Introduction
The publication of the human genome sequence
increased the ability to identify novel diagnostic and therapeutic
methods for the treatment of human diseases. The combination of
morphological methods, the well-established biology of premalignant
and invasive alterations, together with molecular biology
techniques, which enables the expression of several hundred or even
several thousand genes to be evaluated simultaneously, increases
the precision of diagnosis and therapy and leads to the development
of effective targeted molecular therapies for patients. The ability
to use molecular criteria for the selection of an appropriate
therapy for a particular patient has led to the rapid development
of personalized medicine, particularly in oncology (1–3).
Individualization of patient treatment involves adapting therapies
to the individual characteristics of the particular patient and
their disease. Personalized medicine increases the likelihood that
an appropriate, effective and safe therapy is selected. For this
reason, molecular biology techniques are increasingly becoming a
widespread diagnostic tool in clinical practice (4,5).
Molecular diagnostics facilitates the detection of neoplastic
lesions in high-risk individuals at an early stage of disease, as
alterations at the molecular level precede alterations at the
phenotypic level. In addition, it enables the detection of drug
resistance, which allows the precision of the treatment strategy to
be improved.
Various mechanisms are associated with an altered
expression profile of genes involved in the regulation of
angiogenesis (6,7). Understanding these differences may be
important for diagnosis and therapy. One current method of
effective anticancer therapy is an anti-angiogenic therapy that
targets products of genes involved in the regulation of
angiogenesis. This treatment demonstrates a positive effect in
numerous cancer types; however, extended duration of treatment
often results in drug resistance (8,9).
Maintaining anti-angiogenic therapy sometimes requires change of
the drug as well as the molecular target due to drug resistance
(10–12). Consequently, the search for novel
targets for targeted therapy is ongoing (13,14).
Cancer of the uterus lining, also termed the
endometrium, is one of the most commonly diagnosed gynecological
cancers worldwide, and primarily affects postmenopausal women
(15,16). According to the World Health
Organization classification system, there are three histological
grades (G) of endometrial cancer, including G1 (well
differentiated), G2 (moderately differentiated) and G3 (poorly
differentiated) (17). According
to clinical, metabolic and endocrine characteristics, there are two
types of endometrial cancer. Type I are estrogen-associated,
low-grade (G1 and G2) and are often associated with obesity. They
are diagnosed early, and therefore demonstrate a favorable
prognosis. Type II are hormone-independent, high-grade (G3) and
commonly occur in non-obese women. This type of endometrial cancer
metastasizes early, and patients demonstrate a poorer prognosis
when compared with type I endometrial cancers (18,19).
The authors of the present study hypothesized that the expression
profile of genes involved in the regulation of angiogenesis may
present diagnostic, prognostic and therapeutic targets in
endometrial cancer. The aim of the current study was therefore to
identify and select genes involved in the regulation of
angiogenesis, which have altered transcriptional activity depending
on the degree of histopathological differentiation of endometrial
adenocarcinoma, that may present diagnostic, prognostic and
therapeutic targets.
Materials and methods
Patients
The present study enrolled 36 patients, 30 with
endometrial cancer with degrees of histopathological
differentiation between G1 and G3 (the case study group), and a
control group consisting of 6 patients without endometrial cancer
that qualified for the removal of the uterus (hysterectomy) due
pathologies of the uterus and adnexa. Patients in the control group
that agreed to participate in the present study qualified for
surgical removal of the uterus (abdominal hysterectomy) with the
following medical indications: Uterine fibroids, benign tumors of
the appendages, or reproductive organ prolapse. The patients were
admitted into Regional Railway Hospital in Katowice from January to
May 2015. The average age of patients in the study group was 65 and
in the control group, 48. All patients provided written informed
consent for the use of their samples in the present study. The
criteria for exclusion from the case study group included patients
that, according to medical history or postoperative pathological
examination, were diagnosed with a form of cancer other than
endometrial endometrioid adenocarcinoma. Additional exclusion
criteria were the detection of endometrial hyperplasia with or
without atypia during postoperative pathological examination, the
use of hormone replacement therapy in the 5 years prior to the
operation and extreme obesity (body mass index, >40). The
histopathological assessments of tumor samples were performed in
the Department of Pathomorphology, Medical University of Silesia
(Katowice, Poland). A total of 28 samples (control, 3; G1, 7; G2,
12; and G3, 6) were selected for microarray analysis.
Ethical approval
The present study was approved by the Bioethical
Committee of the Medical University of Silesia (Sosnowiec, Poland;
no. KNW/0022/KB1/67/13). The study adhered to the tenets of the
Declaration of Helsinki.
Molecular analysis
Molecular analysis of the endometrial samples from
patients that had undergone a hysterectomy (study case group and
control group) involved the extraction of total RNA using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's
instructions. Extracted total RNA was then purified using an RNeasy
Mini kit (Qiagen GmbH, Hilden, Germany) using columns and an
RNase-Free DNase Set (Qiagen GmbH) according to the manufacturer's
instructions. Analysis of gene expression profiles was determined
using an oligonucleotide microarray technique with
GeneChip® Human Genome U133A plates (Affymetrix, Inc.,
Santa Clara, CA, USA). The first step of the analysis was cDNA
synthesis (8 µg RNA was used as a template) with the use of
SuperScript Choice System (Invitrogen; Thermo Fisher Scientific,
Inc.). Then, the synthesis of biotinylated cRNA was performed using
BioArray HighYield RNA Transcript Labeling kit (Enzo Life Sciences,
Farmingdale, NY, USA). Subsequently, the Sample Cleanup Module kit
(Qiagen GmbH, Germany) was used to perform fragmentation of the
biotin-labeled cRNA. After the cRNA hybridized to the HG-U133A
microarray, it was stained with streptavidin-phycoerythrin and
scanned using GeneArray Scanner G2500A (Agilent Technologies, Inc.,
Santa Clara, CA, USA). The data was processed for signal values
using Microarray Suite version 5.0 software (Affymetrix, Inc.). All
of the procedures were performed as recommended by Affymetrix Gene
Expression Analysis Technical Manual (20). Comparative analysis of the
transcriptome was performed for 691 mRNA sequences of genes
encoding proteins involved in the regulation of angiogenesis, which
were selected based on the results of the literature, the NCBI
database (Gene, http://ncbi.nlm.nih.gov/gene) and an Affymetrix
NetAffx™ Analysis Center database (http://www.affymetrix.com/analysis/index.affx).
Statistical analysis
GeneSpring GX version 12.6.1 software (Agilent
Technologies, Inc.) and PL-Grid Infrastructure (http://www.plgrid.pl/) were used for statistical
analysis of the data after microarrays scanning. Microarray
analysis was performed using the following specialized programs
adapted to the analysis of the matrix experiment results:
Microarray Suite 5.0 software (Affymetrix, Inc.), GeneSpring GX
(version, 12.6.1; Agilent Technologies, Inc.) and PANTHER version
11.1 (Protein Analysis Through Evolutionary Relationships,
http://pantherdb.org) (21), Gene List Analysis tool. Initially,
the degree of RNA degradation was assessed using 3′/5′ ratio
(signal intensity ratio of the 3′ probe set over the 5′ probe set)
of the housekeeping genes GAPDH and β-actin. This parameter
was termed RNA degradation index and is commonly used in Affymetrix
U133 plates for RNA quality assessment. This index reflects the
level of RNA integrity and accuracy of sample processing. A low
index (3′/5′ ratio<3) corresponds to high quality material,
whereas a high index (3′/5′ ratio>3) could indicate low quality
material and/or problems during sample processing. The control of
microarray hybridization with mRNA was based on the 8 exogenous
controls, 8 probes complementary to the exogenous RNA added in
varying concentrations to the hybridization cocktail by the
manufacturer of the microarray (Affymetrix, Inc.). The following
probes were used in the present study: AFFX-BioB_at, AFFX-BioC_at,
AFFX-BioDn_at, AFFX-CreX_at, AFFX-r2-Ec-BioB_at,
AFFX-r2-Ec-BioC_at, AFFX-r2-Ec-BioD_at, AFFX-r2-P1-cre_at.
Following the acceptance of the microarray for comparative
analysis, the obtained results were normalized using the Robust
Multi-array Average (RMA) Express program version 1.1.0 (http://rmaexpress.bmbolstad.com/) (22), and genes that were differentially
expressed in the transcriptome depending on the severity of the
disease (histopathological grade) were selected for molecular
analysis using GeneSpring GX software (version12.6.1). One-way
analysis of variance (ANOVA) with the Benjamini-Hochberg correction
was performed to identify mRNAs that exhibited significantly
altered expression in endometrial adenocarcinoma samples compared
with the controls. The Tukey's test was used for multiple
comparisons. P<0.05 was considered to indicate a statistically
significant difference.
Results
The expression profiles of mRNA in endometrial
samples with varying degrees of histological differentiation of
endometrial adenocarcinoma were determined by microarray expression
analysis using GeneChip® Human Genome U133A plates.
Differentially expressed genes among the groups were first
normalized using the RMA Express program. Hierarchical clustering
of the results was then performed, which allowed a preliminary
assessment of the similarity between mRNA expression profiles in
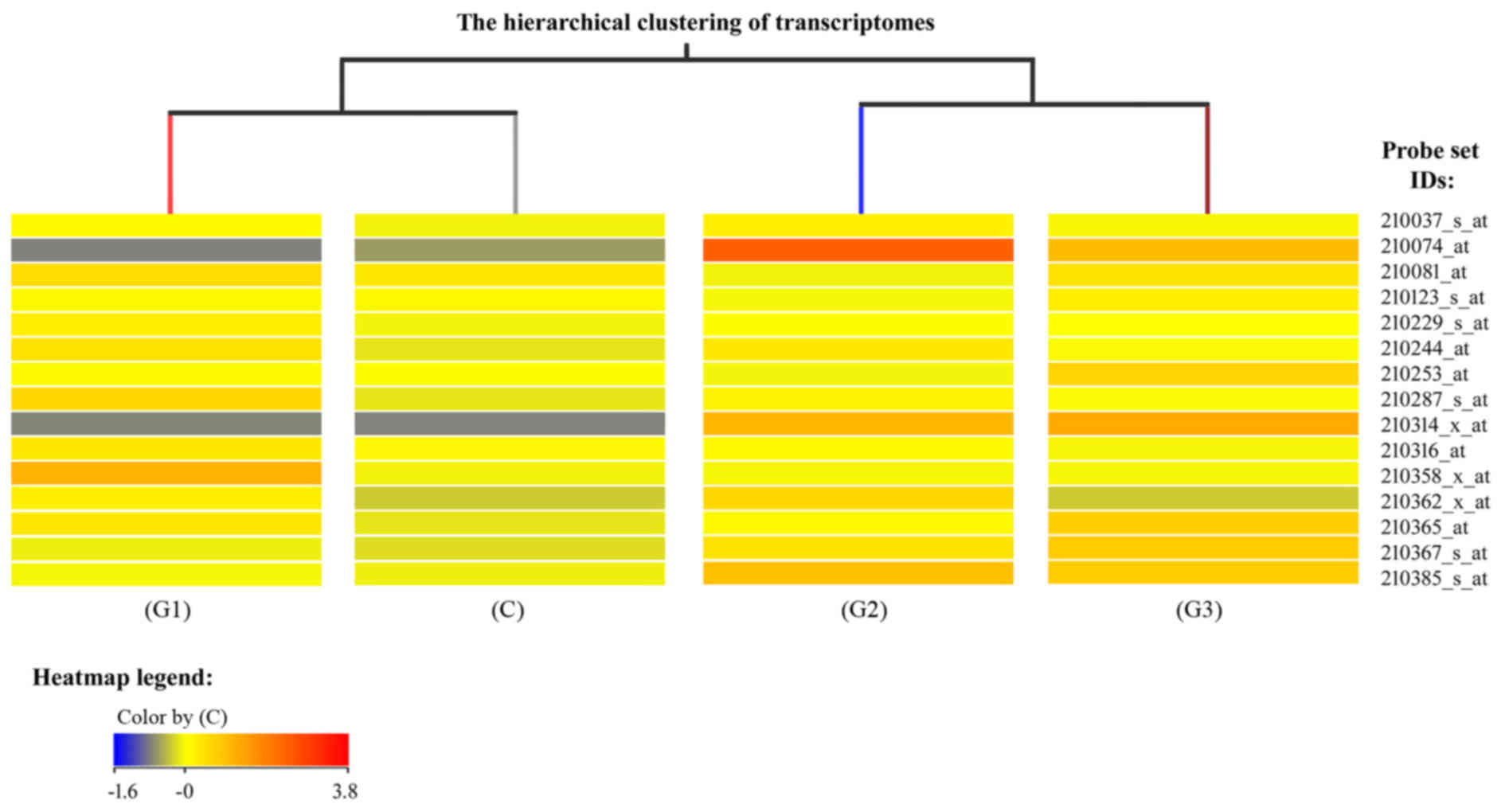
the different groups (Fig. 1).
Fig. 1 is a fragment of a
dendogram showing expression of 15 mRNAs, it demonstrates that the
mRNA expression profile of genes involved in the regulation of
angiogenesis varies depending on the degree of histological
differentiation of endometrial adenocarcinoma. The smallest
differences relative to the control group were observed in the G1
group of endometrial adenocarcinomas, while the transcriptomes of
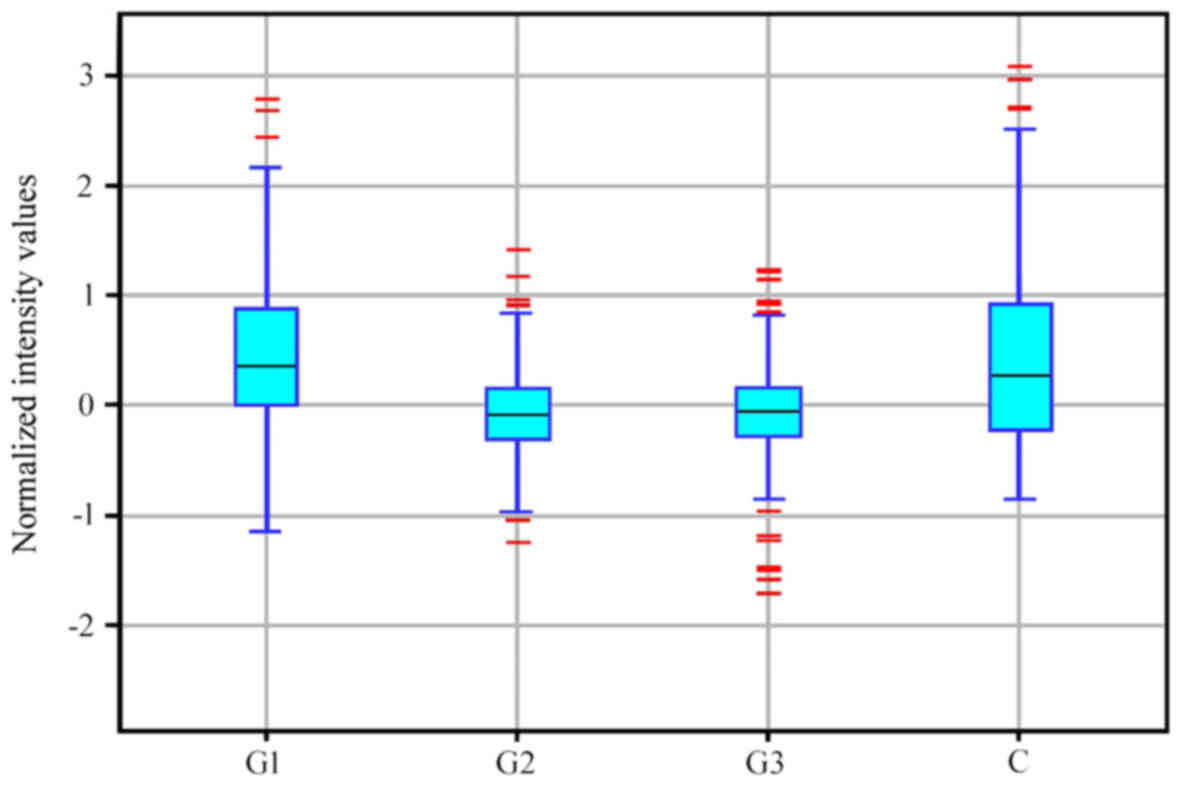
G2 and G3 groups constituted a distinct subgroup (Fig. 1). Similar conclusions were drawn
based on descriptive statistics characterizing the expression
profile of 691 mRNAs associated with the regulation of angiogenesis
in the control and endometrial adenocarcinoma groups (Fig. 2).
To determine which of the observed differences in
mRNA expression were statistically significant, one-way ANOVA was
performed. The results indicated that, out of the 691 mRNAs, 585
mRNAs were significantly differentially expressed in endometrial
adenocarcinoma samples when compared with controls. Table I presents the level of significance
for mRNAs that demonstrated significantly altered expression in
endometrial adenocarcinoma samples when compared with controls.
Multiple comparisons testing was subsequently performed using the
Tukey post hoc test to obtain more detailed information regarding
the differences between mRNA levels among different histological
grades of endometrial adenocarcinoma and controls (Table II). The results indicated that the
number of mRNAs with significantly altered expression when compared
with the controls for each histological grade were as follows: G1
vs. control, 27; G2 vs. control, 113; and G3 vs. control, 81
(P<0.05; Table II). Therefore,
the number of mRNAs that demonstrated significantly altered
expression in endometrial adenocarcinoma samples compared with the
controls varied depending on whether they were G1, G2 or G3
samples.
 | Table I.Number of mRNA sequences associated
with angiogenesis that were significantly, differentially expressed
in patients with endometrial adenocarcinoma when compared with
controls. |
Table I.
Number of mRNA sequences associated
with angiogenesis that were significantly, differentially expressed
in patients with endometrial adenocarcinoma when compared with
controls.
| P-value | Number of
mRNAs |
|---|
| >0.020 to
<0.050 | 203 |
| >0.010 to
<0.020 | 140 |
| >0.005 to
<0.010 | 115 |
| >0.001 to
<0.005 | 82 |
| <0.001 | 45 |
 | Table II.Number of mRNA sequences that were
differentially expressed among the transcriptome groups. |
Table II.
Number of mRNA sequences that were
differentially expressed among the transcriptome groups.
| Transcriptome
group | C | G1 | G2 | G3 |
|---|
| C | 203 | 27a | 113a | 81a |
| G1 | 176 | 203 | 86b | 90b |
| G2 | 90 | 117 | 203 | 32c |
| G3 | 122 | 113 | 171 | 203 |
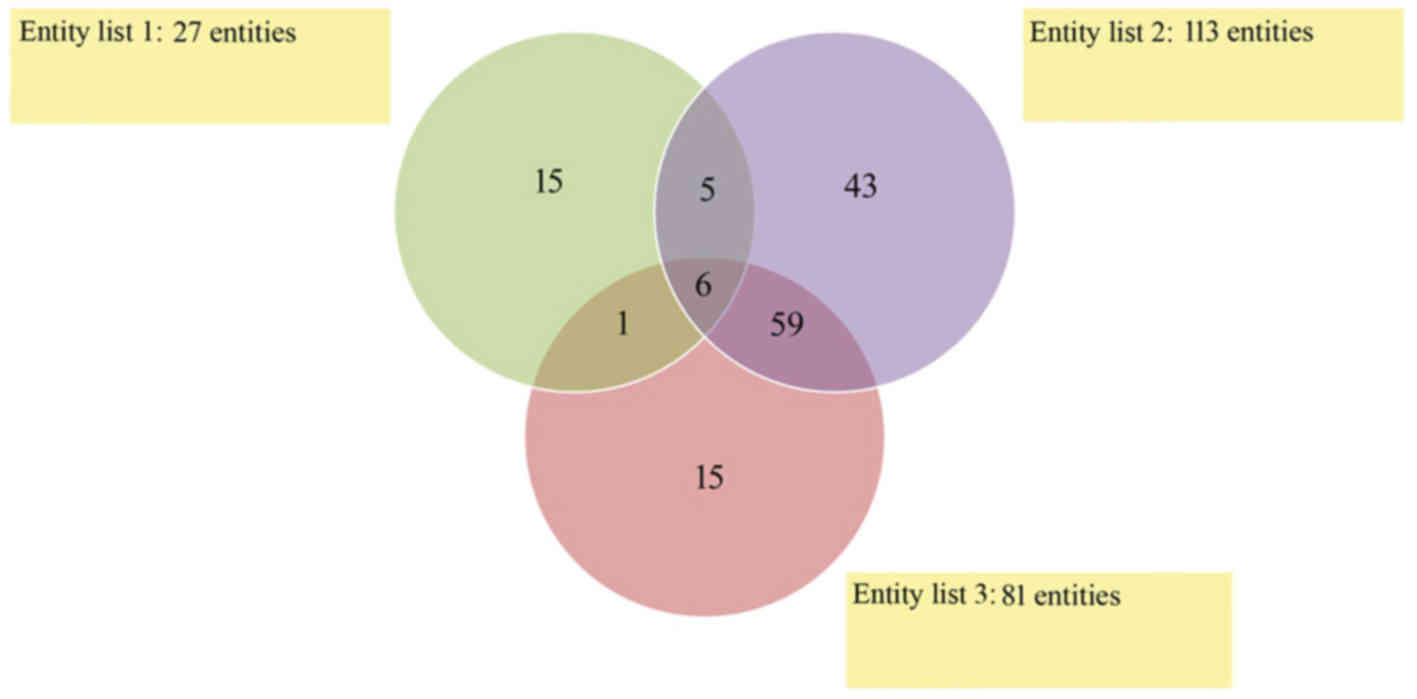
A Venn diagram was constructed to demonstrate the
number of overlapping mRNAs that exhibited significantly altered
expression levels among endometrial adenocarcinoma samples of any
histological grade and the controls (Fig. 3). The results indicate that six
mRNAs differentiated endometrial adenocarcinoma samples from the
control regardless of histological grade (Fig. 3). The number of mRNAs that were
significantly differentiated between specific histological grades
and controls (significantly altered expression in only one
histological grade vs. controls) were as follows: G1 vs. control,
15; G2 vs. control, 43; and G3 vs. control, 15 mRNA (Fig. 3).
This next step of the analysis was completed by
performing an over-representation test using PANTHER software (Gene
List Analysis). This allowed for the identification of
differentially expressed mRNAs that are essential for tumor
angiogenesis (Table III). The
results demonstrated that in G1 endometrial adenocarcinoma, three
genes, including endoglin (ENG), EGF like repeats and discoidin
domains 3 (EDIL3) and neuropilin 2 (NRP2) demonstrated a
significant increase in expression when compared with the control
group (Table III). In addition,
a significant increase in the mRNA expression levels of semaphorin
(SEMA) 3B in G2 endometrial adenocarcinomas, and SEMA3F expression
in G3 endometrial adenocarcinoma was observed when compared with
the control group (Table III).
The remaining mRNAs identified by the over-representation test that
were determined to be important for tumor angiogenesis, occurred in
G2 and G3 endometrial adenocarcinoma at an expression level that
was significantly lower when compared with the control. A
characteristic feature was that the mRNA level was gradually
reduced with the increase of histopathological differentiation of
endometrial adenocarcinoma (Table
III). The overlapping mRNAs between the G2 vs. control and G3
vs. control groups included the transforming growth factor β
receptor 3 (TGFBR3) isoform of ENG, formed due to
post-transcriptional modification of the TEK receptor tyrosine
kinase, vascular endothelial growth factor C (VEGFC), SEMA5A and
homebox A5. The over-representation test did not identify mRNA
sequences that differentiated all histological grades of
endometrial adenocarcinoma (G1 vs. control, G2 vs. control and G3
vs. control) from controls. Similarly, no mRNAs that were common
among G1 and G2 vs. control or G1 and G3 vs. control groups were
identified as essential for tumor angiogenesis (Table III).
 | Table III.Genes essential for the regulation of
angiogenesis in different histological grades of endometrial
adenocarcinoma. |
Table III.
Genes essential for the regulation of
angiogenesis in different histological grades of endometrial
adenocarcinoma.
|
|
| mRNAs important for
tumor angiogenesis |
|---|
| Groups
compared | Total number of
differentially expressed mRNAsa | ID | Symbol | P-value | FC (log2) |
|---|
| G1 vs. C | 15 | 201808_s_at | ENG |
0.0013 | (+) 1.66033 |
|
|
| 207379_at | EDIL3 |
0.0018 | (+) 1.79402 |
|
|
| 214632_at | NRP2 |
0.0130 | (+) 1.85248 |
| G2 vs. C | 43 | 203070_at | SEMA3B |
0.0019 | (+) 1.50104 |
| G3 vs. C | 15 | 35666_at | SEMA3F | <0.001 | (+) 1.55995 |
| G1 vs. C; G2 vs.
C | 5 | – | – | – | – |
| G1 vs. C; G3 vs.
C | 1 | – | – | – | – |
| G2 vs. C; G3 vs.
C | 59 | 206702_at | TEK | <0.001 | (−) 2.15583 |
|
|
| 209946_at | VEGF C | <0.001 | (−) 2.33019 |
|
|
| 201809_s_at | ENG | <0.001 | (−) 2.74506 |
|
|
| 205405_at | SEMA5A | <0.001 | (−) 3.05802 |
|
|
| 213169_at | SEMA5A |
0.0052 | (−) 3.51414 |
|
|
| 213844_at | HOXA5 |
0.0067 | (−) 3.53497 |
| G1 vs. C; G2 vs. C;
G3 vs. C | 6 | – | – | – | – |
Discussion
Microarray technology facilitates detection of the
expression levels of several thousand genes with one experiment,
and is therefore a useful tool for determining the influence of
changes in gene expression levels in the development of human
disease, such as cancer (23). The
results of the present study suggest that the expression of genes
involved in the regulation of angiogenesis may present a useful
diagnostic marker that facilitates differentiation between low and
high grades of endometrial adenocarcinoma.
Neoangiogenesis is associated with the expression of
certain markers of angiogenesis, including VEGF, ENG (transmembrane
glycoprotein receptor for TGF-β) and NRP (a co-receptor for VEGF)
(24–27). Conventionally, the prediction of
malignancy and treatment of cancer patients is based primarily on
the determination of the clinical stage. Understanding the cellular
processes responsible for the metastasis of cancer, as well as the
molecular markers of invasiveness, may be useful to improve the
diagnosis and prognosis of the disease. In addition, this
understanding may facilitate the development of drugs targeting the
factors responsible for the invasiveness of cancer and the enable
the development of novel therapeutic regimens. ENG serves an
important role in tumor progression via the regulation of
angiogenesis, cell migration and metastasis. It is present on the
surface of endothelial cells of tumor blood vessels and particular
types of tumor cells. ENG is an auxiliary co-receptor of TGF-β that
is responsible for cell proliferation, differentiation, migration
and adhesion (28–31). It is known that TGF-β1 functions as
an inhibitory factor during tumor development. In addition, it
induces inflammation and releases angiogenic factors from
inflammatory cells in vivo (32). It was previously demonstrated that
inhibition of ENG stimulates cell growth induced by TGF-β1 and
inhibits cell migration (33). By
contrast, ENG is an identified component of the endothelial nitric
oxide synthase signaling pathway, which modulates the activity of
cyclooxygenase-2 (34,35). ENG appears to modulate the
transition from endothelial progenitor cells to active endothelial
cells (36).
Angiogenesis is essential for numerous physiological
and pathological processes, such as tumor progression, as
vascularization is required for tumor growth and metastasis
(37). When there is an
insufficient blood supply, cancer cells undergo apoptosis/necrosis.
However, due to the observed distribution of ENG in all tissues and
the other proven functional involvement with TGF-β (38,39),
it was demonstrated to be involved in angiogenesis (25,40).
The participation of ENG in angiogenesis corresponds with the
observed death of ENG−/− knockout mice by vascular
malformations (40). An
association between the levels of ENG and cell proliferation
markers, such as Ki-67 were observed (25). Using immunohistochemistry,
increased expression of ENG has been observed in endothelial cells
undergoing active angiogenesis, including those in the tumor, when
compared with normal endothelium (41,42).
The function of ENG may be contradicting as it is required for
tumor neoangiogenesis; however, the overexpression of this gene may
act as a suppressor of invasion and metastasis (43). ENG as a part of the TGF-β receptor
complex modulates TGF-β receptor signaling. The complex relays
contradicting signals from TGF-β that has a paradoxical role in
cancer development. It may inhibit cell growth and induce apoptosis
or differentiation during the premalignant phase of carcinogenesis.
Conversely, TGF-β may modulate processes such as cell invasion,
angiogenesis, immune regulation after the cancer cells lose
inhibitory growth responses, which allows them to become malignant
(44). Previous studies observed
that downregulation of endoglin was associated with malignant
progression in prostate carcinoma cell lines and enhanced migration
and invasion of nontumorigenic prostate cell lines, while
overexpression had the opposite effect (45,46).
The present study, revealed that ENG was overexpressed in the G1
vs. control group [FC=(+)1.66033]. Therefore, it may be assumed
that the increase in ENG mRNA levels may lead to inhibition of cell
growth, and will thus function to inhibit tumor growth. From the G2
vs. control and G3 vs. control transcriptome groups, PANTHER
highlighted ENG as a differentially expressed gene. However, in
this case, decreases in mRNA expression levels [FC=(−)2.7451] were
observed. As a consequence, TGF-β may induce tumor cell growth and
the likelihood of tumor progression may increase.
The expression of NRPs is observed in healthy,
mature and developing vasculature systems, on endothelial cells,
vascular smooth muscle cells (47,48)
and in the vascular endothelium surrounding tumors (47,49–51).
This is an important observation concerning the role of NRPs in the
promotion of angiogenesis in cancer. Previous studies have
indicated that NRPs are proangiogenic mediators that bind to VEGF.
However, they may possess antiangiogenic roles by interacting with
class 3 SEMAs (52). SEMA3 family
proteins bind to the a1/a2 domains, whereas VEGF family proteins
bind to the b1/b2 domains. Previous studies revealed that anti-Nrp1
antibody (anti-Nrp1A), that blocks the SEMA3 binding domain, does
not block VEGF binding, whereas the antibody blocking the VEGF
binding domain (anty-Nrp1B) does not block binding to SEMA3
(9,53). There is sufficient evidence to
indicate that SEMAs are involved in the regulation of apoptosis,
cell migration, tumor growth and angiogenesis (54–57).
SEMA3 particularly affects endothelial cells (54,58,59).
NRP2 preferentially binds to SEMA3B, 3C, 3D and 3F (54,55,60).
By inhibiting the interaction between VEGF-NRP, the SEMA3F isoform
inhibits VEGF-dependent cell proliferation and migration (54). The results of the present study
demonstrated that NRP2 was overexpressed in the G1 group when
compared with the control group [FC=(+)1.8525)], and was the most
over-represented gene. If the information presented is reliable, it
is possible that cancer cells at this stage demonstrate increased
pro-angiogenic interactions between VEGF and NRP2 in G1 compared
with control, which may promote increased angiogenesis and
subsequent cancer progression. SEMA3F is an inhibitor of cell
adhesion and migration, which is partially due to signaling
alterations that affect the activation or stabilization of surface
integrins (61,62). Integrin inhibition by SEMA3F may
explain the inhibition of endothelial and cancer cell migration,
which leads to a reduction of angiogenesis and metastasis (63,64).
It is considered that at least two protein members
of the class 3 SEMA family, one of which includes SEMA3F, exhibit
antiangiogenic effects involving NRPs (65). Microarray analysis performed in the
current study, demonstrated that SEMA3B and 3F of the class 3 SEMA
family, were overexpressed in endometrial adenocarcinoma samples
when compared with controls. SEMA3B was identified as the
over-represented gene in the G2 vs. control groups, and SEMA3F was
the over-represented gene in the G3 vs. control groups.
Overexpression of SEMA3F may inhibit cellular migration, which may
result in a reduction of angiogenesis (61).
It has been previously demonstrated that SEMA3F
inhibits angiogenesis, cell proliferation and cell survival, which
are induced by VEGF and basic fibroblast growth factor (bFGF)
(66). The lack of bFGF-binding
NRPs, coupled with the fact that SEMA3F does not inhibit binding of
bGFG to its receptor, indicate that SEMA3F may function through
NRP2 (66–68). Additional studies have demonstrated
that SEMA3F inhibits tumor angiogenesis in vivo. Previous
studies have suggested that, VEGF promotes angiogenesis via NRP2,
while SEMA3F is an inhibitor of angiogenesis (66). These features of NRP and
SEMA3Findicate that they may be potential targets for
anti-angiogenic therapy. The direct effect of VEGF on endothelial
cells has long been established; however, the role of the NRP in
this process is currently under investigation. Angiogenic induction
through the binding of VEGF to NRP2 has been characterized to a
limited extent, and the evidence presented so far suggests that the
regulation occurs in a different manner to that of VEGF-NRP1
(46). Favier et al
(65) investigated the role of
NRP2 in primary human endothelial cells, and the results indicated
that NRP2 interacts with VEGFR2 and VEGFR3. By assessing the
overexpression or suppression of NRP2, an essential role of NRP2 in
the survival and migration of endothelial cells induced by VEGFA
and VEGFC was demonstrated. In addition, this previous study
demonstrated that NRP2 functions as a co-receptor for VEGFR. As
VEGFR2 is always present on endothelial cells, whereas VEGFR3 is
expressed, it was demonstrated that NRP2 interacts with VEGFR3 and
VEGFR2. VEGFR2 is responsible for the full spectrum of action of
VEGF factors on endothelial cells. Unlike VEGFR1, expression of the
VEGFR2 gene is not dependent on hypoxia (69). Expression of VEGFR2 occurs
primarily in vascular endothelial cells and in megakaryocytes,
platelets and hematopoietic stem cells (70–72).
The expression of VEGFR3 regulates the development and growth of
the lymphatic system (73).
Embryos with a defective VEGFR3 gene succumb at an early stage of
development due to malformation of the cardiovascular system
(74). In adults, expression of
this isoform occurs exclusively in the endothelial lymphatic
vessels. In addition, its expression was identified in the blood
vessels of various tumors during neovascularization, which is
associated with the spread of the tumor process. If the interaction
between VEGFR and NRP2 is supported by a co-ligand, it is possible
that NRP2 may function as a co-receptor for VEGFR2 and VEGFR3. The
results obtained by Favier et al (65) demonstrated that NRP2 functions as a
co-receptor for VEGFR in response to VEGFC. The present study
demonstrated that VEGFC was strongly silenced in G2 and G3
endometrial adenocarcinomas. This may be due to the marked decrease
in the concentration of a specific mRNA, which affects the decrease
in survival and migration of endothelial cells.
SEMA5A demonstrates oncogenic and tumor suppressive
functions in a number of cancers. High expression of SEMA5A and its
receptor, plexin-B3, is associated with the aggressiveness of
pancreatic and prostate cancers. It was demonstrated that their
expression was the primary factor underlying the involvement of
SEMA5A in the invasion, migration, proliferation and growth of
tumors (64,75,76).
In vitro studies have demonstrated that one of the
consequences of SEMA5A expression is a decrease in the level of
apoptosis of endothelial cells (75,77).
Furthermore, downregulation of SEMA5A at the transcriptional and
translational levels was observed in lung cancer samples, which is
generally associated with low survival rates (78). The use of microarrays in the
present study demonstrated that SEMA5A is silenced [FC=(−)3.5141]
in G2 and G3 endometrial adenocarcinomas when compared with
controls. Based on these observations, it is possible that SEMA5A
may not be associated with cancer aggressiveness.
EDIL3, also termed DEL-1, was one of the first
extracellular matrix proteins determined to be involved in vascular
morphogenesis. EDIL3 has been investigated extensively, and is
associated with the regulation of angiogenesis and cell adhesion.
It is an embryonic endothelial cell protein, which is not expressed
following birth; however, it is expressed in numerous tumor types
(79,80). The current study demonstrated that
EDIL3 was overexpressed in the G1 vs. control groups
(FC=(+)1.7940). Overexpression of EDIL3 reduces apoptosis of tumor
cells and leads to increased tumor vascularization, which promotes
tumor growth (80,81). The potential modulation of tumor
vasculature by EDIL3 may be a potential target for anti-angiogenic
therapy of cancer (82,83).
In conclusion, the results of the present study
provide potential novel results that may facilitate diagnosis and
treatment of patients with endometrial cancer. The scientific value
of the present study has been discussed; however, certain
limitations have to be considered. The small total number of
collected and selected samples analyzed, may affect the impact of
the current study. In order to improve the results further, a study
that includes a higher number of patients would be desirable. The
results of the present study indicate that NRP2 may demonstrate an
important role in the activity of endothelial cells, including
VEGF, and potentially plexins and integrins. An understanding of
how these proteins interact remains to be established. In the
future, this may lead to an improved understanding of the
mechanisms underlying angiogenesis. The results of the present
study indicate that NRP2 may be a worthwhile target for future
pharmacological studies.
References
|
1
|
Ong FS, Das K, Wang J, Vakil H, Kuo JZ,
Blackwell WL, Lim SW, Goodarzi MO, Bernstein KE, Rotter JI and
Grody WW: Personalized medicine and pharmacogenetic biomarkers:
Progress in molecular oncology testing. Expert Rev Mol Diagn.
12:593–602. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Biankin VA, Piantadosi S and Hollingsworth
SJ: Patient-centric trials for therapeutic development in precision
oncology. Nature. 526:361–370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Le Tourneau C, Kamal M, Tsimberidou AM,
Bedard P, Pierron G, Callens C, Rouleau E, Vincent-Salomon A,
Servant N, Alt M, et al: Treatment algorithms based on tumor
molecular profiling: The essence of precision medicine trials. J
Natl Cancer Inst. 108:djv3622015. View Article : Google Scholar :
|
|
4
|
Yamaguchi T, Bando H, Mori T, Takahashi K,
Matsumoto H, Yasutome M, Weich H and Toi M: Over expression of
soluble vascular endothelial growth factor receptor 1 in colorectal
cancer: Association with progression and prognosis. Cancer Sci.
98:405–410. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lesslie DP, Summy JM, Parikh NU, Fan F,
Trevino JG, Sawyer TK, Metcalf CA, Shakespeare WC, Hicklin DJ,
Ellis LM and Gallick GE: Vascular endothelial growth factor
receptor-1 mediates migration of human colorectal carcinoma cells
by activation of Src family kinases. Br J Cancer. 94:1710–1717.
2006.PubMed/NCBI
|
|
6
|
Ziyad S and Iruela-Arispe ML: Molecular
mechanisms of tumor angiogenesis. Genes Cancer. 2:1085–1096. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lanara Z, Giannopoulou E, Fullen M,
Kostantinopoulos E, Nebel JC, Kalofonos HP, Patrinos GP and
Pavlidis C: Comparative study and meta-analysis of meta-analysis
studies for the correlation of genomic markers with early cancer
detection. Hum Genomics. 7:142013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Caunt M, Mak J, Liang WC, Stawicki S, Pan
Q, Tong RK, Kowalski J, Ho C, Reslan HB, Ross J, et al: Blocking
neuropilin-2 function inhibits tumor cell metastasis. Cancer Cell.
13:331–342. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pan Q, Chanthery Y, Liang WC, Stawicki S,
Mak J, Rathore N, Tong RK, Kowalski J, Yee SF, Pacheco G, et al:
Blocking neuropilin-1 function has an additive effect with
anti-VEGF to inhibit tumor growth. Cancer Cell. 11:53–67. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rajaganeshan R, Prasad R, Guillou PJ,
Chalmers CR, Scott N, Sarkar R, Poston G and Jayne DG: The
influence of invasive growth pattern and microvessel density on
prognosis in colorectal cancer and colorectal liver metastases. Br
J Cancer. 96:1112–1127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gee MG, Procopio WN, Makonnen S, Feldman
MD, Yeilding NM and Lee WM: Tumor vessel development and maturation
impose limits on the effectiveness of anti-vascular therapy. Am J
Pathol. 162:183–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xian X, Håkansson J, Ståhlberg A, Lindblom
P, Betsholtz C, Gerhardt H and Semb H: Pericytes limit tumor cell
metastasis. J Clin Invest. 116:642–651. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Romani AA, Borghetti AF, Del Rio P,
Sianesi M and Soliani P: The risk of developing metastatic disease
in colorectal cancer is related to CD105-positive vessel count. J
Surg Oncol. 93:446–455. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saad RS, Liu YL, Nathan G, Celebrezze J,
Medich D and Silverman JF: Endoglin (CD105) and vascular
endothelial growth factor as prognostic markers in colorectal
cancer. Mod Pathol. 17:197–203. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kölbl AC, Birk AE, Kuhn C, Jeschke U and
Andergassen U: Influence of VEGFR and LHCGR on endometrial
adenocarcinoma. Oncol Lett. 12:2092–2098. 2016.PubMed/NCBI
|
|
16
|
Trimble CL, Method M, Leitao M, Lu K,
Ioffe O, Hampton M, Higgins R, Zaino R, Mutter GL, et al: Society
of Gynecologic Oncology Clinical Practice Committee: Management of
endometrial precancers. Obstet Gynecol. 120:1160–1175.
2012.PubMed/NCBI
|
|
17
|
Klemba A, Kukwa W, Bartnik W, Krawczyk T,
Scińska A, Golik P and Czarnecka AM: Molecular biology of
endometrial carcinoma. Postepy Hig Med Dosw (online). 62:420–432.
2008.(In Polish). PubMed/NCBI
|
|
18
|
Colombo N, Preti E, Landoni F, Carinelli
S, Colombo A, Marini C, Sessa C, et al: ESMO Guidelines Working
Group: Endometrial cancer: ESMO clinical practice guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 22:(Suppl 6).
vi35–vi39. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Murali R, Soslow RA and Weigelt B:
Classification of endometrial carcinoma: More than two types.
Lancet Oncol. 15:e268–e278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Witek Ł, Janikowski T, Bodzek P, Olejek A
and Mazurek U: Expression of tumor suppressor genes related to the
cell cycle in endometrial cancer patients. Adv Med Sci. 61:317–324.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mi H, Huang X, Muruganujan A, Tang H,
Mills C, Kang D and Thomas PD: PANTHER version 11: Expanded
annotation data from gene ontology and reactome pathways and data
analysis tool enhancements. Nucleic Acids Res. 45:D185–D189. 2017.
View Article : Google Scholar
|
|
22
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on bias and variance.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chrominski K and Tkacz M: Comparison of
high-level microarray analysis methods in the context of result
consistency. PLoS One. 10:e01288452015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim WH, Lee SH, Jung MH, Seo JH, Kim J,
Kim MA and Lee YM: Neuropilin2 expressed in gastric cancer
endothelial cells increases the proliferation and migration of
endothelial cells in response to VEGF. Exp Cell Res. 315:2154–2164.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gluzman-Poltorak Z, Cohen T, Shibuya M and
Neufeld G: Vascular endothelial growth factor receptor-1 and
neuropilin-2 form complexes. J Biol Chem. 276:18688–18694. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nassiri F, Cusimano MD, Scheithauer BW,
Rotondo F, Fazio A, Yousef GM, Syro LV, Kovacs K and Lloyd RV:
Endoglin (CD105): A review of its role in angiogenesis and tumor
diagnosis, progression and therapy. Anticancer Res. 31:2283–2290.
2011.PubMed/NCBI
|
|
27
|
Bernabeu C, Lopez-Novoa JM and Quintanilla
M: The emerging role of TGF-beta superfamily coreceptors in cancer.
Biochim Biophys Acta. 1792:954–973. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wong SH, Hamel L, Chevalier S and Philip
A: Endoglin expression on human microvascular endothelial cells
association with betaglycan and formation of higher order complexes
with TGF-beta signaling receptors. Eur J Biochem. 267:5550–5560.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Blobe GC, Schiemann WP and Lodish HF: Role
of transforming growth factor β in human disease. N Engl J Med.
342:1350–1358. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Govinden R and Bhoola KD: Genealogy,
expression, and cellular function of transforming growth
factor-beta. Pharmacol Ther. 98:257–265. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu HJ and Burgess AW: Regulation of
transforming growth factor-beta signaling. Mol Cell Biol Res
Commun. 4:321–330. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hata A, Shi Y and Massagué J: TGF-beta
signaling and cancer: Structural and functional consequences of
mutations in Smads. Mol Med Today. 4:257–262. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li C, Hampson IN, Hampson L, Kumar P,
Bernabeu C and Kumar S: CD105 antagonizes the inhibitory signaling
of transforming growth factor beta1 on human vascular endothelial
cells. FASEB J. 14:55–64. 2000.PubMed/NCBI
|
|
34
|
Jerkic M, Rivas-Elena JV, Prieto M, Carrón
R, Sanz-Rodríguez F, Pérez-Barriocanal F, Rodríguez-Barbero A,
Bernabéu C and López-Novoa JM: Endoglin regulates nitric
oxide-dependent vasodilatation. FASEB J. 18:609–611.
2004.PubMed/NCBI
|
|
35
|
Jerkic M, Rodríguez-Barbero A, Prieto M,
Toporsian M, Pericacho M, Rivas-Elena JV, Obreo J, Wang A,
Barriocanal F Pérez, Arévalo M, et al: Reduced angiogenic responses
in adult Endoglin heterozygous mice. Cardiovasc Res. 69:845–854.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alev C, McIntyre BA, Ota K and Sheng G:
Dynamic expression of Endoglin, a TGF-beta co-receptor, during
pre-circulation vascular development in chick. Int J Dev Biol.
54:737–742. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Folkman J, Watson K, Ingber D and Hanahan
D: Induction of angiogenesis during the transition from hyperplasia
to neoplasia. Nature. 339:58–61. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheifetz S, Bellón T, Calés C, Vera S,
Bernabeu C, Massagué J and Letarte M: Endoglin is a component of
the transforming growth factor-beta receptor system in human
endothelial cells. J Biol Chem. 267:19027–19030. 1992.PubMed/NCBI
|
|
39
|
Barbara NP, Wrana JL and Letarte M:
Endoglin is an accessory protein that interacts with the signaling
receptor complex of multiple members of the transforming growth
factor-beta superfamily. J Biol Chem. 274:584–594. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li DY, Sorensen LK, Brooke BS, Urness LD,
Davis EC, Taylor DG, Boak BB and Wendel DP: Defective angiogenesis
in mice lacking endoglin. Science. 284:1534–1537. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Burrows FJ, Derbyshire EJ, Tazzari PL,
Amlot P, Gazdar AF, King SW, Letarte M, Vitetta ES and Thorpe PE:
Up-regulation of endoglin on vascular endothelial cells in human
solid tumors: Implications for diagnosis and therapy. Clin Cancer
Res. 1:1623–1634. 1995.PubMed/NCBI
|
|
42
|
Wang JM, Kumar S, Pye D, Haboubi N and
al-Nakib L: Breast carcinoma: Comparative study of tumor
vasculature using two endothelial cell markers. J Natl Cancer Inst.
86:386–388. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Duff SE, Li C, Garland JM and Kumar S:
CD105 is important for angiogenesis: Evidence and potential
applications. FASEB J. 17:984–992. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pérez-Gómez E, Del Castillo G, Francisco S
Juan, López-Novoa JM, Bernabéu C and Quintanilla M: The role of the
TGF-β coreceptor endoglin in cancer. ScientificWorldJournal.
10:2367–2384. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Craft C, Romero D, Vary C and Bergan R:
Endoglin inhibits prostate cancer motility via activation of the
ALK2-Smad1 pathway. Oncogene. 26:7240–7250. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Oh H, Takagi H, Otani A, Koyama S,
Kemmochi S, Uemura A and Honda Y: Selective induction of
neuropilin-1 by vascular endothelial growth factor (VEGF): A
mechanism contributing to VEGF-induced angiogenesis. Proc Natl Acad
Sci USA. 99:383–388. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Stephenson JM, Banerjee S, Saxena NK,
Cherian R and Banerjee SK: Neuropilin-1 is differentially expressed
in myoepithelial cells and vascular smooth muscle cells in
preneoplastic and neoplastic human breast: A possible marker for
the progression of breast cancer. Int J Cancer. 101:409–414. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang H, Li M, Chai H, Yan S, Lin P,
Lumsden AB, Yao Q and Chen C: Effects of cyclophilin A on cell
proliferation and gene expressions in human vascular smooth muscle
cells and endothelial cells. J Surg Res. 123:312–319. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Broholm H and Laursen H: Vascular
endothelial growth factor (VEGF) receptor neuropilin-1′s
distribution in astrocytic tumors. APMIS. 112:257–263. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fakhari M, Pullirsch D, Abraham D, Paya K,
Hofbauer R, Holzfeind P, Hofmann M and Aharinejad S: Selective
upregulation of vascular endothelial growth factor receptors
neuropilin-1 and −2 in human neuroblastoma. Cancer. 94:258–263.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Straume O and Akslen LA: Increased
expression of VEGF-receptors (FLT1, KDR, NRP-1) and
thrombospondin-1 is associated with glomeruloid microvascular
proliferation, an aggressive angiogenic phenotype, in malignant
melanoma. Angiogenesis. 6:295–301. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Miao HQ and Klagsbrun M: Neuropilin is a
mediator of angiogenesis. Cancer Metastasis Rev. 19:29–37. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Appleton BA, Wu P, Maloney J, Yin J, Liang
WC, Stawicki S, Mortara K, Bowman KK, Elliott JM, Desmarais W, et
al: Structural studies of neuropilin/antibody complexes provide
insights into semaphorin and VEGF binding. EMBO J. 26:4902–4912.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yazdani U and Terman JR: The semaphorins.
Genome Biol. 7:2112006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Janssen BJ, Malinauskas T, Weir GA, Cader
MZ, Siebold C and Jones EY: Neuropilins lock secreted semaphorins
onto plexins in a ternary signaling complex. Nat Struct Mol Biol.
19:1293–1299. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lepelletier Y, Moura IC, Hadj-Slimane R,
Renand A, Fiorentino S, Baude C, Shirvan A, Barzilai A and Hermine
O: Immunosuppressive role of semaphorin-3A on T cell proliferation
is mediated by inhibition of actin cytoskeleton reorganization. Eur
J Immunol. 36:1782–1793. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Delgoffe GM, Woo SR, Turnis ME, Gravano
DM, Guy C, Overacre AE, Bettini ML, Vogel P, Finkelstein D,
Bonnevier J, et al: Stability and function of regulatory T cells is
maintained by a neuropilin-1-semaphorin-4a axis. Nature.
501:252–256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Uniewicz KA and Fernig DG: Neuropilins: A
versatile partner of extracellular molecules that regulate
development and disease. Front Biosci. 13:4339–4360. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Roth L, Nasarre C, Dirrig-Grosch S, Aunis
D, Crémel G, Hubert P and Bagnard D: Transmembrane domain
interactions control biological functions of neuropilin-1. Mol Biol
Cell. 19:646–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Raimondi C and Ruhrberg C: Neuropilin
signalling in vessels, neurons and tumours. Semin Cell Dev Biol.
24:172–178. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Potiron VA, Roche J and Drabkin HA:
Semaphorins and their receptors in lung cancer. Cancer Lett.
273:1–14. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Potiron V and Roche J: Class 3 semaphorin
signaling: The end of a dogma. Sci STKE. 285:pe242005.
|
|
63
|
Kruger RP, Aurandt J and Guan KL:
Semaphorins command cells to move. Nat Rev Mol Cell Biol.
6:789–800. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Serini G, Napione L, Arese M and Bussolino
F: Besides the adhesion: New perspectives of integrin functions in
angiogenesis. Cardiovasc Res. 78:213–222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Favier B, Alam A, Barron P, Bonnin J,
Laboudie P, Fons P, Mandron M, Herault JP, Neufeld G, Savi P, et
al: Neuropilin-2 interacts with VEGFR-2 and VEGFR-3 and promotes
human endothelial cell survival and migration. Blood.
108:1243–1250. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kessler O, Shraga-Heled N, Lange T,
Gutmann-Raviv N, Sabo E, Baruch L, Machluf M and Neufeld G:
Semaphorin-3F is an inhibitor of tumor angiogenesis. Cancer Res.
64:1008–1015. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chen H, Chédotal A, He Z, Goodman CS and
Tessier-Lavigne M: Neuropilin-2, a novel member of the neuropilin
family, is a high affinity receptor for the semaphorins Sema E and
Sema IV but not Sema III. Neuron. 19:547–559. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Giger RJ, Urquhart ER, Gillespie SK,
Levengood DV, Ginty DD and Kolodkin AL: Neuropilin-2 is a receptor
for semaphorin IV: Insight into the structural basis of receptor
function and specificity. Neuron. 21:1079–1092. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Karkkainen MJ and Petrova TV: Vascular
endothelial growth factor receptors in the regulation of
angiogenesis and lymphangiogenesis. Oncogene. 19:5598–5605. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Rahimi N: Vascular endothelial growth
factor receptors: Molecular mechanisms of activation and
therapeutic potentials. Exp Eye Res. 83:1005–1016. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kowanetz M and Ferrara N: Vascular
endothelial growth factor signaling pathways: Therapeutic
perspective. Clin Cancer Res. 12:5018–5022. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cebe-Suarez S, Zehnder-Fjallman A and
Ballmer-Hofer K: The role of VEGF receptors in angiogenesis;
complex partnerships. Cell Mol Life Sci. 63:601–15. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Iljin K, Karkkainen MJ, Lawrence EC, Kimak
MA, Uutela M, Katri JT, Alhonen PL, Halmekytö M, Finegold DN,
Ferrell RE and Alitalo K: VEGFR3 gene structure, regulatory region,
and sequence polymorphisms. FASEB J. 15:1028–1036. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Saharinen P, Tammela T, Karkkainen MJ and
Alitalo K: Lymphatic vasculature: Development, molecular regulation
and role in tumor metastasis and inflammation. Trends Immunol.
25:387–95. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sadanandam A, Varney ML, Singh S, Ashour
AE, Moniaux N, Deb S, Lele SM, Batra SK and Singh RK: High gene
expression of semaphorin 5A in pancreatic cancer is associated with
tumor growth, invasion and metastasis. Int J Cancer. 127:1373–1383.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Pan G, Lv H, Ren H, Wang Y, Liu Y, Jiang H
and Wen J: Elevated expression of semaphorin 5A in human gastric
cancer and its implication in carcinogenesis. Life Sci. 86:139–144.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Pan GQ, Ren HZ, Zhang SF, Wang X and Wen
J: Expression of semaphorin 5A and its receptor plexin B3
contributes to invasion and metastasis of gastric carcinoma. World
J Gastroenterol. 15:2800–2804. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Resende C, Ristimaki A and Machado JC:
Genetic and epigenetic alteration in gastric carcinogenesis.
Helicobacter. 15:(Suppl 1). S34–S39. 2010. View Article : Google Scholar
|
|
79
|
Jiang SH, Wang Y, Yang JY, Li J, Feng MX,
Wang YH, Yang XM, He P, Tian GA, Zhang XX, et al: Overexpressed
EDIL3 predicts poor prognosis and promotes anchorage-independent
tumor growth in human pancreatic cancer. Oncotarget. 7:4226–4240.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Aoka Y, Johnson FL, Penta K, Hirata K Ki,
Hidai C, Schatzman R, Varner JA and Quertermous T: The embryonic
angiogenic factor Del1 accelerates tumor growth by enhancing
vascular formation. Microvasc Res. 64:148–161. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sadanandam A, Rosenbaugh EG, Singh S,
Varney M and Singh RK: Semaphorin 5A promotes angiogenesis by
increasing endothelial cell proliferation, migration, and
decreasing apoptosis. Microvasc Res. 79:1–9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC,
Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC and Chuang EY:
Identification of a novel biomarker, SEMA5A, for non-small cell
lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers
Prev. 19:2590–2597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ferrara N and Kerbel RS: Angiogenesis as a
therapeutic target. Nature. 438:967–974. 2005. View Article : Google Scholar : PubMed/NCBI
|