our finding confirms the mechanism that autophagy is a
surveillance mechanism used by normal cells to protect them from
transformation to malignancy by removing damaged organelles and
reducing reactive oxygen species. Besides, the autophagy mechanisms
are also involved in cancer cell.
As we known E2F1 activity is negatively regulated by
CDKIs, such as p16INK4a and p27 and the E2F1 pathway is
crucial in regulating cell growth and apoptosis. So we asked
whether suppression of E2F1 by CKDIs would result in autophagy.
Here, we report that p27 positively regulates autophagy by
repressing E2F1 activity in IMR90 cells, while p16INK4a
don't take part in this pathway. This may coincide with that p27 is
a downstream gene of HIF-1 under lack of energy conditions
(32). As the expression of E2F1
induces apoptosis and we discovered that p27 silence inhibits
HIF-1α induced autophagy. Our results conclude that p27 silence
caused a higher percent of the cells to undergo apoptosis. Thus,
our results suggest that HIF-1α regulates autophagy through
p27-E2F1 pathway. P27 and E2F1 proteins play opposite roles in the
control of both autophagy and apoptosis.
In summary, we report here that young IMR90 cells
present an increased of autophagy responses than senescence cells
when exposed to hypoxia and HIF-1α could also induce autophagy in
EC109 cells through p27-E2F1 pathway. Ablation of p27 inhibits
HIF-1α induced autophagy and promotes E2F1 induced apoptosis. Our
results may shed light on understanding the compulsive biological
mechanism of autophagy.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81501200), the China
Postdoctoral Science Foundation (funded project no. 2015M572122)
and the Project of Medical Science and Technology Research of Henan
Provincial Health and Family Planning Commission (grant no.
201503029).
References
|
1
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cao J, Ying M, Xie N, Lin G, Dong R, Zhang
J, Yan H, Yang X, He Q and Yang B: The oxidation states of DJ-1
dictate the cell fate in response to oxidative stress triggered by
4-hpr: Autophagy or apoptosis? Antioxid Redox Signal. 21:1443–1459.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rubinsztein DC: The roles of intracellular
protein-degradation pathways in neurodegeneration. Nature.
443:780–786. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Z and Klionsky DJ: Eaten alive: A
history of macroautophagy. Nat Cell Biol. 12:814–822. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mijaljica D, Prescott M and Devenish RJ:
Microautophagy in mammalian cells: Revisiting a 40-year-old
conundrum. Autophagy. 7:673–682. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang P and Mizushima N: Autophagy and
human diseases. Cell Res. 24:69–79. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
White E: Deconvoluting the
context-dependent role for autophagy in cancer. Nat Rev Cancer.
12:401–410. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Burada F, Nicoli ER, Ciurea ME, Uscatu DC,
Ioana M and Gheonea DI: Autophagy in colorectal cancer: An
important switch from physiology to pathology. World J Gastrointest
Oncol. 7:271–284. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cecconi F and Jäättelä M: Targeting
ions-induced autophagy in cancer. Cancer cell. 26:599–600. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shintani T and Klionsky DJ: Autophagy in
health and disease: A double-edged sword. Science. 306:990–995.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sena P, Mariani F, Mancini S, Benincasa M,
Magnani G, Pedroni M, Palumbo C and Roncucci L: Autophagy is
upregulated during colorectal carcinogenesis, and in DNA
microsatellite stable carcinomas. Oncol Rep. 34:3222–3230.
2015.PubMed/NCBI
|
|
13
|
Shelby SJ, Angadi PS, Zheng QD, Yao J, Jia
L and Zacks DN: Hypoxia inducible factor 1α contributes to
regulation of autophagy in retinal detachment. Exp Eye Res.
137:84–93. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang H, Bosch-Marce M, Shimoda LA, Tan
YS, Baek JH, Wesley JB, Gonzalez FJ and Semenza GL: Mitochondrial
autophagy is an HIF-1-dependent adaptive metabolic response to
hypoxia. J Biol Chem. 283:10892–10903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu H, Wang D, Zhang L, Xie X, Wu Y, Liu
Y, Shao G and Su Z: Upregulation of autophagy by hypoxia-inducible
factor-1α promotes EMT and metastatic ability of CD133+
pancreatic cancer stem-like cells during intermittent hypoxia.
Oncol Rep. 32:935–942. 2014.PubMed/NCBI
|
|
16
|
Wang GL, Jiang BH, Rue EA and Semenza GL:
Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS
heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci
USA. 92:5510–5514. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Blagosklonny MV: Antiangiogenic therapy
and tumor progression. Cancer cell. 5:13–17. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Semenza GL: HIF-1 and mechanisms of
hypoxia sensing. Curr Opin Cell Biol. 13:167–171. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Seeber LM, Horrée N, Vooijs MA, Heintz AP,
van der Wall E, Verheijen RH and van Diest PJ: The role of hypoxia
inducible factor-1alpha in gynecological cancer. Crit Rev Oncol
Hematol. 78:173–184. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tittarelli A, Janji B, van Moer K, Noman
MZ and Chouaib S: The selective degradation of synaptic connexin 43
protein by Hypoxia-induced autophagy impairs natural killer
cell-mediated tumor cell killing. J Biol Chem. 290:23670–23679.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang H, Martin V, Gomez-Manzano C,
Johnson DG, Alonso M, White E, Xu J, McDonnell TJ, Shinojima N and
Fueyo J: The RB-E2F1 pathway regulates autophagy. Cancer Res.
70:7882–7893. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen T, Xue L, Niu J, Ma L, Li N, Cao X,
Li Q, Wang M, Zhao W, Li G, et al: The retinoblastoma protein
selectively represses E2F1 targets via a TAAC DNA element during
cellular senescence. J Biol Chem. 287:37540–37551. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Polager S and Ginsberg D: E2F-at the
crossroads of life and death. Trends Cell Biol. 18:528–535. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pattingre S, Tassa A, Qu X, Garuti R,
Liang XH, Mizushima N, Packer M, Schneider MD and Levine B: Bcl-2
antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell.
122:927–939. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo S and Rubinsztein DC: Apoptosis blocks
Beclin 1-dependent autophagosome synthesis: An effect rescued by
Bcl-xL. Cell Death Differ. 17:268–277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chagin AS: Effectors of mTOR-autophagy
pathway: Targeting cancer, affecting the skeleton. Curr Opin
Pharmacol. 28:1–7. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pei Y, Wang P, Liu H, He F and Ming L:
FOXQ1 promotes esophageal cancer proliferation and metastasis by
negatively modulating CDH1. Biomed Pharmacother. 74:89–94. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kirschner K, Samarajiwa SA, Cairns JM,
Menon S, Pérez-Mancera PA, Tomimatsu K, Bermejo-Rodriguez C, Ito Y,
Chandra T, Narita M, et al: Phenotype specific analyses reveal
distinct regulatory mechanism for chronically activated p53. PLoS
Genet. 11:e10050532015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vaupel P: The role of hypoxia-induced
factors in tumor progression. Oncologist. 9 Suppl 5:S10–S17. 2004.
View Article : Google Scholar
|
|
30
|
Unruh A, Ressel A, Mohamed HG, Johnson RS,
Nadrowitz R, Richter E, Katschinski DM and Wenger RH: The
hypoxia-inducible factor-1 alpha is a negative factor for tumor
therapy. Oncogene. 22:3213–3220. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Clarke HJ, Chambers JE, Liniker E and
Marciniak SJ: Endoplasmic reticulum stress in malignancy. Cancer
Cell. 25:563–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu Y, Zhao T, Itasaka S, Zeng L, Yeom CJ,
Hirota K, Suzuki K, Morinibu A, Shinomiya K, Ou G, et al:
Involvement of decreased hypoxia-inducible factor 1 activity and
resultant G1-S cell cycle transition in radioresistance of
perinecrotic tumor cells. Oncogene. 32:2058–2068. 2013. View Article : Google Scholar : PubMed/NCBI
|