Introduction
Ovarian cancer is the most common malignant
carcinoma in the world (1).
Currently, paclitaxel (PTX) is the most popular chemotherapy for
ovarian carcinoma (2). But
multidrug resistance (MDR) is a great problem of PTX treatment.
There is a large percentage of patients that become resistant to
PTX, causing relapse (3–5). Unfortunately, the mechanisms of MDR
remain poorly understood. Many studies demonstrated that
chemoresistance in cancer cells facilitated
epithelial-to-mesenchymal transition (EMT) (6,7). EMT
works as a crucial physiological process that serve a key function
in cancer cell progression and metastasis (8). The characteristics of EMT are a loss
of epithelial traits and a gain of mesenchymal traits. At the
molecular level, several epithelial markers, for example α-catenin
and E-cadherin, are significantly decreased, whereas mesenchymal
markers, for example vimentin and N-cadherin, are obviously
increased (9,10).
Histone modification is an important mechanism that
regulates cell processes (11).
KDM5A is a histone demethylase which is specific for H3K4 and it is
required for cell development (12–14).
Initially, KDM5A was identified as a retinoblastoma associated
protein (15). In addition, the
KDM5A homolog gene in Drosophila mutation cause
differentiation and cell growth defects (16). Gene abnormal expression is closely
correlated with human cancer development. Previously, there have
been multiple reports revealing that KDM5A is highly expressed in
gastric cancer (17), acute
myeloid leukemia (18) and breast
cancer (19).
However, the function of KDM5A in ovarian cancer is
not quite clear. In the present study, the authors identified that
KDM5A was highly expressed in ovarian cancer tissues and cell
lines. In addition, KDM5A promoted EMT and metastasis in ovarian
cell lines. Cell proliferation also regulated by KDM5A, but
detailed mechanisms of how KDM5A regulates cell proliferation is
still unknown. Moreover, the authors indicated that a high
expression level of KDM5A was associated with PTX resistance. The
above work indicated that KDM5A was a novel target of ovarian
cancer.
Materials and methods
Ovarian tissue and cell lines
All ovarian patient tissue experiments were approved
by Ethics Committee of the Wuhan University (Wuhan, China). A total
of 47 pairs of ovarian tissue samples and adjacent normal tissue
samples were collected from 47 patients who were diagnosed with
ovarian cancer.
Human normal ovarian cell line IOSE80 and ovarian
cancer cell line SKOV3 and OVCA429 were purchased from American
Type Culture Collection (Manassas, VA, USA). SKOV3/PTX, which is
paclitaxel-resistant, was obtained from the Zhongnan Hospital of
Wuhan University (Wuhan, China). All cells were cultivated in
Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare
Life Sciences, Logan, UT, USA) supplemented with 1%
penicillin/streptomycin and 10% FBS at 37°C under 5%
CO2. In order to maintain the PTX resistance, SKOV3/PTX
cells were treated with 2 nM PTX (Abcam, Cambridge, UK).
Cell transfection
Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was used to transfect cells.
Overexpressed or silenced KDM5A in ovarian cancer cells was created
using KDM5A or KDM5A small interfering (si)RNA respectively.
Following 48 h transfection, cells were collection and subsequent
experiments were performed. The siRNA sequences were as follows:
KDM5A siRNA#1, 5′-AAGAGCUACAACAGGCUCGGU-3′ (sense); KDM5A siRNA#2,
5′-AAGUCCUCUAGUAGUCUUGAA-3′ (sense); scramble siRNA (SCR),
5′-UUCUCCGAAC-GUGUCACGUTT-3′ (sense).
Reverse transcription-quantitative
polymerase chain reaction analysis (RT-qPCR)
RNA was extracted by TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), then synthesized cDNA through
TransScript First-Strand cDNA Synthesis SuperMix (Transgen Biotech,
Beijing, China). Relative mRNA expression was detected by
SYBR-Green Mix (Roche Diagnostics, Basel, Switzerland). Primer
pairs for cDNA amplification were as follows:
5′-ATGATCCCTGCTCTTCTGTG-3′ (forward) and
5′-GATACCATCTTCCACAACTTTCAG-3′ (reverse) for α-catenin;
5′-AATAAAGACCAAGTGACCACC-3′ (forward) and
5′-GCAGAATCAGAATTAGCAAAGC-3′ (reverse) for E-cadherin;
5′-TCATTAATGAGGGCCTTAAAGC-3′ (forward) and
5′-GTTCAGGTAATCATAGTCCTGCT-3′ (reverse) for N-cadherin;
5′-GTGAATACCAAGACCTGCTC-3′ (forward) and
5′-ATCCAGATTAGTTTCCCTCAG-3′ (reverse) for vimentin;
5′-GAACCATGAGAAGTATGACAACAG-3′ (forward) and
5′-ATGGACTGTGGTCATGAGTC' (reverse) for GAP DH. GAP DH was used to
as internal control. All experiments were performed three
independent times.
Western blot analysis
Cells were initially lysed using
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) and the protein concentration was
quantified using the Bio-Rad protein assay kit (cat no. 5000001;
Bio-Rad Laboratories, Inc., Hercules, CA, USA). Protein (40 µg) was
separated using 12% SDS-PAGE gels and then transferred to a
nitrocellulose filter membrane. The membrane was then blocked using
skimmed milk and washed with PBS-1% Tween-20 solution. Membranes
were incubated with indicated antibody at 4°C overnight and washed
with PBS-1% Tween-20 solution. Then, membranes were incubated with
secondary antibodies conjugated with horseradish peroxidase
(1:5,000; cat nos. A21010 and A21020; Abbkine Scientific Co., Ltd.,
Wuhan, China) at room temperature for 1 h. Protein bands were
visualized using an enhanced chemiluminescence kit (Biorbyt, Ltd.,
Cambridge, UK). The primary antibodies were as follows: anti-KDM5A
(1:2,500; cat no. SAB4301220; Sigma-Aldrich, Merck KGaA, Darmstadt,
Germany); anti-E-cadherin (1:2,000; cat no. ab1416);
anti-N-cadherin (1:1,500; cat no. ab76057); anti-α-catenin
(1:2,000; cat no. ab51032); anti-vimentin (1:3,000; cat no.
ab92547); anti-P-glycoprotein (P-gp; 1:500; cat no. ab103477) (all
from Abcam); and anti-β-actin (1:5,000; cat no. A1978;
Sigma-Aldrich; Merck KGaA).
Cell Counting Kit (CCK)-8 assay
A CCK-8 assay was used to assess cell growth. In
brief, cells were placed at a density of 5×103
cells/well in 6-well plates. A total of 10 µl CCK-8 solution was
added per 100 µl medium and incubated 30 min at 37°C. Absorbance
was detected at 450 nm. All experiments were performed three
times.
Anchorage-independent growth
assay
The method was conducted according to a previously
described method (20). In brief,
the culture dish was coated with 2X DMEM supplemented with 2%
penicillin/streptomycin, 20% FBS and 1.2% Bacto agar (BD
Biosciences, Franklin Lakes, NJ, USA) at a ratio of 1:1. Following
ectopic expression of KDM5A in SKOV3 cells and after knocking down
KDM5A in SKOV3/PTX cells, 1×104 cells were added to 2X
DMEM medium supplemented with 2% penicillin/streptomycin, 20% FBS
and 0.7% Bacto agar at a ratio of 1:1. Cells were incubated for 3
weeks, stained with 0.1% crystal violet at room temperature for 15
min and counted under a light microscope (magnification, ×20). All
experiments were performed three times independently.
Transwell assay
The Transwell chamber (Corning, Inc., Corning, NY,
USA) was coated with Matrigel basement membrane matrix (BD
Biosciences). Cells (density, 4×103) were added to the
upper chamber and incubated with DMEM, which was serum-free, and
the lower well contained 10% FBS. Cells were incubated at 37°C for
24 h. Cells were then fixed in methanol for 20 min, follow by
staining with 0.1% crystal violet at room temperature for 15 min
and washed with PBS solution. The number of invasive cells was
counted under a light microscope (magnification, ×20). All
experiments were performed three independent times.
Wound healing assay
To detect cell migration, a wound healing assay was
performed. In brief, 5×105 cells were placed in a 6-well
plate and a pipette tip was used to scratch when the cells had
reached 100% confluence. This was photographed immediately (time 0)
and at 48 h. The ratio of cell migration into the scratch area was
measured. All experiments were performed three independent
times.
Drug sensitivity assay
Following ectopic expression of KDM5A in SKOV3
cells, and inhibition of KDM5A in SKOV3/PTX cells, 5×103
cells were plated in 96-well plates for 24 h at 37°C. Consequently,
PTX was added at various concentrations (0, 5, 10, 20 and 40 nM) in
DMEM medium. Following addition of PTX for 48 h, a CCK-8 assay was
performed to assess cell viability. All experiments were performed
three independent times.
Apoptosis assay
Following the ectopic expression of KDM5A in SKOV3
cells and the knocking down of KDM5A in SKOV3/PTX cells, 10 nM PTX
was added for 48 h. Then, FACS flow cytometry was used to detect
the percentage of apoptotic cells by Annexin V-FITC kit. All
experiments were performed three independent times.
Statistical analysis
All data analysis was performed by SPSS software
(version, 19.0; IBM PSS, Armonk, NY, USA). Expression of KDM5A in
tissue samples or ovarian cell lines and human normal ovarian cell
lines were analyzed by Chi-squared test. Student's t-test was
performed to analyzing the results of metastasis, apoptosis assay
and cell growth. P<0.05 was considered to indicate a
statistically significant difference.
Results
KDM5A is highly expressed in ovarian
cancer and is associated with PTX resistance
Aberrant gene expression contributes to tumor
development. A previous report indicated that KDM5A is amplified in
multiple tumors, including breast cancer (19). In order to investigate whether
KDM5A serves a role in ovarian cancer, the authors collected 47
pairs of ovarian carcinoma tissues and adjacent normal tissues.
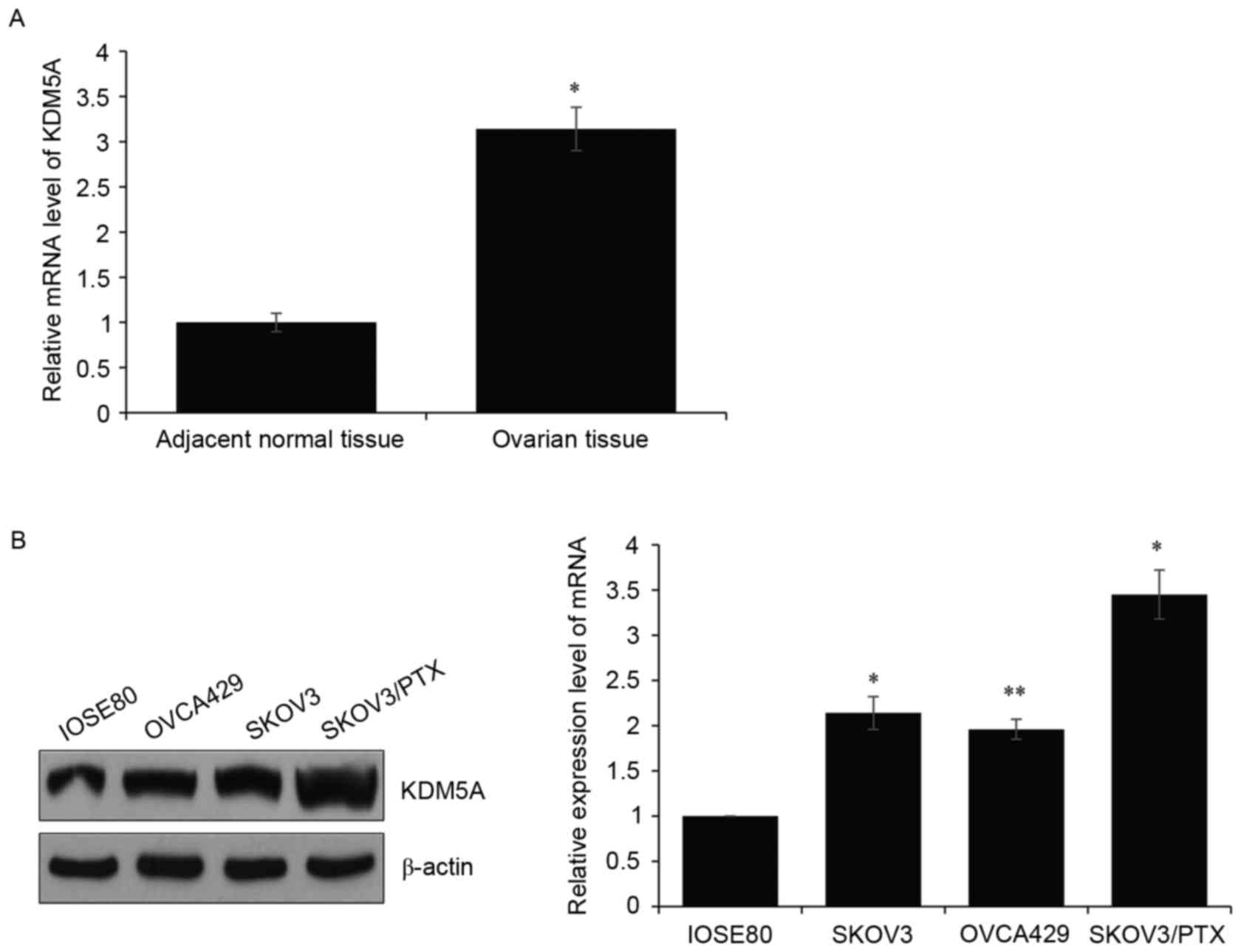
Consequently, RT-qPCR was carried out to determine KDM5A expression
level. It was observed that the KDM5A level in ovarian tissues was
dramatically higher than that in normal tissues (Fig. 1A). Meanwhile, KDM5A expression
levels were measured in various ovarian cell lines (SKOV3, OVCA429
and SKOV3/PTX), compared with the human normal ovarian cell line
IOSE80. The RT-qPCR and western blotting assays revealed both mRNA
and protein levels of KDM5A were amplified in ovarian cell lines
(Fig. 1B). Furthermore, expression
of KDM5A was higher in SKOV3/PTX then other ovarian cell lines.
These results indicated that KDM5A was associated with PTX
resistance in ovarian cancer.
KDM5A facilitates EMT in ovarian
cells
Numerous studies demonstrated that chemoresistance
in cancer cells facilitated EMT (6,7).
Detected EMT markers were identified in SKOV3 and SKOV3/PTX cells
via western blotting. The expression of E-cadherin in SKOV3/PTX
cells was lower than in SKOV3 cells, whereas the expression of
N-cadherin in SKOV3/PTX cells was higher than in SKOV3 cells
(Fig. 2A). Then, the authors
further explored whether KDM5A regulated EMT in ovarian cancer. Two
different KDM5A siRNAs were used to knockdown KDM5A in SKOV3 cells,
and siRNA efficiency was detected by RT-qPCR and western blotting.
As present in Fig. 2B, siKDM5A#1
and KDM5A#2 downregulated KMD5A ~80%, and siKDM5A#1 was more
efficient, so siKDM5A#1 was used for further experiments (Fig. 2B). RT-qPCR and western blotting was
again used to determine the EMT marker in SKOV3/PTX cells that were
transfected with SCR or siKDM5A. The results indicated that, while
there was a knockdown of KDM5A in SKOV3/PTX cells, E-cadherin and
α-catenin were significantly increased. In addition, N-cadherin and
vimentin were obviously deceased (Fig.
2C). Consequently, the authors expressed KDM5A ectopically in
SKOV3 cells and detected the EMT markers. Results indicated that
E-cadherin expression was decreased and N-cadherin expression was
increased (Fig. 2D). These results
suggested that KDM5A modulates the EMT in ovarian cells. EMT has
been reported to promoted cancer cell metastasis, therefore wound
healing and Transwell assays were performed to explore whether
KDM5A regulated cancer cell metastasis. The wound healing assay
revealed that the cells in which KDM5A was depleted migrated to
~50% of the wounded area, whereas the control groups migrated to
~100% of the area (Fig. 2E). In
addition, the Transwell assay revealed that depletion of KDM5A
obviously suppressed cancer cell metastasis, and ectopic expression
of KDM5A significant facilitated cancer cell metastasis (Fig. 2F).
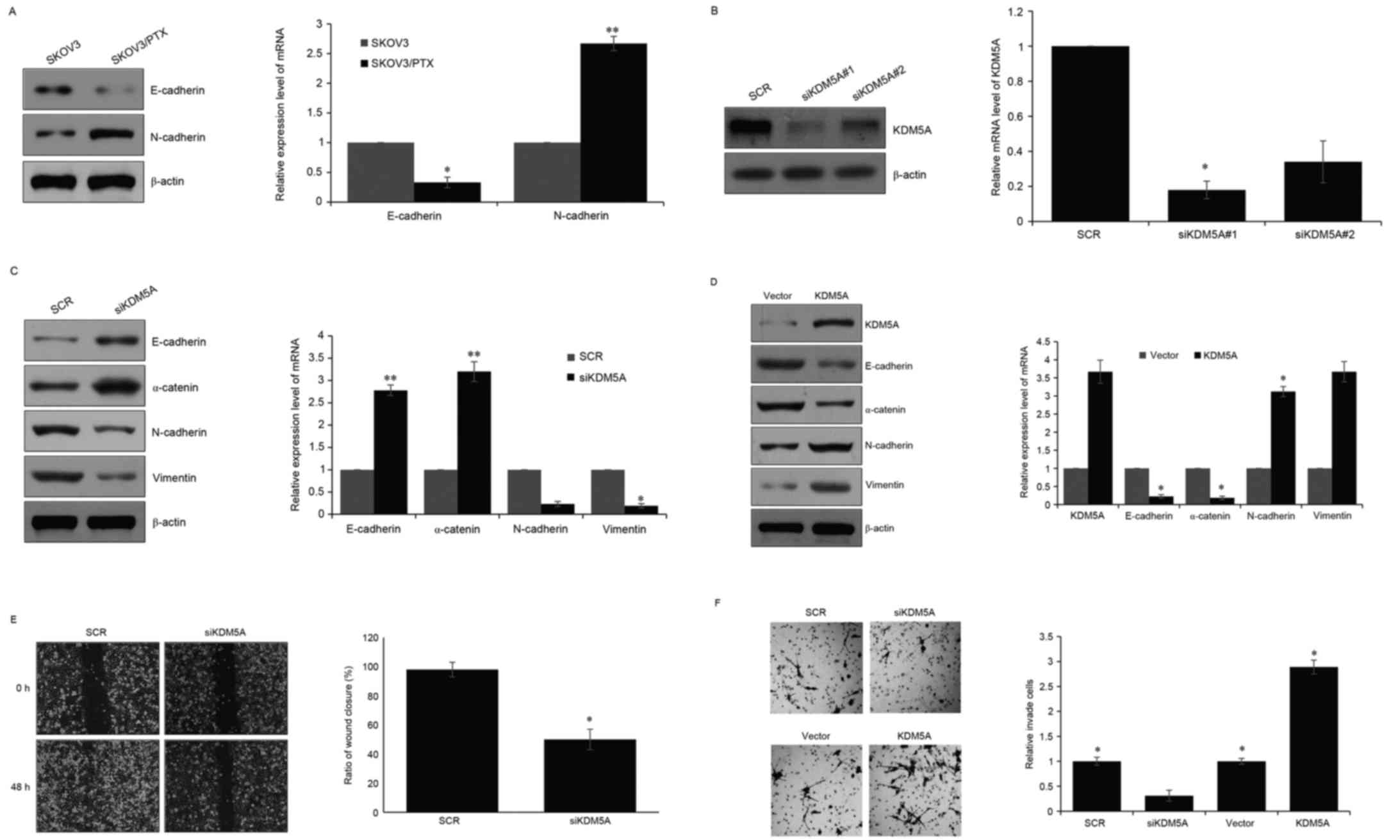 | Figure 2.KDM5A facilitates the EMT in ovarian
cells. (A) Western blotting and RT-qPCR were performed to determine
the protein level and mRNA level of E-cadherin and N-cadherin in
SKOV3 and SKOV3/PTX cells. (B) Two different KDM5A siRNA were
transfected into SKOV3/PTX cells. Following 48 h transfection,
western blotting and RT-qPCR were used to detect the efficiency of
the siRNA. (C) KDM5A was then silenced in SKOV3/PTX cells, and,
after a 48 h transfection, western blot and RT-qPCR were used to
detect the protein level and mRNA level of EMT-associated protein.
(D) KDM5A was expressed ectopically in SKOV3 cells, following 48 h
transfection, western blotting and RT-qPCR were used to detect the
protein level and mRNA level of EMT associated protein. (E) Wound
healing assays was performed on SKOV3/PTX cells in which KDM5A was
depleted to assess the migration ability of ovarian cells. The
distance migrated was measured. All experiments were performed in
three independent times. (F) Transwell assay was performed to
determine the function of KDM5A in ovarian cells. All experiments
were performed in three independent times. *P<0.05, **P<0.01.
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; EMT, epithelial-to-mesenchymal transition; PTX,
paclitaxel; siRNA, small interfering RNA. |
KDM5A modulates PTX sensitivity in
ovarian cells
Previously, KDM5A has been reported to serve an
important role in erlotinib-resistant lung cancer (21). Moreover, KDM5A is more highly
expressed in SKOV3/PTX cells than in other ovarian cells.
Therefore, the authors assumed that KDM5A was also associated with
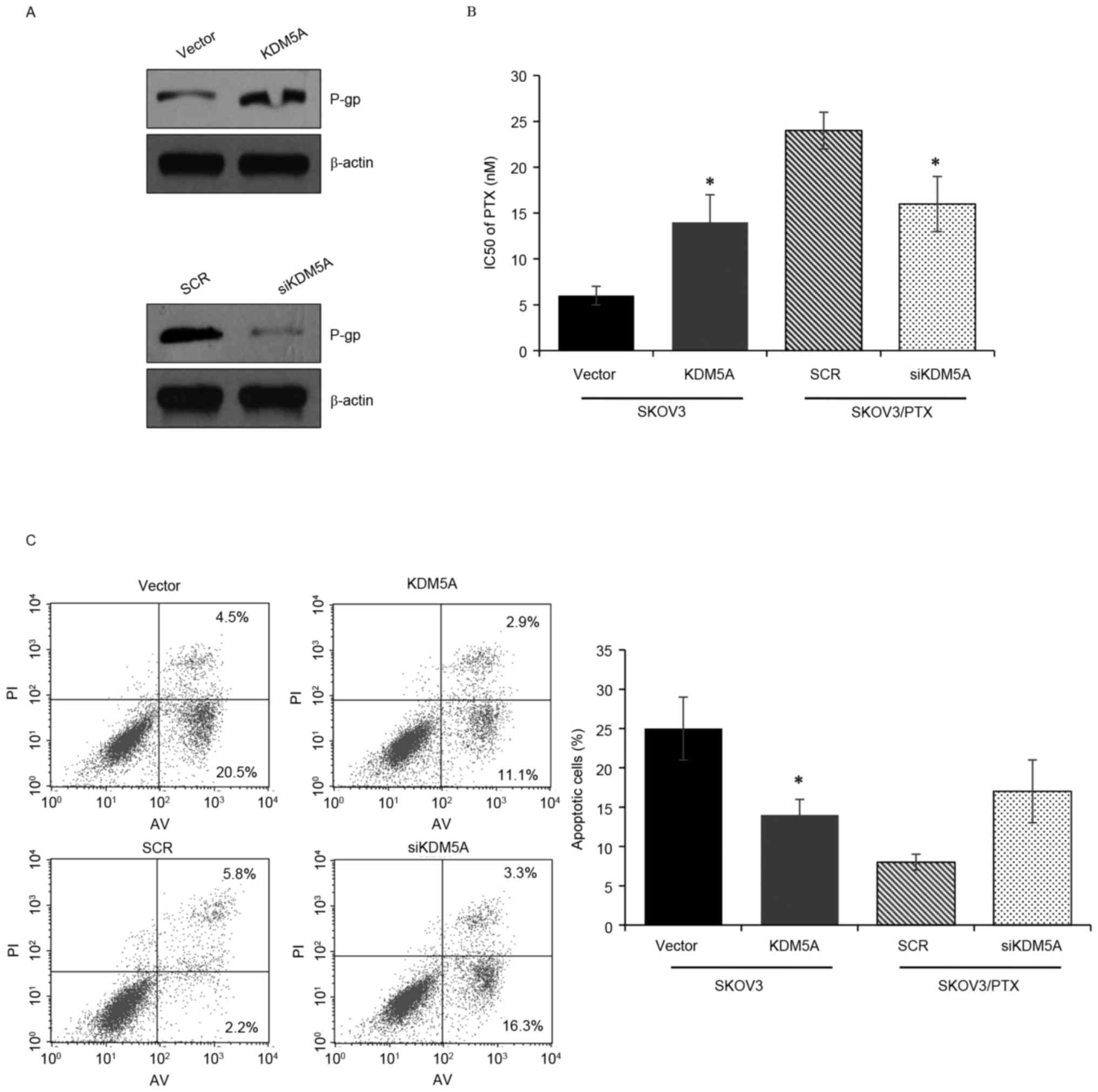
PTX resistance in ovarian carcinoma. P-gp is a protein that is
associated with drug transformation, and high expression of P-gp
indicates drug resistance. The authors initially expressed KDM5A
ectopically in SKOV3 cells and silenced KDM5A in SKOV3/PTX cells.
Following overexpression of KDM5A in SKOV3 cells, P-gp was
increased, whereas when KDM5A was knocked down in SKOV3/PTX cells,
P-gp was decreased (Fig. 3A). The
CCK-8 assay was used to demonstrate the IC50 value of
ovarian carcinoma cells. The results revealed that KDM5A
suppression obviously increased the sensitivity of SKOV3/PTX cells
to PTX, while ectopic expression of KDM5A decreased the sensitivity
of SKOV3 cells to PTX (Fig. 3B).
Consequently, FACS flow cytometry analysis was performed to
investigate how KDM5A regulates cell apoptosis. The results
demonstrated that overexpression of KDM5A decreased the percentage
of apoptotic SKOV3 cells treated with PTX, while knocked down KDM5A
obviously increased the percentage of apoptotic SKOV3/PTX cells
treated with PTX (Fig. 3C).
KDM5A promotes proliferation of
ovarian cancer cells
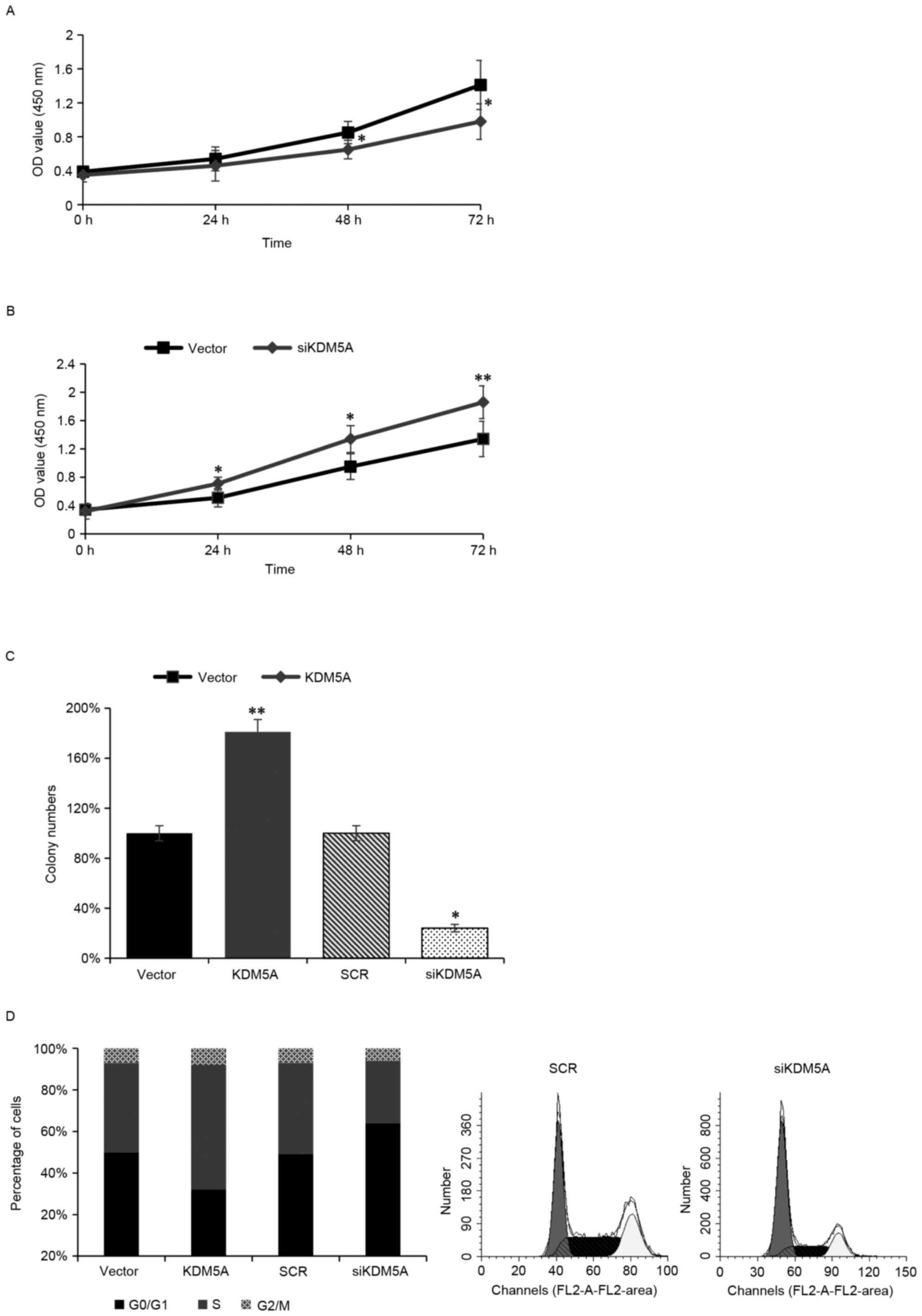
To further assess the function of KDM5A on ovarian
carcinoma transformation, KDM5A was knocked down in SKOV3 cells and
a CCK-8 assay was performed to identify the role of KDM5A in cell
growth. When KDM5A was depleted, cell growth was significantly
inhibited (Fig. 4A). In contrast,
when KDM5A was overexpressed in SKOV3 cells, the cell growth was
obviously increased (Fig. 4B).
Furthermore, anchorage-independent growth assay also confirmed that
overexpression of KDM5A facilitated cell anchorage-independent
growth (Fig. 4C). Next, FACS flow
cytometry was used to assess whether KDM5A regulated the cell
cycle. When KDM5A was expressed ectopically in SKOV3 cells, the
number of S phase cells was obviously increased (Fig. 4C). When KDM5A was silenced in SKOV3
cells, the number of S phase cells was obviously decreased and the
number of G0/G1 phase cells was increased
(Fig. 4D). The present study
indicated that KDM5A facilitated cell proliferation via promoted
cell into S phase.
Discussion
Ovarian cancer is the most common malignant
carcinoma in the world, seriously threatening women's health
(1). In the past decade, multiple
chemotherapy drugs have emerged along with the development of
cytoreductive surgery; however, the 5-year survival rate remains
unsatisfactory. Drug resistance during chemotherapy is a crucial
reason for the constant search for new drugs.
Some previous studies demonstrated that KDM5A serves
a crucial role in drug-resistance in glioblastoma or breast cancer
(19,22). Abnormal expression of KDM5A has
been reported in breast cancer (19) and glioblastoma (22). Although KDM5A regulated
drug-resistance and other cell processes in several cancers, the
effect of KDM5A in ovarian cancer remains unknown, especially in
PTX-resistant ovarian cancer.
Compared with normal ovarian tissues, the authors
indicated that ovarian cancer tissues had an obviously high
expression of KDM5A. As a result, the authors decided to further
investigate its expression in ovarian cell lines. As expected,
KDM5A was highly expressed in ovarian cell lines, compared with
normal ovarian cell lines. Moreover, the expression level of KDM5A
in SKOV3/PTX was higher than other ovarian cell lines that lack PTX
resistance. The authors hypothesized that KDM5A effected PTX
sensitivity in the ovarian cancer cells. Meanwhile, there are many
previous studies demonstrated that chemoresistance in cancer cells
facilitate EMT (6,7). The authors first investigated whether
KDM5A regulates the EMT in ovarian cancer cells. The results
revealed that overexpression of KDM5A promoted the EMT, epithelial
marker levels were decreased, and the mesenchymal markers were
increased. While KDM5A was silenced, the EMT was suppressed. The
EMT has been indicated to facilitate cancer cell metastasis
(23,24). The authors next detected whether
KDM5A regulates ovarian cell metastasis. Transwell and wound
healing assays confirmed that KDM5A served a crucial role in
ovarian cell metastasis. The study's previous results indicated
that KDM5A was expressed more highly in SKOV3/PTX cells than in
other ovarian cell lines; therefore, the further investigated the
function of KDM5A on PTX sensitivity. The expression of P-gp was
increased following ectopic expression of KDM5A in SKOV3 cells,
however the expression of P-gp was decreased following silencing
KDM5A in SKOV3/PTX cells. FACS flow cytometry analysis and CCK-8
assay both confirmed that KDM5A inhibition facilitated the PTX
sensitivity of SKOV3/PTX cells, however, KDM5A overexpression
inhibited the PTX sensitivity of SKOV3 cells. In addition, CCK-8
and anchorage-independent growth assays demonstrated that KDM5A
promoted cell proliferation. In addition, the FACS flow cytometer
assay revealed that KDM5A promoted cell proliferation through
facilitating cell cycle progression. However, the detailed
molecular mechanisms of KDM5A have not been elucidated. KDM5A as a
histone demethylase specific for H3K4 and it is required for cell
development (12–14). The authors hypothesized that there
may be several downstream target genes of KDM5A, and that these
genes may regulate cell proliferation and the cell cycle.
Currently, cancer treatment options include
chemotherapy, radiation therapy and surgical resection (25). For ovarian and lung cancer
patients, PTX is one of the most commonly used anticancer drugs.
However, there are multiple cancers are resistant to it (26). The authors explored the role of
KDM5A in SKOV3/PTX cells. Although the target genes regulated by
KDM5A that are involved in drug-resistance have not yet been
identified, the expression of P-gp and the phenomenon of PTX
sensitivity all conformed that KDM5A serve a key function in
drug-resistance. Moreover, histone demethylases have been
identified as an active frontier in epigenetic drug development
(27,28). A previous study suggested that the
drug-resistant mechanisms of carcinoma included anti-oncogene
inactivation, oncogene activation, drug target molecular changes,
reduced intracellular drug concentration, enhanced DNA damage
repair function, metabolism detoxification and inhibition of tumor
cells apoptosis. Drug-resistance is resulted by multiple genes,
factors and a series of complex processes (29). The detail mechanisms of KDM5A in
PTX-resistance remain unknown, therefore, the authors have decided
to next identify the downstream target gene of KDM5A, and further
decipher the function of KDM5A in drug-resistance.
To the best of the authors' knowledge, the present
study is the first to report that KDM5A mediates drug resistance in
ovarian cancer, and increases ovarian carcinoma cell invasion
ability and growth. The present work suggested that KDM5A was a
critical regulator in ovarian cancer and may be a novel therapeutic
target of ovarian cancer, especially in PTX-resistant cancer.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morgan RJ Jr, Alvarez RD, Armstrong DK,
Burger RA, Chen LM, Copeland L, Crispens MA, Gershenson DM, Gray
HJ, Hakam A, et al: Ovarian cancer, version 2.2013. J Natl Compr
Canc Netw. 11:1199–1209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Di Francia R, De Lucia L, Di Paolo M, Di
Martino S, Del Pup L, De Monaco A, Lleshi A and Berretta M:
Rational selection of predictive pharmacogenomics test for the
Fluoropyrimidine/Oxaliplatin based therapy. Eur Rev Med Pharmacol
Sci. 19:4443–4454. 2015.PubMed/NCBI
|
|
5
|
Ding L, Wang XQ, Zhang J, Mu ZL, Zhou XX
and Liu PS: Underlying mechanism of 2-methoxyestradiol-induced
apoptosis and growth arrest in SKOV3 human ovarian cancer cells.
Eur Rev Med Pharmacol Sci. 19:2084–2090. 2015.PubMed/NCBI
|
|
6
|
Sestito R, Cianfrocca R, Rosanò L, Tocci
P, Semprucci E, Di Castro V, Caprara V, Ferrandina G, Sacconi A,
Blandino G and Bagnato A: miR-30a inhibits endothelin A receptor
and chemoresistance in ovarian carcinoma. Oncotarget. 7:4009–4023.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu X, Shen H, Yin X, Long L, Xie C, Liu
Y, Hui L, Lin X, Fang Y, Cao Y, et al: miR-186 regulation of Twist1
and ovarian cancer sensitivity to cisplatin. Oncogene. 35:323–332.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang J, Yin XJ, Xu CJ, Ning YX, Chen M,
Zhang H, Chen SF and Yao LQ: The histone deacetylase SIRT6 inhibits
ovarian cancer cell proliferation via down-regulation of Notch 3
expression. Eur Rev Med Pharmacol Sci. 19:818–824. 2015.PubMed/NCBI
|
|
9
|
Moisan F, Francisco EB, Brozovic A, Duran
GE, Wang YC, Chaturvedi S, Seetharam S, Snyder LA, Doshi P and
Sikic BI: Enhancement of paclitaxel and carboplatin therapies by
CCL2 blockade in ovarian cancers. Mol Oncol. 8:1231–1239. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu S, Sun J, Cai B, Xi X, Yang L, Zhang
Z, Feng Y and Sun Y: NANOG regulates epithelial-mesenchymal
transition and chemoresistance through activation of the STAT3
pathway in epithelial ovarian cancer. Tumour Biol. 37:9671–9680.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Christensen J, Agger K, Cloos PA, Pasini
D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE and Helin
K: RBP2 belongs to a family of demethylases, specific for tri-and
dimethylated lysine 4 on histone 3. Cell. 128:1063–1076. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lopez-Bigas N, Kisiel TA, Dewaal DC,
Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA and
Benevolenskaya EV: Genome-wide analysis of the H3K4 histone
demethylase RBP2 reveals a transcriptional program controlling
differentiation. Mol Cell. 31:520–530. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pasini D, Hansen KH, Christensen J, Agger
K, Cloos PA and Helin K: Coordinated regulation of transcriptional
repression by the RBP2 H3K4 demethylase and polycomb-repressive
complex 2. Genes Dev. 22:1345–1355. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Defeo-Jones D, Huang PS, Jones RE, Haskell
KM, Vuocolo GA, Hanobik MG, Huber HE and Oliff A: Cloning of cDNAs
for cellular proteins that bind to the retinoblastoma gene product.
Nature. 352:251–254. 1991. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gildea JJ, Lopez R and Shearn A: A screen
for new trithorax group genes identified little imaginal discs, the
Drosophila melanogaster homologue of human retinoblastoma binding
protein 2. Genetics. 156:645–663. 2000.PubMed/NCBI
|
|
17
|
Zeng J, Ge Z, Wang L, Li Q, Wang N,
Björkholm M, Jia J and Xu D: The histone demethylase RBP2 Is
overexpressed in gastric cancer and its inhibition triggers
senescence of cancer cells. Gastroenterology. 138:981–992. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin W, Cao J, Liu J, Beshiri ML, Fujiwara
Y, Francis J, Cherniack AD, Geisen C, Blair LP, Zou MR, et al: Loss
of the retinoblastoma binding protein 2 (RBP2) histone demethylase
suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl
Acad Sci USA. 108:13379–13386. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou J, Wu J, Dombkowski A, Zhang K,
Holowatyj A, Boerner JL and Yang ZQ: Genomic amplification and a
role in drug- resistance for the KDM5A histone demethylase in
breast cancer. Am J Transl Res. 4:247–256. 2012.PubMed/NCBI
|
|
20
|
Liu G, Bollig-Fischer A, Kreike B, van de
Vijver MJ, Abrams J, Ethier SP and Yang ZQ: Genomic amplification
and oncogenic properties of the GASC1 histone demethylase gene in
breast cancer. Oncogene. 28:4491–4500. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sharma SV, Lee DY, Li B, Quinlan MP,
Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach
MA, et al: A chromatin-mediated reversible drug-tolerant state in
cancer cell subpopulations. Cell. 141:69–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Banelli B, Carra E, Barbieri F, Würth R,
Parodi F, Pattarozzi A, Carosio R, Forlani A, Allemanni G, Marubbi
D, et al: The histone demethylase KDM5A is a key factor for the
resistance to temozolomide in glioblastoma. Cell Cycle.
14:3418–3429. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cao H, Xu E, Liu H, Wan L and Lai M:
Epithelial-mesenchymal transition in colorectal cancer metastasis:
A system review. Pathol Res Pract. 211:557–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reck M, Heigener DF, Mok T, Soria JC and
Rabe KF: Management of non-small-cell lung cancer: Recent
developments. Lancet. 382:709–719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao F, Siu MK, Jiang L, Tam KF, Ngan HY,
Le XF, Wong OG, Wong ES, Gomes AR, Bella L, et al: Overexpression
of forkhead box protein M1 (FOXM1) in ovarian cancer correlates
with poor patient survival and contributes to paclitaxel
resistance. PLoS One. 9:e1134782014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grant S: Targeting histone demethylases in
cancer therapy. Clin Cancer Res. 15:7111–7113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Natoli G, Testa G and De Santa F: The
future therapeutic potential of histone demethylases: A critical
analysis. Curr Opin Drug Discov Devel. 12:607–615. 2009.PubMed/NCBI
|
|
29
|
Agarwal R and Kaye SB: Ovarian cancer:
Strategies for overcoming resistance to chemotherapy. Nat Rev
Cancer. 3:502–516. 2003. View
Article : Google Scholar : PubMed/NCBI
|