Introduction
Allergenic pollens that cause allergic diseases, for
example, asthma (1) and rhinitis
(2,3), are those from trees or plants that
pollinate through the air. P. acerifolia trees are
frequently used as ornamental trees in the urban environment. P.
acerifolia pollen has been described to cause airway allergies
worldwide (4), in particular in
the early spring from March to April. The reported prevalence of
sensitization to P. acerifolia pollen in Mediterranean
Europe ranges between 3 and 52% (5).
So far, three major allergens including the
invertase inhibitor Pla a 1 (6–8), the
polygalacturonase Pla a 2 (9,10)
and the non-specific lipid transfer protein (nsLTP) Pla a 3
(9,11), have been identified from P.
acerifolia pollen. Pla a 1 (18 kDa) and Pla a 2 (43 kDa) have
been reported as major allergens with 80–90% sensitization
frequencies among Spanish patients with plane pollen allergy
(5,12), whereas 45% of plane pollen
allergies were sensitized to purified natural Pla a 3 (9).
Allergen-specific immunotherapy (AIT) is an
effective treatment for allergic diseases. There is evidence that
AIT can relieve symptoms in patients with asthma (13,14),
allergic conjunctivitis (15) and
allergic rhinitis (16). The
administration of increasing doses of allergen extracts to patients
is the most commonly applied method. However, the use of crude
extracts has several disadvantages. It may increase the risk of
allergic side effects or result in the sensitization towards new
allergens present in crude extracts (17,18).
Compared with allergen extracts, recombinant allergens can be
formulated with high quality and precise quantity.
In the present study, the authors expressed and
purified the Pla a 3 allergen in the E. coli system, which
will provide a foundation for further study of this allergen in
diagnosis and treatment of plane pollen allergy. Moreover, the
identification of B cell and T cell epitopes of allergens will
benefit to the diagnosis, therapy and development of effective
vaccines for immunotherapy (19,20).
Until now, there has been no report of the epitope of the Pla a 3
allergen. In the present study, the authors identified the B and T
cell epitopes of Pla a 3 allergen, which contribute to the
potential utility in a peptide-based vaccine design for pollen
allergy.
Materials and methods
Patients and samples
A total of 12 subjects were included in the study,
comprising two patient groups: (A) 6 allergic rhinitis patients
(aged 15–34; 3 males and 3 females; recruited between January and
June 2015) with positive skin prick test (allergens were supplied
by ALK-Abelló, Copenhagen, Denmark) and positive serum IgE test to
the P. acerifolia pollen extract by using ImmunoCAP assay
(Phadia AB, Uppsala, Sweden); (B) 6 healthy controls (aged 20–45; 3
males and 3 females; recruited in June 2015). The study protocol
was approved by the ethical committee of the First Affiliated
Hospital of Nanjing Medical University (Nanjing, China). Written
informed consent for the use of blood samples were obtained from
all participants before study entry according to the Declaration of
Helsinki.
Sequence retrieval
The complete nucleotide acid and amino acid
sequences of Pla a 3 allergen were retrieved from the GenBank
database. The open reading frames (ORF) of Pla a 3 allergen had 354
bases pairs, encoding 118 amino acids. Because this ORF contained a
26 amino acid signal peptide, the mature Pla a 3 allergen had 276
bases pairs, encoding 92 amino acids.
Expression and purification of Pla a 3
allergen in E. coli
The nucleotide acid of mature Pla a 3 allergen was
synthesized by GenScript (Piscataway, NJ, USA) and sub-cloned into
pSUMO-Mut vector using Stu I and Xho I sites
and verified by DNA sequencing. The recombinant pSUMO-Mut-Pla a 3
plasmid was transformed into Arctic Express™ (DE3) RP E.
coli host strain.
A positive colony was selected to inoculate in a 3
ml lysogeny broth (LB)-kanamycin broth (Shanghai Sanger Biotech
Technology, Co., Ltd., Shanghai, China), and incubated at 37°C
overnight. A total of 1% of the overnight culture of the
transformant was transferred into fresh LB-kanamycin broth and
cultured at 37°C with shaking at 200 × g up to the absorbance
0.6–0.8 at 600 nm. The culture was induced with 1 mM
isopropyl-b-D-thiogalactopyranoside (IPTG) and harvested following
an additional 4 h incubation. The culture was disrupted by
sonication at 60 kHz, 4 sec pulse-on, 8 sec pulse-off and
centrifuged at 10,000 × g for 30 min at 4°C. The results indicated
that recombinant Pla a 3 was mainly in supernatant fraction
(Fig. 1) and purified by Nickel
affinity chromatography (GenScript). The washing buffer contains
100 mM NaH2PO4, 20 mM Tris-HCl, 10 mM
imidazole, pH 8.0, and the eluting buffer contains 100 mM
NaH2PO4, 20 mM Tris-HCl, 250 mM imidazole, pH
8.0.
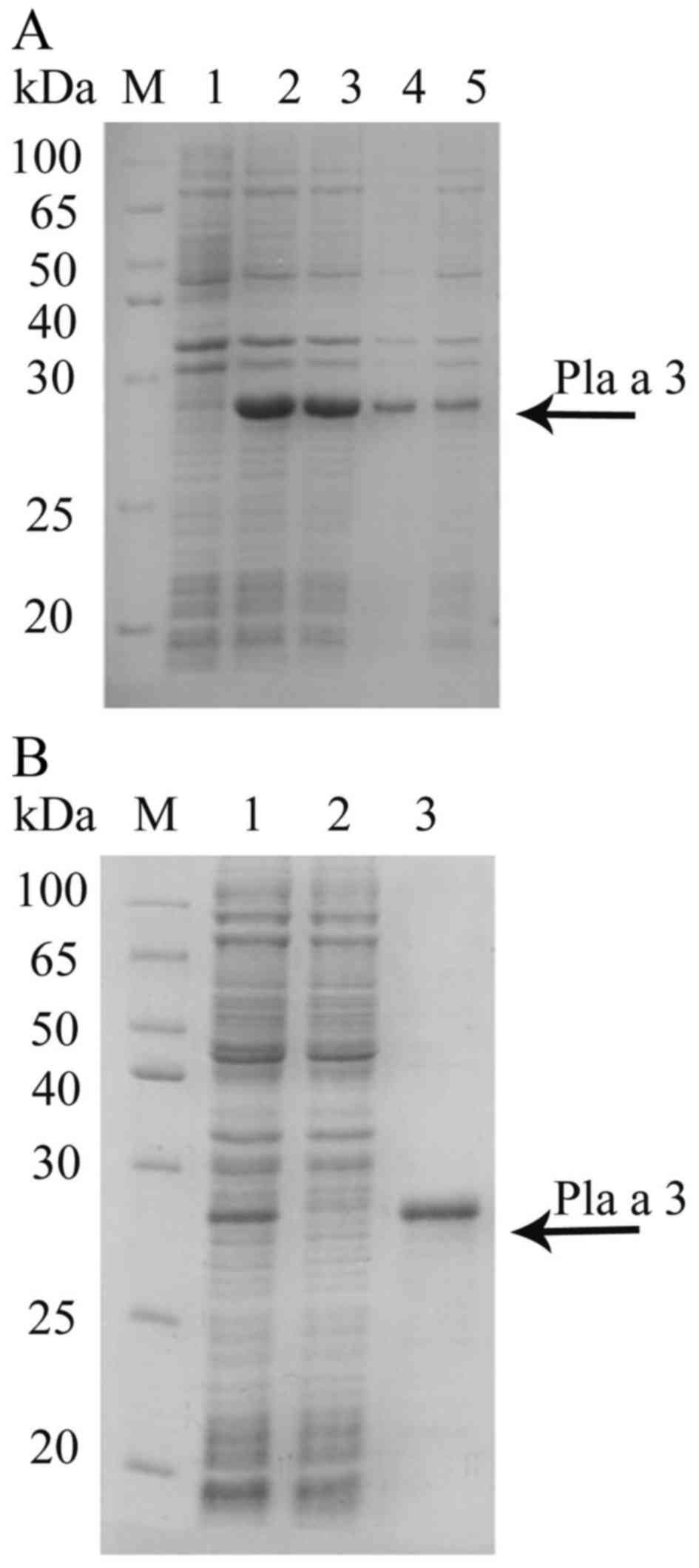 | Figure 1.Expression and purification of the
Pla a 3 allergen in E. coli. (A) Lane M, protein molecular
weight standard; lane 1, before induction; lanes 2 and 3,
isopropyl-b-D-thiogalactopyranoside-induced recombinant Pla a 3
allergen; lane 4, precipitation fraction following ultrasonication;
lane 5, supernatant fraction following ultrasonication. The Pla a 3
allergen is denoted with an arrow. (B) SDS-PAGE following affinity
chromatography of the Pla a 3 allergen. Lane M, protein molecular
weight standard; lane 1, no-binding protein; lane 2, elute washed
with 10 mM imidazole, 100 mM NaH2PO4, 20 mM
Tris-HCl; lane 3, elute with 250 mM imidazole, 100 mM
NaH2PO4, 20 mM Tris-HCl. The arrow points to
the purified Pla a 3 allergen. |
IgE binding activity of recombinant
Pla a 3 allergen
An IgE binding assay was conducted as previously
described. Briefly, proteins were analyzed via 12% sodium
dodecylsulfatc-polyacrylamide gel electrophoresis (SDS-PAGE)
(21). The gel was transferred to
polyvinylidene difluoride membranes (22). The blots were blocked in 5% skim
milk for 2 h, then incubated with serum of P. acerifolia
pollen allergic patients (diluted 1:40 in PBS) as the first
antibody overnight at 4°C. Then, IgE that bound to the allergen was
detected with a horseradish peroxidase-conjugated goat anti-human
IgE mAb (1:3,000, catalog no. A9667, Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) at 4°C for 1 h and then detected using a
ImageQuant LAS 4000 mini detection system (GE Healthcare Life
Sciences, Chalfont, UK). A total of 6 healthy individuals were used
as a negative serum control.
Homology modeling
The Pla a 3 allergen protein sequence was searched
for homology in the PDB. As well, the homologous templates suitable
for Pla a 3 allergen were selected from SWISS-MODEL server
(http://swissmodel.expasy.org/) (23). SWISS-MODEL Repository is a database
of 3D protein structure models generated by The Pla a 3 allergen
protein sequence Homology modeling. The Pla a 3 allergen
protein sequence was searched for homology in the Protein Data
Bank. In addition, the homologous templates suitable for the Pla a
3 allergen were selected from the SWISS-MODEL server (http://swissmodel.expasy.org/) (23). The SWISS-MODEL Repository is a
database of 3D protein structure models generated by the
SWISS-MODEL homology-modeling pipeline (24). The best template was retrieved from
the results of previous methods and used for homology modeling. Pla
a 3 allergen modeled protein structure was built through alignment
mode in SWISS-MODEL using the complete amino acid sequence.
Prediction of B cell epitopes
A total of three immunoinformatics tools including
the DNAStar protean system, the Bioinformatics Predicted Antigenic
Peptides (BPAP) system (http://imed.med.ucm.es/Tools/antigenic.pl) and the
BepiPred 1.0 server (http://www.cbs.dtu.dk/services/BepiPred/) were used to
predict the B cell epitopes of the Pla a 3 allergen. The ultimate
consensus epitope results were obtained by combining the results of
the three tools together (25). In
the DNAStar protean system, four properties (hydrophilicity,
flexibility, accessibility and antigenicity) of the amino acid
sequence were chosen as parameters for epitopes prediction
(26). The peptide regions with
good hydrophilicity, high flexibility, surface accessibility and
high antigenic index were chosen as candidate epitopes for further
investigation.
Prediction of T cell epitopes
T cell epitopes are principally predicted indirectly
by identifying the binding of peptide fragments to the major
histocompatibility complexes (MHCs). The binding significance of
each peptide to the given MHC molecule was based on the estimated
strength of binding exhibited by a predicted nested core peptide at
a set threshold level. SYFPEITHI and NetMHCII-2.2 (http://www.cbs.dtu.dk/services/NetMHC
II-2.2) (27) were used to predict
T cell epitopes of the Pla a 3 allergen. In the current study,
HLA-DR10101 and HLADR50101 were used to predict HLA-DR-based T cell
epitope prediction. As a result, the ultimate results of T epitopes
were obtained by combining the results of the SYFPEITHI and
NetMHCII-2.2. B cell and T cell epitopes identified by
immunoinformatics tools were mapped onto a linear sequence and
presented on the three dimensional model of the Pla a 3 allergen to
determine their position.
Results
Expression and purification of the Pla
a 3 allergen in E. coli
The Pla a 3 allergen was subcloned into a
pSUMO-Mut vector and transformed into the E. coli
host strain. The culture was induced with IPTG and expressed at
37°C for 4 h and the resulting protein presented a band with a
apparent molecular weight of 28 kDa (Fig. 1A; lane 2 and lane 3), and no such
band was seen in the non-induced cells (Fig. 1A; lane 1). Protein was produced
primarily in the supernatant fraction following sonication
(Fig. 1A; lane 5) and purified by
Ni column. Finally, the purity of the purified Pla a 3 allergen was
identified by SDS-PAGE. As presented in Fig. 1B, the recombinant Pla a 3 allergen
was successfully purified by elution buffer.
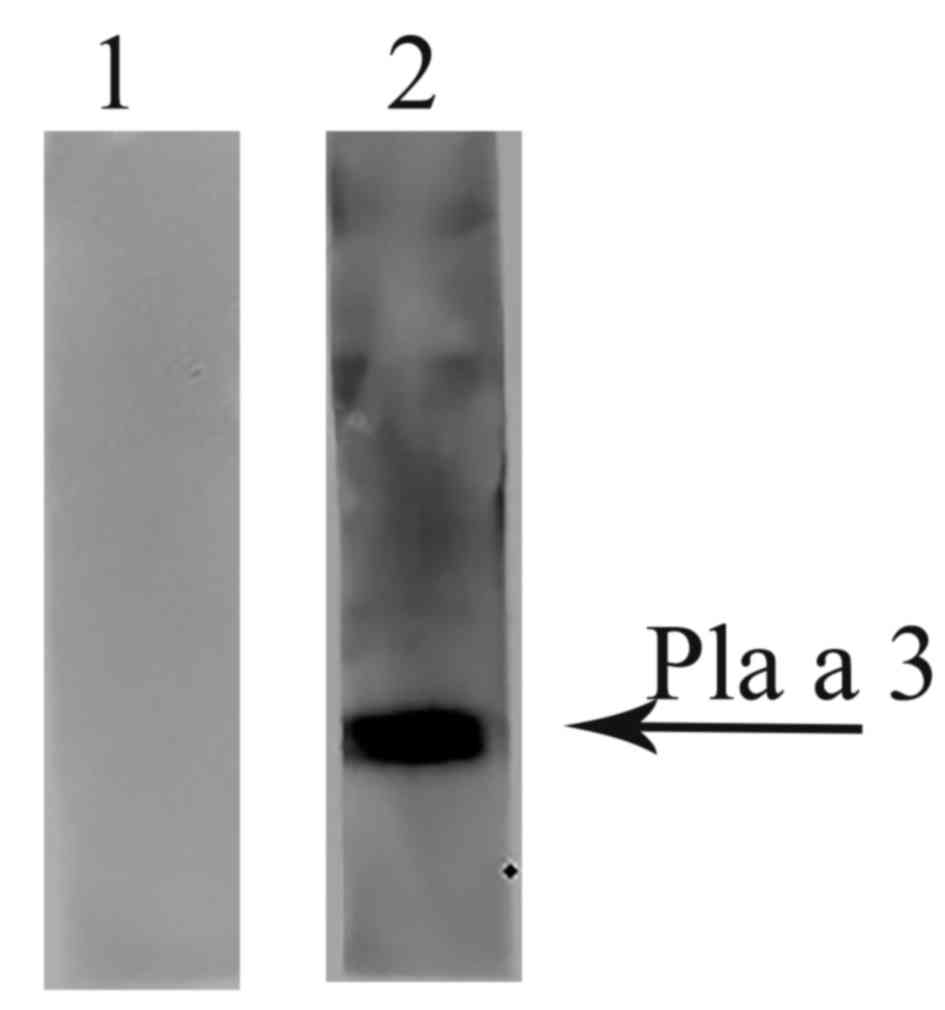
IgE binding activity of the
recombinant Pla a 3 allergen
The ability of recombinant Pla a 3 allergen to bind
IgE from the P. acerifolia pollen allergy patients' serum
was determined by western blotting. Fig. 2 demonstrated that mixed P.
acerifolia pollen allergy patients' serum revealed positive IgE
reactivity to the Pla a 3 allergen, but healthy controls failed
to.
Homology modeling
Searching for the proteins with known tertiary
structure in the Protein Data Bank, 1t12.1.A (PDB accession number)
reported the highest sequence identity (62.64%) to the Pla a 3
allergen. Therefore, the best template 1t12.1.A was used for
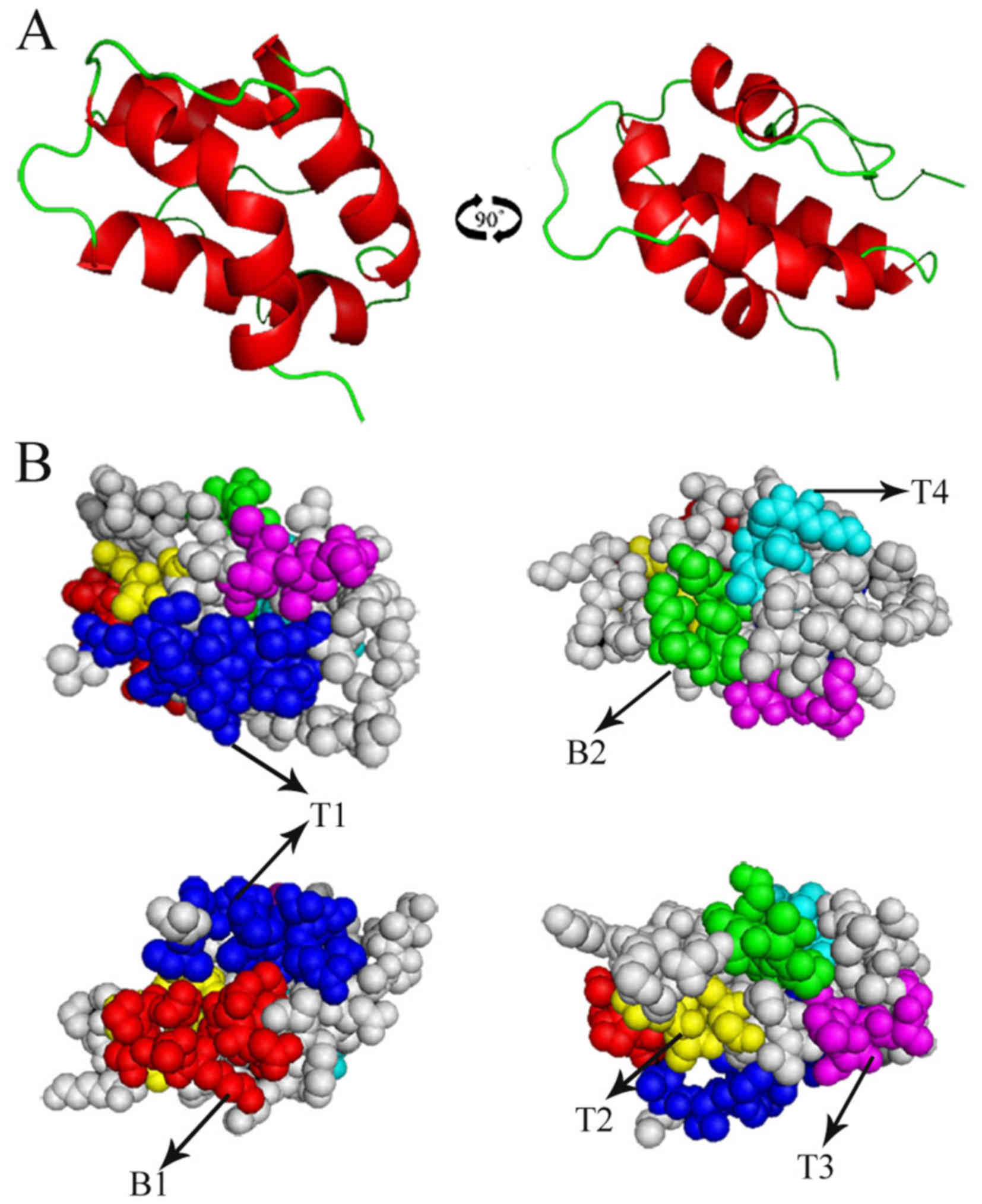
homology modeling. Fig. 3A
demonstrated the overall 3D structure of the Pla a 3 allergen, and
Fig. 3B demonstrated the predicted
B cell and T cell epitopes superimposition on the surface of the
Pla a 3 allergen structure.
B cell epitope prediction
Surface accessibility and fragment flexibility are
important features for predicting antigenic epitopes. In addition,
the existence of regions with high hydrophobicity provides strong
evidence for epitope identification. Antigenic index directly
indicated the epitope forming capacity of the Pla a 3 allergen
sequence. Based on these sequence properties, the final predicting
regions of the Pla a 3 allergen by DNAstar were obtained as: 35–45
and 81–86 (Table I). The predicted
results of the BPAP system were 4–17, 19–34, 45–54 and 63–88. The
predicted results of the BepiPred 1.0 server were 22–26, 35–47,
57–59 and 81–87. Furthermore, the final potential B cell epitopes
of the Pla a 3 allergen were selected on the basis of the results
of these three tools. The ultimate results of the three
immunoinformatics tools finally predicted two peptides (35–45 and
81–86) and these peptides are presented in Fig. 3.
 | Table I.Location of the B and T cell epitopes
of the Pla a 3 allergen predicted by immunoinformatics tools. |
Table I.
Location of the B and T cell epitopes
of the Pla a 3 allergen predicted by immunoinformatics tools.
|
| Tools | Location of the
prediction results |
|---|
| B cell epitope
prediction | DNAStar
protean |
|
|
| Hydrophilicity | 33-47, 83–92 |
|
| Flexible
regions | 10-12, 19–22,
35–45, 53–61, 81–86 |
|
| Antigenic
index | 33–48, 50–57,
86–92 |
|
| Surface
probability | 35–45, 54–55,
81–82, 91–92 |
|
| BPAP | 4–17, 19–34, 45–54,
63–88 |
|
| BepiPred | 22–26, 35–47,
57–59, 81–87 |
| T cell epitope
prediction | SYFPEITHI | 4–18, 29–37, 57–63,
67–73, 77–85 |
|
| NetMHCII
(HLA-DR) |
|
|
| DRB1*01:01 | 2–16, 45–50,
55–61 |
|
| DRB5*01:01 | 2–15, 47,
56–59 |
T cell epitope prediction
SYFPEITHI and NetMHCII-2.2 were used to identify the
T cell epitope of the Pla a 3 allergen. The predicted results of
SYFPEITHI were 4–18, 29–37, 57–63, 67–73 and 77–85 (Table I). For HLA-DR-based T cell epitope
prediction, the final predicting regions of HLA-DR10101 and HLA-DR
50101 are presented in Table I. As
a result, the Pla a 3 allergen was predicted to have four T cell
epitope sequences including 2–15, 45–50, 55–61 and 67–73, as
presented in Table II.
 | Table II.Predicted B and T cell epitopes of
the Pla a 3 allergen. |
Table II.
Predicted B and T cell epitopes of
the Pla a 3 allergen.
| Number | Type of
epitope | Position | Sequence |
|---|
| P1 | B1 | 35–45 | LNNDAKTTPDR |
| P2 | B2 | 81–86 | KISPTI |
| P3 | T1 |
2–15 | ITCGTVVTRLTPCL |
| P4 | T2 | 45–50 | RQAACG |
| P5 | T3 | 55–61 | ASTSISG |
| P6 | T4 | 67–73 | AASLAGK |
Discussion
P. acerifolia pollen is the most frequently
present in pollen counts (28,29)
and the contribution of it in pollen allergies has increased over
the past decade. A high percentage of patients with seasonal
pollinosis are sensitive to it. The quantification of major
allergens has become a crucial goal for the standardization of
allergen products intended for clinical use (30,31).
Therefore, the determination of Pla a 3 allergen content is very
important for the development of a P. acerifolia pollen
allergen vaccine since it is the major allergen in P.
acerifolia pollen. In the present study, the Pla a 3 allergen
was expressed successfully in the soluble form in E. coli.
The purified Pla a 3 allergen was analyzed by western blotting and
the results revealed that the Pla a 3 allergen has the ability to
bind IgE in the P. acerifolia pollen allergic patients' sera
(5,32,33).
Although used in clinical settings for more than a
century, AIT is currently the only known causal treatment of
allergic diseases (34). To
relieve allergic symptoms, AIT serves a significant role in
preventing new allergies and shows long-term effects following
termination of treatment. The safety and efficacy of AIT has been
demonstrated in a large number of clinical trials. However, AIT is
not effective in all allergic individuals, and it carries the risk
of adverse effects. For this reason, there is a strong requirement
to increase its safety and effectiveness by further research.
Establishing the optimal route and dose of administration may
benefit to a develop better clinical desensitization effect than
before. Growing evidence on the immunological effects of AIT,
especially B cell and T cell epitopes related to allergen
tolerance, provide novel concepts for safer and more effective
vaccination. Therefore, the authors predicted B and T cell epitopes
of the Pla a 3 allergen in the present study. Firstly, in order to
better understand the structure and function of the Pla a 3
allergen, the basic sequence properties were analyzed and the 3D
structures of the Pla a 3 allergen were studied. The 3D structure
of the Pla a 3 allergen was performed by homology modeling which is
widely used in many areas of structure-based analysis. The Protein
Data Bank server was used to search templates of the Pla a 3
allergen and reported that the structure of 1t12.1.A was the best
template with the highest identity (62.64%). An earlier study
demonstrated that the use of bioinformatics approach to predict B
cell epitopes correlated well with the experimental approach
(35). In the current study, the
authors used three immunoinformatics tools (DNAStar protean system,
BPAP and the BepiPred 1.0 server) to predict the B cell epitopes.
As a result, two B cell epitopes (35–45 and 81–86) were predicted
as the B cell epitopes. However, these B cell epitopes need to be
confirmed in further clinical samples. On the other hand, SYFPEITHI
and NetMHCII-2.2 were used to predict the T cell epitopes and
predicted four potential T cell epitopes including 2–15, 45–50,
55–61 and 67–73. Despite the high accuracy of these predictions,
this approach has not yet been applied to peptide-based vaccine
development for allergic diseases.
In summary, the successful production of the
recombinant Pla a 3 allergen revealed the importance of the Pla a 3
allergen in P. acerifolia pollen allergy, and provides a
foundation for further study of the allergen in diagnosis and
treatment of plane pollen allergy. Moreover, the authors have
predicted two B cell epitopes (35–45 and 81–86) and four T cell
epitopes including 2–15, 45–50, 55–61 and 67–73 of the Pla a 3
allergen, which can be used to contribute to allergen
immunotherapies and is useful in peptide-based vaccine designs of
plane pollen allergy.
Acknowledgements
The current project was sponsored by a grant from
the Special Fund for Forestry-scientific Research in the Public
Interest (grant no. 201304103), grants from the National Natural
Science Foundation of China (grant nos. 81571568, 31340073 and
81273274) and the Jiangsu Province's Key Provincial Talents Program
(grant no. RC201170).
References
|
1
|
Subiza J, Cabrera M, Valdivieso R, Subiza
JL, Jerez M, Jiménez JA, Narganes MJ and Subiza E: Seasonal asthma
caused by airborne platanus pollen. Clin Exp Allergy. 24:1123–1129.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Desai MB, Gavrilova T, Liu J, Patel SA,
Kartan S, Greco SJ, Capitle E and Rameshwar P: Pollen-induced
antigen presentation by mesenchymal stem cells and T cells from
allergic rhinitis. Clin Transl Immunology. 2:e72013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dondi A, Tripodi S, Panetta V, Asero R,
Businco AD, Bianchi A, Carlucci A, Ricci G, Bellini F, Maiello N,
et al: Pollen-induced allergic rhinitis in 1360 Italian children:
comorbidities and determinants of severity. Pediatr Allergy
Immunol. 24:742–751. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alcázar P, García-Mozo H, Trigo Mdel M,
Ruiz L, González-Minero FJ, Hidalgo P, de la Díaz Guardia C and
Galán C: Platanus pollen season in Andalusia (southern Spain):
Trends and modeling. J Environ Monit. 13:2502–2510. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fernández-González D, González-Parrado Z,
Vega-Maray AM, Valencia-Barrera RM, Camazón-Izquierdo B, De Nuntiis
P and Mandrioli P: Platanus pollen allergen, Pla a 1:
Quantification in the atmosphere and influence on a sensitizing
population. Clin Exp Allergy. 40:1701–1708. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Asturias JA, Ibarrola I, Eraso E, Arilla
MC and Martínez A: The major platanus acerifolia pollen allergen
Pla a 1 has sequence homology to invertase inhibitors. Clin Exp
Allergy. 33:978–985. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arilla MC, Ibarrola I, Mir A, Monteseirín
J, Conde J, Martínez A and Asturias JA: Development of a
sandwich-type ELISA for measuring Pla a 1, the major allergen of
platanus acerifolia pollen. Int Arch Allergy Immunol. 138:127–133.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fernández-González M, Guedes A, Abreu I
and Rodríguez-Rajo FJ: Pla a_1 aeroallergen immunodetection related
to the airborne platanus pollen content. Sci Total Environ.
463–464:855–860. 2013. View Article : Google Scholar
|
|
9
|
Lauer I, Miguel-Moncin MS, Abel T,
Foetisch K, Hartz C, Fortunato D, Cistero-Bahima A, Vieths S and
Scheurer S: Identification of a plane pollen lipid transfer protein
(Pla a 3) and its immunological relation to the peach
lipid-transfer protein, Pru p 3. Clin Exp Allergy. 37:261–269.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ibarrola I, Arilla MC, Martínez A and
Asturias JA: Identification of a polygalacturonase as a major
allergen (Pla a 2) from platanus acerifolia pollen. J Allergy Clin
Immunol. 113:1185–1191. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wangorsch A, Larsson H, Messmer M,
García-Moral A, Lauer I, Wolfheimer S, Schülke S, Bartra J, Vieths
S, Lidholm J and Scheurer S: Molecular cloning of plane pollen
allergen Pla a 3 and its utility as diagnostic marker for peach
associated plane pollen allergy. Clin Exp Allergy. 46:764–774.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iglesias I, Rodriguez-Rajo FJ and Méndez
J: Behavior of platanus hispanica pollen, an important spring
aeroallergen in northwestern Spain. J Investig Allergol Clin
Immunol. 17:145–156. 2007.PubMed/NCBI
|
|
13
|
Abramson MJ, Puy RM and Weiner JM:
Allergen immunotherapy for asthma. Cochrane Database Syst Rev.
Cd0011862003.PubMed/NCBI
|
|
14
|
Penagos M, Passalacqua G, Compalati E,
Baena-Cagnani CE, Orozco S, Pedroza A and Canonica GW: Metaanalysis
of the efficacy of sublingual immunotherapy in the treatment of
allergic asthma in pediatric patients, 3 to 18 years of age. Chest.
133:599–609. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dahl R, Kapp A, Colombo G, de Monchy JG,
Rak S, Emminger W, Rivas MF, Ribel M and Durham SR: Efficacy and
safety of sublingual immunotherapy with grass allergen tablets for
seasonal allergic rhinoconjunctivitis. J Allergy Clin Immunol.
118:434–440. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calderon MA, Alves B, Jacobson M, Hurwitz
B, Sheikh A and Durham S: Allergen injection immunotherapy for
seasonal allergic rhinitis. Cochrane Database Syst Rev.
CD0019362007.PubMed/NCBI
|
|
17
|
Eifan AO, Calderon MA and Durham SR:
Allergen immunotherapy for house dust mite: Clinical efficacy and
immunological mechanisms in allergic rhinitis and asthma. Expert
Opin Biol Ther. 13:1543–1556. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frati F, Scurati S, Puccinelli P, Justicia
JL, Adamec T, Sieber HJ, Ras L, David M, Marcucci F and Incorvaia
C: Development of an allergen extract for sublingual
immunotherapy-evaluation of staloral. Expert Opin Biol Ther.
9:1207–1215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alvarez-Cuesta E, Bousquet J, Canonica GW,
Durham SR, Malling HJ, Valovirta E, et al: EAACI, Immunotherapy
Task Force: Standards for practical allergen-specific
immunotherapy. Allergy. 61:(Suppl 82). S1–S20. 2006. View Article : Google Scholar
|
|
20
|
Sharma V, Singh BP, Gaur SN, Pasha S and
Arora N: Bioinformatics and immunologic investigation on B and T
cell epitopes of Cur l 3, a major allergen of curvularia lunata. J
Proteome Res. 8:2650–2655. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Towbin H, Staehelin T and Gordon J:
Electrophoretic transfer of proteins from polyacrylamide gels to
nitrocellulose sheets: Procedure and some applications. Proc Natl
Acad Sci USA. 76:4350–4354. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arnold K, Bordoli L, Kopp J and Schwede T:
The SWISS-MODEL workspace: A web-based environment for protein
structure homology modelling. Bioinformatics. 22:195–201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kiefer F, Arnold K, Künzli M, Bordoli L
and Schwede T: The SWISS-MODEL repository and associated resources.
Nucleic Acids Res. 37:(Database Issue). D387–D392. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang X and Yu X: An introduction to
epitope prediction methods and software. Rev Med Virol. 19:77–96.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zheng LN, Lin H, Pawar R, Li ZX and Li MH:
Mapping IgE binding epitopes of major shrimp (penaeus monodon)
allergen with immunoinformatics tools. Food Chem Toxicol.
49:2954–2960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nielsen M and Lund O: NN-align. An
artificial neural network-based alignment algorithm for MHC class
II peptide binding prediction. BMC Bioinformatics. 10:2962009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Garrido-Lestache J Subiza: Allergenic
pollens in Spain. Allergol Immunopathol (Madr). 32:121–124.
2004.(In Spanish). PubMed/NCBI
|
|
29
|
Miralles JC, Caravaca F, Guillén F,
Lombardero M and Negro JM: Cross-reactivity between platanus pollen
and vegetables. Allergy. 57:146–149. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Boluda L, Alonso C and Fernández-Caldas E:
Purification, characterization, and partial sequencing of two new
allergens of Olea europaea. J Allergy Clin Immunol. 101:(2 Pt 1).
210–216. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
van Ree R: CREATE Partnership: The CREATE
project: EU support for the improvement of allergen standardization
in Europe. Allergy. 59:571–574. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Asero R, Mistrello G, Amato S and Villalta
D: Monosensitization to a novel plane pollen allergen. Eur Ann
Allergy Clin Immunol. 44:167–169. 2012.PubMed/NCBI
|
|
33
|
Enrique E, Alonso R, Bartolomé B,
Miguel-Moncín M San, Bartra J, Fernández-Parra B, Tella R, Asturias
JA, Ibarrola I, Martínez A and Cisteró-Bahíma A: IgE reactivity to
profilin in platanus acerifolia pollen-sensitized subjects with
plant-derived food allergy. J Investig Allergol Clin Immunol.
14:335–342. 2004.PubMed/NCBI
|
|
34
|
Jutel M, Kosowska A and Smolinska S:
Allergen immunotherapy: Past, present, and future. Allergy Asthma
Immunol Res. 8:191–197. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nair S, Kukreja N, Singh BP and Arora N:
Identification of B cell epitopes of alcohol dehydrogenase allergen
of curvularia lunata. PLoS One. 6:e200202011. View Article : Google Scholar : PubMed/NCBI
|