Introduction
Male infertility can be caused by several genetic
factors, including chromosomal abnormalities, Y chromosome
microdeletions and gene mutations (1). Cytogenetic anomalies, especially
structural chromosomal aberrations are an important cause of male
infertility (2). Deletion of the
azoospermic factor on the long arm of the Y chromosome is known to
be involved with spermatogenesis defects (3). Cystic fibrosis genes mutations have
previously been reported in male infertility (4).
Y-autosome translocations are rare in humans, and
they may be identified in both fertile and sterile males (5). The rate of chromosomal rearrangement
ranges from 10–15% in azoospermic males (6). The frequency of Y-autosome
translocations in the general population is approximately 1 in
2,000 (7,8). However, previous studies mainly
basing G-banding with little analysis on the etiology of fertility
(9–13).
In the present study, a whole genome SNP microarray
and a genetic analysis of the Y chromosome with 20 sequence-tagged
sites were performed on three adult azoospermic male with de
novo Y-autosome translocations. The etiology and clinical
features of this rare disease were briefly discussed.
Patients and methods
Cases
The first patient (P1) was a 34-year-old man with a
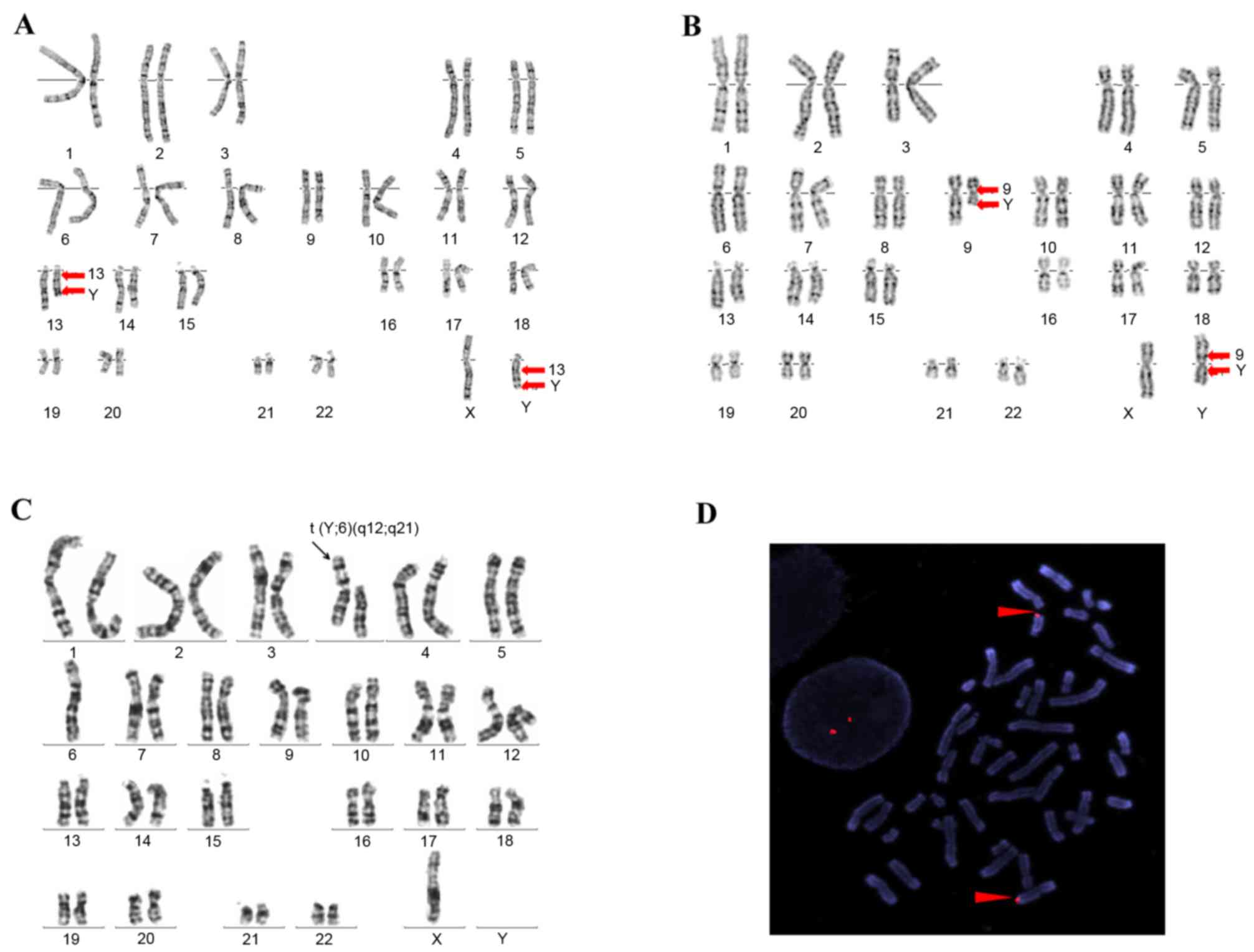
balanced translocation t(Y;13)(q12;q21) (Fig. 1A). His wife possessed a normal
karyotype. He was a well-developed male weighing 79 kg with a
height of 175 cm. Physical examinations revealed normal adult pubic
and axillary hair. The penis, epididymides, spermatic cords and
prostate were normal. The plasma levels of lactate dehydrogenase,
follicle stimulating hormone, prolactin and testosterone were
within normal limits. Repeated semen analyses indicated
azoospermia.
The second patient (P2) was another 34-year-old man
with a balanced translocation t(Y;9)(p11.2;q21) (Fig. 1B). The patient, who was
phenotypically normal and had normal clinical examinations, was
ascertained for infertility. Sperm counts presented severe
oligozoospermia azoospermia. The wife of P2 was able to conceive
naturally once but spontaneous abortion occurred.
The third patient (P3) was a 26-year-old man with a
balanced translocation t(Y;6)(q12;q21) (Fig. 1C). He was a well-developed male
weighing 65 kg with a height of 170 cm. His physical examination
was normal, and had an unremarkable family history. No sperm were
identified in any of the three routine semen analyses.
Ethical approval
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
ethics committee of The Third Affiliated Hospital of Guangzhou
Medical University (Guangzhou, China) and with the 1964 Helsinki
Declaration and its later amendments or comparable ethical
standards. All patients gave their informed consent to participate
in the present study.
Karyotype
Cytogenetic investigations were performed on the
patients' chromosomes obtained from peripheral blood lymphocytes.
The lymphocytes were cultured for 72 h in RPMI medium 1640 (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA),
phytohemagglutinin (Shanghai Yihua Medical Science & Technology
Co., Ltd., Shanghai, China), and fetal bovine serum (Beijing
Dingguo Changsheng Biotechnology Co., Ltd., Beijing, China)
following treatment with 50 µg/ml colcemid (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). The cells were made hypotonic using
potassium chloride (0.075 M), fixed in a fixation medium [acetic
acid combined with methanol (1:3)] at room temperature for 1 h,
dropped on precooled glass and incubated at 60°C overnight.
Metaphase chromosome spreads were studied by standard G-banding
procedures, using trypsin and Giemsa for G-banding. FISH analysis
of the rearrangement of the human Y chromosome was conducted using
specific Xq/Yq probes (Vysis; Abbott Pharmaceutical Co., Ltd., Lake
Bluff, IL, USA).
Chromosome microarray analysis
(CMA)
CMA-SNP array analysis was performed on the
probands' and their parents' peripheral blood samples using the
Affymetrix Cyto HD Array (Affymetrix, Inc., Santa Clara, CA, USA).
The DNA was amplified, labeled and hybridized to the CytoScan HD
Array platform according to the manufacturer's protocol. The array
was designed specifically for cytogenetic research. It includes
>2 million markers across the genome, in addition to SNP probes
and probes for detecting copy-number variations. The raw data files
were obtained by scanning the arrays, which were then analyzed with
the Chromosome Analysis Suite software 33.1 (Affymetrix, Inc.), and
the reference genome GRCh37 (hg19) was used for the annotations.
Only those signals meeting the manufacturer's quality cut-off
criteria were included in the present analysis. Gains and losses
that affected a minimum of 50 markers over 100 kb lengths were
initially considered.
Genetic analysis of Y chromosome
A total of 20 sites were analyzed with regards to
the Y chromosome microdeletion by the Y chromosome Deletion
Detection System kit (version 2.0; Promega Corporation, Madison,
WI, USA). This system is designed to detect deletions occurring in
the azoospermia factor region on the Y chromosome long arm (YqAZF).
This system consists of 20 primer pairs that are homologous to
previously identified and mapped sequence-tagged sites. The primers
have been combined into five sets for use in multiplex polymerase
chain reactions (PCR): Multiplex A Master Mix for the detection of
deleted in azoospermia (DAZ; SY254), DYS240 (SY157), DYS271 (SY81),
DYS221 (SY130) and anosmin 2, pseudogene (SY182) microdeletions,
with lysine demethylase 5C (SMCX) as control; Multiplex B Master
Mix for the detection of lysine demethylase 5D (SYPR3), DYS218
(SY127) and DAZ (SY242 and SY208) microdeletions, with SMCX as
control; Multiplex C Master Mix for the detection of DYS219
(SY128), DYS212 (SY121), DYF51S1 (SY145) and DAZ (SY255)
microdeletions, with SMCX as control; Multiplex D Master Mix for
the detection of DYS223 (SY133), DYS236 (SY152) and DYS215 (SY124)
microdeletions, with SMCX as control; Multiplex E Master Mix for
the detection of sex determining region Y (SY14), DYS224 (SY134),
DYS148 (SY86) and DYS273 (SY84) microdeletions, with zinc finger
protein, X linked/zinc finger protein, Y-linked as control. The
sequence tagged site number is presented in brackets. This makes it
possible to determine the presence or absence of all 20
sequence-tagged sites by performing five parallel PCR
amplifications.
Results
Cytogenetic studies
All the patients, their spouses and their parents
had normal karyotypes based on G-banded metaphase chromosomes. G
banding indicated that all metaphase cells revealed an apparently
balanced reciprocal translocation (Fig. 1A). By performing FISH, the
translocation between chromosome 13 and the Y chromosome were
clearly defined in P1 (Fig. 1D).
G-banding demonstrated an apparent Y autosomal translocation, and a
derivative Y chromosome was observed in P2 (Fig. 1B). The presence of a whole
chromosome translocation was revealed in P3 (Fig. 1C).
Molecular analysis
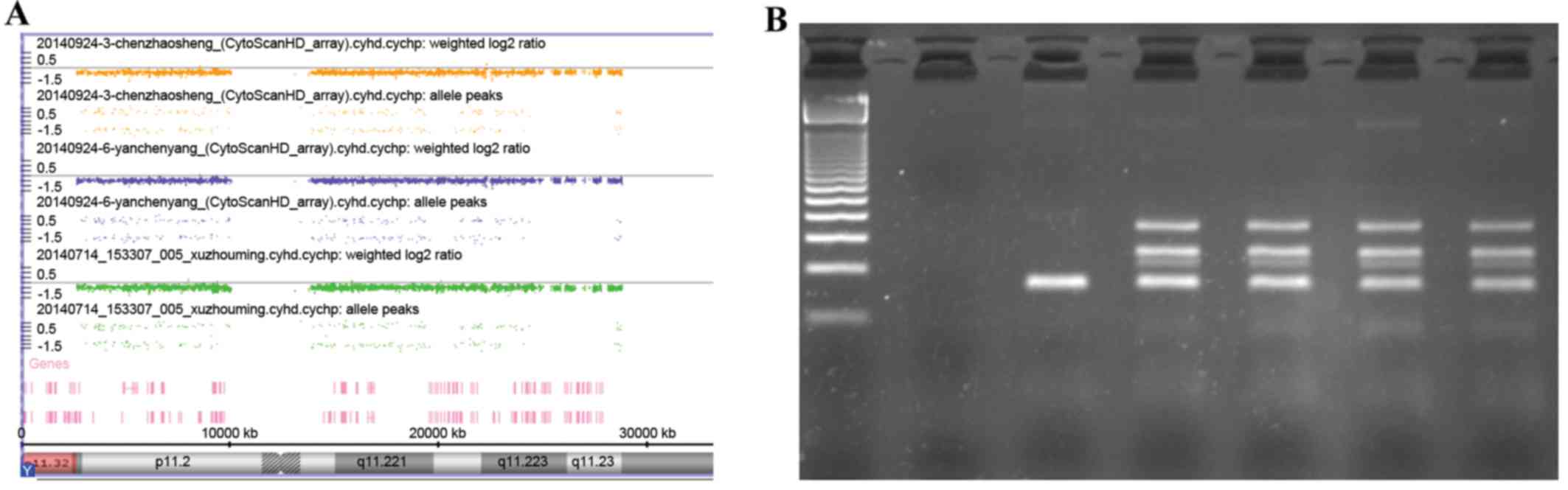
The SNP-array of three patients using the Affymetrix
Cyto HD array did not reveal any abnormalities, especially within
the Y chromosome (Fig. 2A). At a
molecular level, by means of PCR, no microdeletions were detected
in the AZF region of the Y chromosome in the infertile man (data
not shown). Y chromosome microdeletion analysis performed by using
the Y chromosome Deletion Detection system did not identify any
deletion (Fig. 2B).
Discussion
Reciprocal translocations between the Y and a
non-acrocentric chromosome, such as the de novo balance
translocation described in the present study are rarely observed
(17–20). A review of previous report
concerning a Y-autosome carrier indicated that most of the
translocations led to azoospermia or oligozoospermia (Table I), and the phenotype was associated
with the localization of the breakpoint and the nature of the lost
Yq material (21–23). In the current study, three new
cases of balanced de novo Y autosomal translocation are
described in infertile men. Our patients had normal development and
normal phenotype besides presenting severe oligozoospermia. The
cytogenetic G-banding analysis revealed Y autosomal translocation
in all metaphase cells.
 | Table I.Genotype–phenotype correlation in
adult males with Y;13,Y;6 translocation. |
Table I.
Genotype–phenotype correlation in
adult males with Y;13,Y;6 translocation.
| Karyotype | Origin | Molecular
analysis | Sperm count | (Refs.) |
|---|
|
t(Y;13)(q11.2;q12) | NP | Interval 5L
deleted | Azoospermia | (14) |
|
t(Y;13)(p11:32;p12) | de novo | Intact SRY and AZF
deletion | Oligozoospermia | (15) |
|
t(Y;6)(Yp6p;Yq6q) | NP | NP | Azoospermia | (16) |
|
t(Y;6)(q11.23;p11.1) | NP | Retention of the DAZ
gene | Oligozoospermia | (17) |
Since the sterile phenotype associated with Yq
breakpoint localization and Yq deletion, it is of great importance
to fully understand the Yq breakpoint by DNA molecular studies
(22). However, in the present
study, CMA-SNP array analysis and molecular deletion analysis
didn't reveal any deletion/duplication in the patients, as well as
microdeletions in the AZF region. This assignment and the retention
of the DAZ gene could not explain the infertility of the patients.
In addition, Y chromosome microdeletion demonstrated that no
deletion occurred in any of the patients. On this basis, the
authors propose the following hypothesis that a position effect of
unknown spermatogenesis regulatory gene(s) serve a key role in male
infertility and rearrangement of a chromosome can regulate gene
expression without microdeletion in the AZF region or other
conventional genetic factors. Alternatively, as a previous study
reported, the translocation regulates gene expression by performing
a disturbance to the heterochromatin region of chromosomes
(5).
In conclusion, the present study of three carriers
of Y-autosome translocations highlights the importance of
chromosomal rearrangement and position effect of susceptibility
genes, and may help to improve genetic counseling in male
infertility therapy.
Acknowledgements
The present study is supported by Guangdong
Provincial Natural Science Foundation (grant no. S2013040012649)
the New Teacher Project of Education Ministry (grant no.
20134423120005), the National Natural Science Funds of China (grant
nos. 81202604 and 81401205) and the Scientific Research Project of
Guangzhou Science and Information Bureau (grant no.
2014J4100024).
References
|
1
|
Giltay JC, Tiemessen CH, van Inzen WG and
Scheres JM: One normal child and a chromosomally balanced/normal
twin after intracytoplasmic sperm injection in a male with a
de-novo t(Y;16) translocation. Hum Reprod. 13:2745–2747. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Barasc H, Mary N, Letron R, Calgaro A,
Dudez AM, Bonnet N, Lahbib-Mansais Y, Yerle M, Ducos A and Pinton
A: Y-autosome translocation interferes with meiotic sex
inactivation and expression of autosomal genes: A case study in the
pig. Sex Dev. 6:143–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reijo R, Lee TY, Salo P, Alagappan R,
Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et
al: Diverse spermatogenic defects in humans caused by Y chromosome
deletions encompassing a novel RNA-binding protein gene. Nat Genet.
10:383–393. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van der Ven K, Messer L, van der Ven H,
Jeyendran RS and Ober C: Cystic fibrosis mutation screening in
healthy men with reduced sperm quality. Hum Reprod. 11:513–517.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang YT, Zhang HG, Wang RX, Yu Y, Zhang
ZH and Liu RZ: Novel Y chromosome breakpoint in an infertile male
with a de novo translocation t(Y;16): A case report. J Assist
Reprod Genet. 29:1427–1430. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martin RH: Cytogenetic determinants of
male fertility. Hum Reprod Update. 14:379–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schmid M: Demonstration of Y/autosomal
translocations using distamycin A. Hum Genet. 53:107–109. 1979.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nielsen J and Rasmussen K: Y/autosomal
translocations. Clin Genet. 9:609–617. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen CP, Lin SP, Tsai FJ, Wang TH, Chern
SR and Wang W: Characterization of a de novo unbalanced Y;autosome
translocation in a 45,X mentally retarded male and literature
review. Fertil Steril. 90:1198.e11–e18. 2008. View Article : Google Scholar
|
|
10
|
Chen CP, Fu CH, Chern SR, Wu PS, Su JW,
Lee CC, Lee MS and Wang W: De novo unbalanced translocation
resulting in monosomy for distal 5p (5p14.1 → pter) and 14q
(14q32.31 → qter) associated with fetal nuchal edema, microcephaly,
intrauterine growth restriction and single umbilical artery:
Prenatal diagnosis and molecular cytogenetic characterization.
Taiwan J Obstet Gynecol. 52:401–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen-Shtoyerman R, Ben-Yehoshua S
Josefsberg, Nissani R, Rosensaft J and Appelman Z: A prevalent Y;15
translocation in the Ethiopian Beta Israel community in Israel.
Cytogenet Genome Res. 136:171–174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dutrillaux B, Descailleaux J,
Viegas-Pequignot E and Couturier J: Y-autosome translocation in
Cacajao calvus rubicundus (Platyrrhini). Ann Genet. 24:197–201.
1981.PubMed/NCBI
|
|
13
|
Sasagawa I, Nakada T, Adachi Y, Kato T,
Sawamura T, Ishigooka M and Hashimoto T: Y-autosome translocation
associated with azoospermia. Scand J Urol Nephrol. 27:285–268.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui YX, Xia XY, Pan LJ, Wang YH, Yao B and
Huang YF: An infertile male with apparent 45,X turned out to have
45,X,der(Y)t(Y;13) (q11.2;q12),-13: Clinicopathologic and
cytogenomic studies. Fertil Steril. 88:1676.e7–e11. 2007.
View Article : Google Scholar
|
|
15
|
Alves C, Carvalho F, Cremades N, Sousa M
and Barros A: Unique (Y;13) translocation in a male with
oligozoospermia: Cytogenetic and molecular studies. Eur J Hum
Genet. 10:467–474. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Viguie F, Romani F and Dadoune JP: Male
infertility in a case of (Y;6) balanced reciprocal translocation.
Mitotic and meiotic study. Hum Genet. 62:225–227. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Delobel B, Djlelati R, Gabriel-Robez O,
Croquette MF, Rousseaux-Prevost R, Rousseaux J, Rigot JM and
Rumpler Y: Y-autosome translocation and infertility: Usefulness of
molecular, cytogenetic and meiotic studies. Hum Genet. 102:98–102.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moreau N, Teyssier M and Rollet J: A new
case of (Y;1) balanced reciprocal translocation in an infertile man
with Hodgkin's disease. J Med Genet. 24:379–380. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yumura Y, Murase M, Katayama K, Segino M,
Aizawa Y, Kuroda SN and Noguchi K: Y-autosome translocation
associated with male infertility: A case report. Hinyokika Kiyo.
58:307–310. 2012.PubMed/NCBI
|
|
20
|
Vogt PH, Edelmann A, Kirsch S, Henegariu
O, Hirschmann P, Kiesewetter F, Köhn FM, Schill WB, Farah S, Ramos
C, et al: Human Y chromosome azoospermia factors (AZF) mapped to
different subregions in Yq11. Hum Mol Genet. 5:933–943. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Krausz C, Forti G and McElreavey K: The Y
chromosome and male fertility and infertility. Int J Androl.
26:70–75. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krausz C, Chianese C, Giachini C,
Guarducci E, Laface I and Forti G: The Y chromosome-linked copy
number variations and male fertility. J Endocrinol Invest.
34:376–382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vogt PH: Genomic heterogeneity and
instability of the AZF locus on the human Y chromosome. Mol Cell
Endocrinol. 224:1–9. 2004. View Article : Google Scholar : PubMed/NCBI
|