Introduction
The human gut harbors a wide range of microorganisms
that determine the hemostasis of the host and enable various
metabolic functions, such as biosynthesis of vitamins that humans
are not born with (1). The gut
microbiota are vital in host health development by interacting with
the host (2). A general dysbiosis
in microbiota composition and abnormal interactions in gut
microbiota may result in various types of disorder. Current gut
microbiota studies contribute to the understanding of the complex
interactions between biological processes of the microbiota and
host. These interactions characterize the underlying mechanism of
the association between human health and gut microbiota.
Inflammatory bowel diseases (IBD) is characterized
as a group of chronic gastrointestinal inflammation disorders,
including Crohn's disease and ulcerative colitis (3). A recent study revealed that IBD is
caused by the alterations in gut microbial communities and abnormal
interactions between the immune system and the gut microbiota
(4). A broad range of microbiota
reside in the gastrointestinal tract, including the phylum
Firmicutes, Bacteroidetes, Proteobacteria and
Actinobacteria, which are dominant in the gut microbial
community (5). Normally in a
healthy gut, Firmicutes occupy ~60% of the microbiota,
whereas Bacteroidetes constitute ~20% of the normal human
microbiota (6,7). However, the abundance of microbiota
is imbalanced in IBD and the diversity is also reduced (8,9).
Microbiota dysbiosis of IBD includes the increased abundance of the
phylum Proteobacteria and Bacteroidetes, while the
phyla Firmicutes is decreased (10,11).
The microbiota composition is associated with gastrointestinal
inflammation, therefore the majority of therapeutic strategies for
IBD are focused on reconstructing the normal microbiota community
of the host gut.
Probiotics are reported to benefit the host, and are
non-digestible and fermentable (12). Functional studies of probiotics
have been performed in the treatment of a series of inflammatory
conditions, including ulcerative colitis and Crohn's disease
(13–15). Via stimulating the growth of
commensal flora, probiotics alter the composition of the intestinal
microbes and enhance resistance to detrimental bacteria
localization, therefore contributing to colitis reduction (16,17).
Administration of various probiotic strains has been identified as
an effective treatment method for IBD (18). Our previous work demonstrated the
administration of Lactobacillus plantarum LP-Onlly (LP) may
attenuate inflammation of colitis in knockout (IL-10−/−)
mice (19), however, the
underlying mechanism remains unknown.
The aim of the present study was to reveal the
alteration of gut microbiota under the influence of LP
administration in colitis and clarify the underlying mechanism of
LP treatment in experimental colitis. Metagenomic sequencing was
performed to investigate the diverse microbiota in IL-10 deficient
(IL-10−/−) mice with and without LP administration. In
addition, a group of wild type (WT) mice and another group of mice
with LP treatment (WT + LP) were sequenced to serve as a control.
The abundance of microbiota in the LP treated mice
(IL-10−/− + LP) and mice without LP treatment
(IL-10−/−) was compared. De novo assembly
revealed the taxonomic classification, and further characterized
the functional activities of colitis and LP treatment in the gut
microbiota of mice.
Materials and methods
Animals
Homozygous IL-10−/− mice (weight, 220±12
g; age, 8 weeks; sex, female) generated on a 129 Sv/Ev background
(n=12), and normal 129 Sv/Ev controls (n=12) (The Jackson
Laboratory, Bar Harbor, ME, USA) were housed under
specific-pathogen-free conditions (temperature, 25°C; humidity,
70%) in Shanghai Jiao Tong University Medical School (Shanghai,
China). Mice were fed a standard sterile diet and filtered water ad
libitum under a 12-h light/dark cycle. The animal studies were
approved by the Ethical Committee of the Affiliated Sixth People's
Hospital of Shanghai Jiao Tong University. Scoring of the disease
activity index was performed by an individual blinded to the
treatment.
Microbiome genomic DNA extraction and
sequencing
Microbiome genomic DNA from mouse stools was
prepared using a QIAamp Fast DNA Stool Mini kit (Cat No. 51604,
Qiagen GmbH, Hilden, Germany). All samples were sequenced in the
Illumina HiSeq2000 instrument at SciLifeLab (Stockholm, Sweden)
with up to 10 samples pooled in one lane. Libraries were prepared
with a fragment length of 300 bp. Paired-end reads were generated
with 100 bp in the forward and reverse direction. Sequencing
adapter sequences were removed with cutadapt (http://code.google.com/p/cutadapt/). The length
of each read was trimmed using SolexaQA (http://solexaqa.sourceforge.net/) with the options ‘-b
-p 0.05’. Read pairs with either reads <35 bp were removed with
a custom Python script. The high-quality reads were then aligned to
the human genome (National Center for Biotechnology Information;
NCBI version 37) with Bowtie using ‘-n 2-l 35-e 200-best-p
8-chunkmbs 1024-X 600-tryhard’. This set of high-quality reads was
subsequently used for further analysis.
Alignment to reference genomes and
taxonomical analysis
A set of 2,797 microbial reference genomes were
obtained from the NCBI and Human Microbiome Project (20,21)
on 02 August 2011. The reference genomes were combined into two
Bowtie indexes and the metagenomic sequence reads were aligned to
the reference genomes using Bowtie with parameters ‘-n 2-l 35-e
200-best-p 8-chunkmbs 1024-X 600-tryhard’. Mapping results were
merged by selecting the alignment with fewest mismatches; if a read
was aligned to a reference genome with the same number of
mismatches, each genome was assigned half to each genome. The
relative abundance of each genome was calculated by summing the
number of reads aligned to that genome divided by the genome size.
In each subject, the relative abundance was scaled to sum to one.
The taxonomic rank for every genome was downloaded from NCBI
taxonomy to assign each genome to a species, genus and phyla. The
relative abundance for each taxonomical rank was calculated by
summing the relative abundance of all its members.
Statistical analysis
The high-quality reads were used for de novo
assembly with Velvet (22) into
contigs (length, ≥500 bp) using 3 as the coverage cutoff and a kmer
length of 31. To obtain long contigs with high specificity,
parameter values for the kmer length and coverage cutoff were
iteratively investigated to balance the total assembly length and
the N50 value to be used in the final de novo assembly.
Reads from each subject were used in separate assemblies and
unassembled reads were subsequently used in a global final
assembly. Genes were predicted on the contigs using MetaGeneMark
(23). All genes were then aligned
on the contigs with Bowtie using the above-mentioned parameters.
The abundance of a predicted gene was calculated by counting the
number of reads that align to the gene, and then the read counts
were normalized by the gene length and the total mapped reads. The
genes were annotated to the eggCOG database (24) with hidden Markov models (HMMs).
Protein sequences for microbial orthologs were downloaded and
aligned with MUSCLE (25). HMMs
were generated with HMMer3 (26)
for each KO mouse.
To determine the differential abundance of
metagenomic features, an unpaired t-test was applied. Strains and
genera with a relative abundance in any subject >10 were
included in the analysis. The R package ade4 (27) (https://www.r-project.org/) using instrumental
principal component analysis was used to determine the global
analysis of species abundance between each group.
The relative abundance of a given taxon in a
community was calculated as: Relative abundance = a/b × 100%.
Where ‘a’ is the number of sequences assigned to the
taxon and ‘b’ is the total number of sequences assigned to all the
taxa in the community). Similar calculations were performed for
relative abundance of a given gene, Clusters of Orthologous Groups
(COG), COG category, Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway and KEGG subcategory (28). Statistical enrichment of a given
gene or COG between two data sets was determined by pairwise
comparisons using two-tailed Fisher's exact test, with confidence
intervals at 99% significance and Benjamini-Hochberg correction. In
all analyses, P<0.05 was considered to indicate a significant
difference.
Results
LP treatment reduces inflammation in
the mouse gut
To investigate the important role of LP in gut
microbiota homeostasis, four groups of mice models were
established: WT (WT mice without LP treatment), WT + LP (WT mice
with LP treatment), IL-10−/− (IL-10−/− mice
without LP treatment), and IL-10−/− + LP
(IL-10−/− mice with LP treatment) (n=6/group).
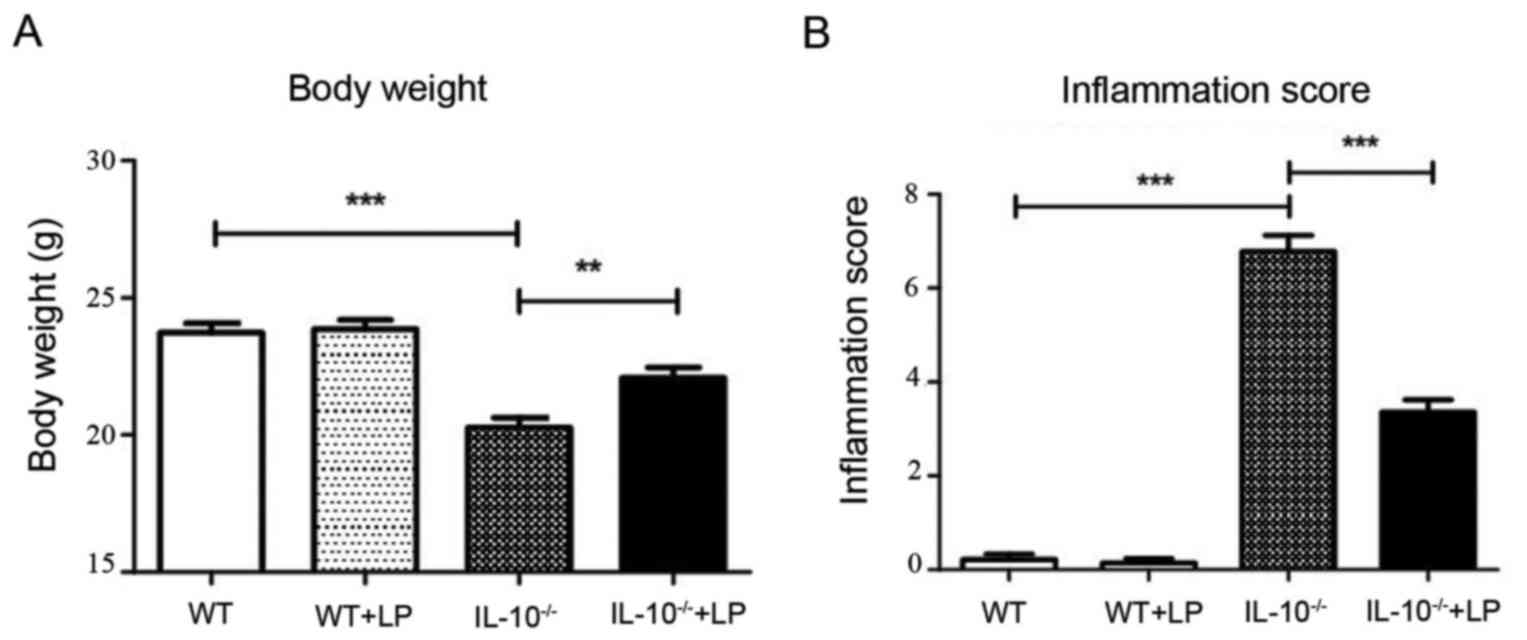
Consistent with a previous study (19), we observed that LP administration
attenuated the inflammation syndrome of gut colitis. The body
weight of IL-10−/− mice decreased markedly after 4
weeks; however, the body weight of the IL-10−/− mice
with LP treatment was significantly greater than that of mice
without LP treatment (Fig. 1A).
Consistently, the inflammation score of IL-10−/− mice
was significantly greater than that of IL-10−1− mice
with LP feeding, and no inflammation syndrome was observed in the
WT groups (Fig. 1B).
LP administration profoundly affected
the gut microbiome
To clarify the influence of LP administration on the
gut microbial environment in the gut of colitis mouse models, the
gut metagenome of the four groups of mice was sequenced. A total of
~411 million 101-bp paired-end clean reads were generated, the
sequencing adapter and low quality reads were removed. The reads
from mice genomes were identified and filtered. To reveal the
composition of the gut microbiota, all the remaining reads were
aligned to a catalog of 2,797 non-redundant NCBI microbial
reference genomes (29). On
average, 23% of the reads in a sample could be aligned to the
reference genome, this ratio was close to the previous metagenome
studies (29).
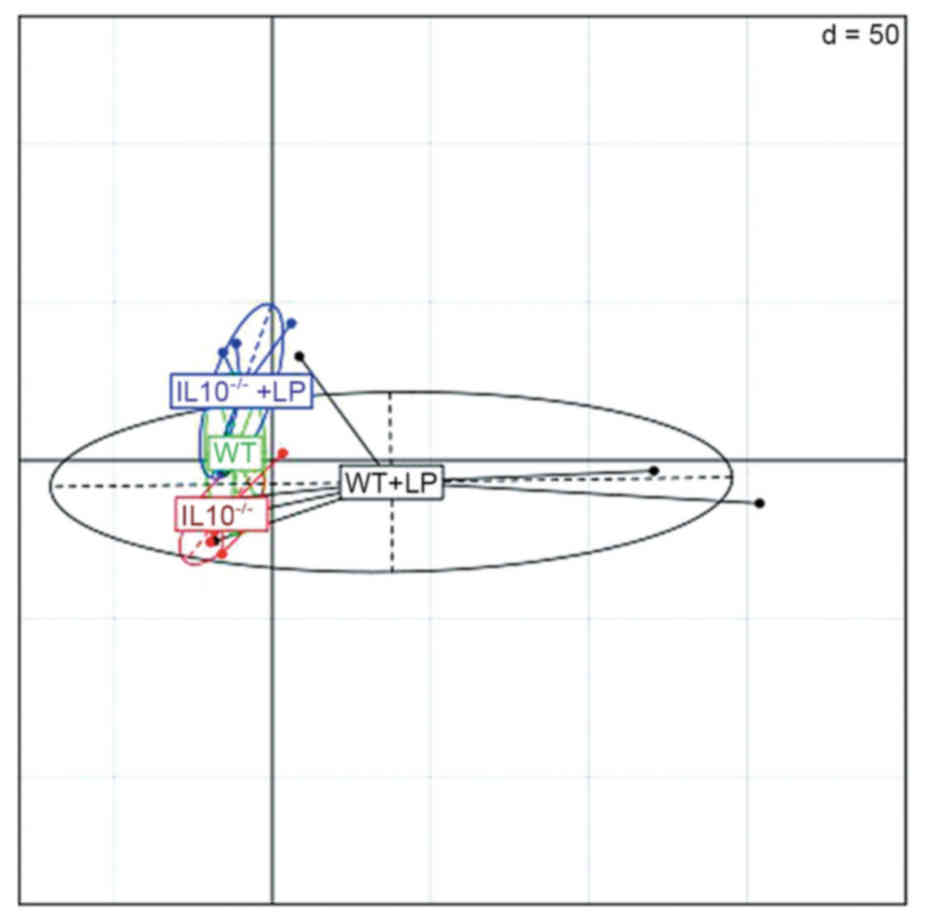
The principal component analysis confirms that the
WT samples and IL-10−/− samples were differentiated by
the abundance of microbial species. As presented in Fig. 2, samples of control group with LP
administration were clustered together and the IL-10−/−+
LP samples were clustered in another group. However, the
IL-10−/−samples without LP administration were
dispersed, this may indicate that variation existed among the
microbes in the most seriously inflamed gut samples.
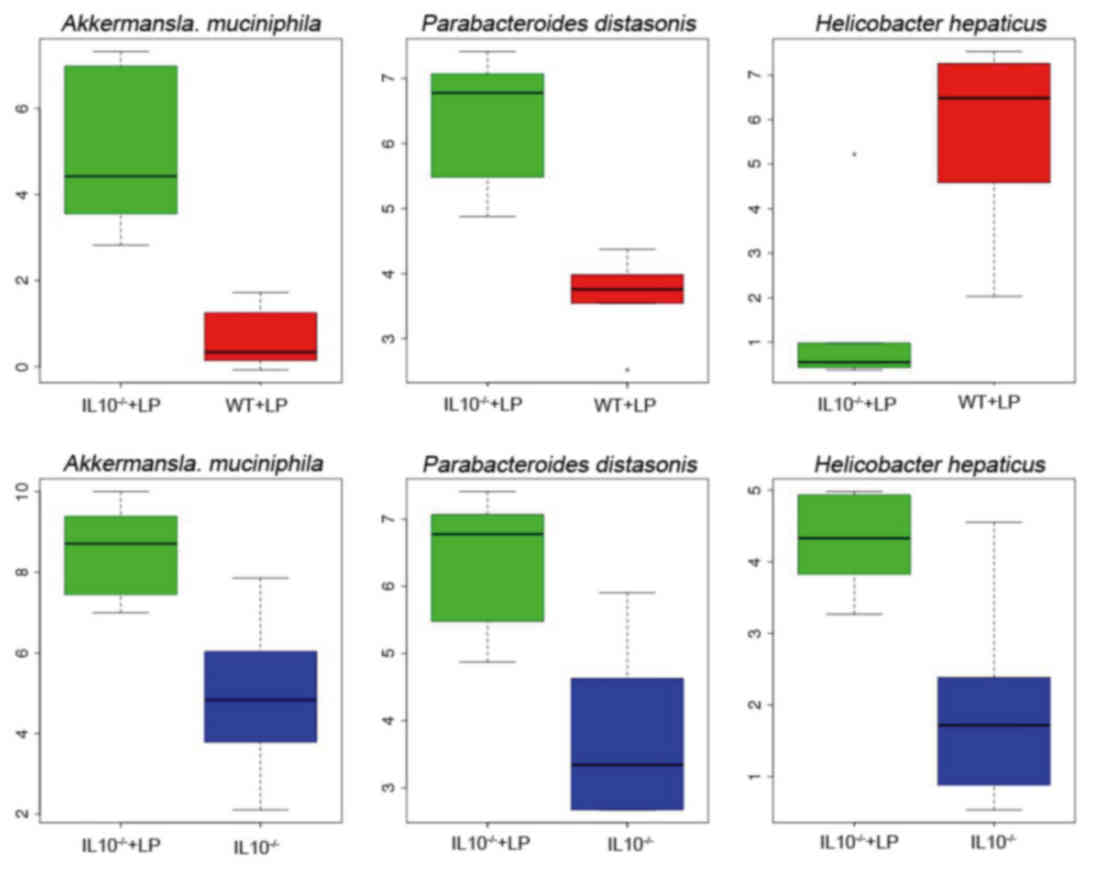
To investigate the influence of inflammation on gut
microbial communities and regulation of LP administration, the
change of identified microbial abundance was compared between the
different experiment groups. The species A. muciniphila and
Parabacteroides distasonis were enriched in
IL-10−/−mice, whereas Helicobacter hepaticus was
enriched in the control groups (Fig.
3). Previous studies have confirmed the association between
these three types of species and IBD. A. muciniphila
exacerbates gut inflammation by disturbing host mucus homeostasis
(30). However, the oral
administration of P. distasonis has been reported as a novel
therapeutic strategy for IBD (31). In addition, the H. hepaticus
was associated with IBD.
L. plantarum is frequently used as a
probiotic, and has been associated with the maintenance of
intestinal homeostasis and modulation of the immune system. It
regulates the quantity of pathogenic bacteria. Following L.
plantarum administration, the abundance of three types of
microbes, namely, Bacteroides uniformis, P. distasonis and
Bacteroides salanitronis, were downregulated compared with
the IL-10−/− control groups. Bacteroides
uniformis have been identified as essential members of gut
microbiota, and are enriched in the gut of healthy individuals
without IBD (32,33). Furthermore, P. distasonis
exerts beneficial effects on gut health; Kverka et al
identified that the oral administration of P. distasonis
attenuated the inflammation of IBD by modulation of immunity
(31). To the best of our
knowledge, our study is the first to report the association between
Bacteroides salanitronis and IBD. These findings may
indicate that the loss of probiotic in the IL-10−/− mice
gut may aggravate the inflammation and that L.
plantarum may increase the abundance of probiotic.
Taxonomic characterization in the mice
gut microbiomes
To further identify novel genes in the mice gut
metagenome and investigate the variation in IL-10−/−
mice gut microbiota communities with and without LP administration,
the de novo assembly was performed for the sequence data. In
total 0.32 Gbp of contigs >500 bp were assembled with an N50
value of 0.8 kbp and 5 as the coverage cutoff. Genes were predicted
using the assembled contigs and 0.2 million non-redundant genes
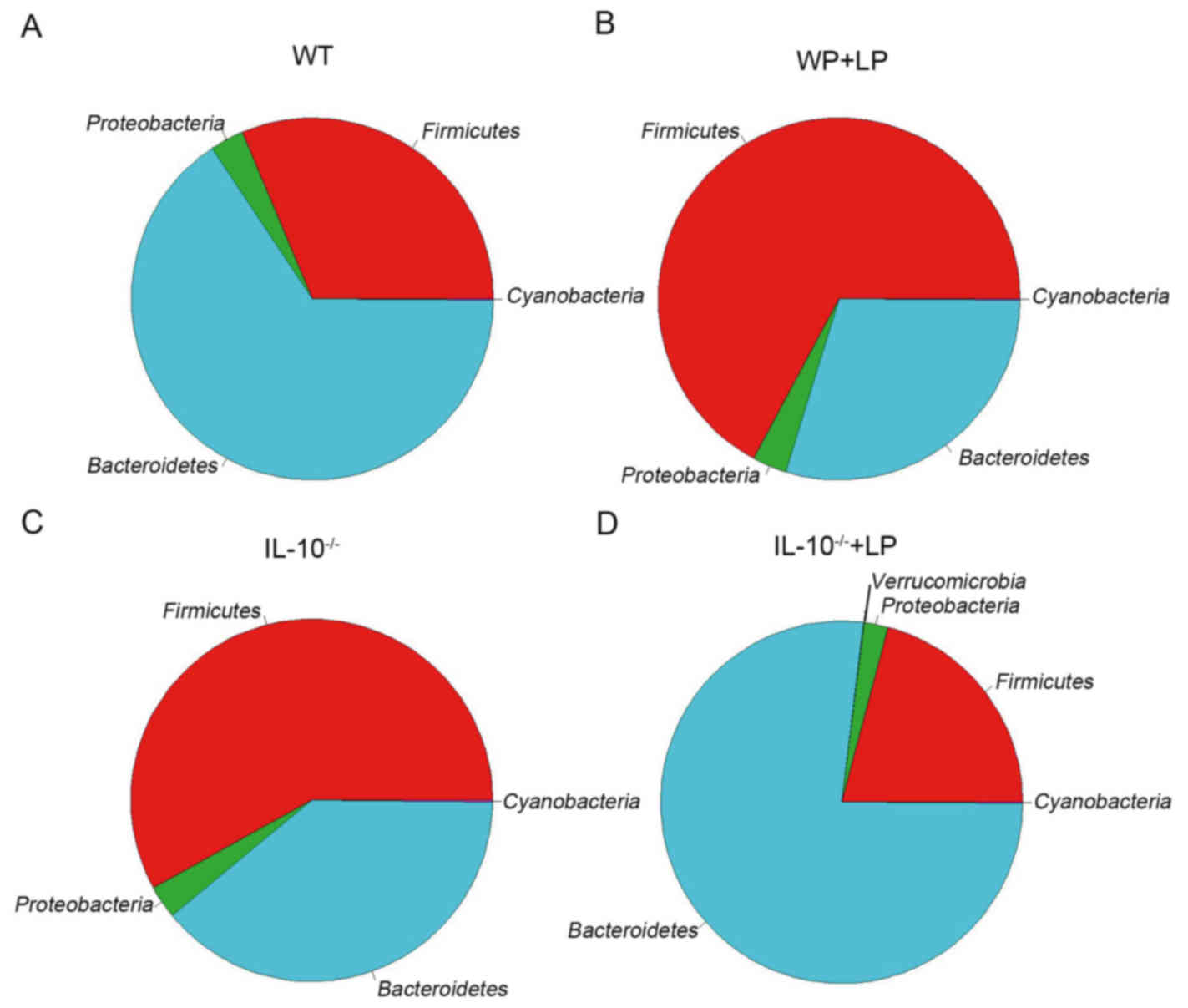
were identified. For the phylum level, as presented in Fig. 4, the gut inflammation and LP
administration greatly affected the phyla proportion of
Firmicutes and Bacteroidetes, as well as the
abundance of various other phyla. This was consistent with previous
studies, that the mice gut microbiome was greatly dominated by
Firmicutes, Bacteroidetes and Proteobacteria.
The ratio of Bacteroidetes/Firmicutes was markedly decreased
in IL-10−/− mice, which may be associated with the
inflammation of mice gut; however, following LP administration, the
Bacteroidetes/Firmicutes ratio in IL-10−/− + LP
group was increased compared with the IL-10−/− group,
and was comparable with the control group. In addition, a small
influence on Cyanobacteria and Proteobacteria was
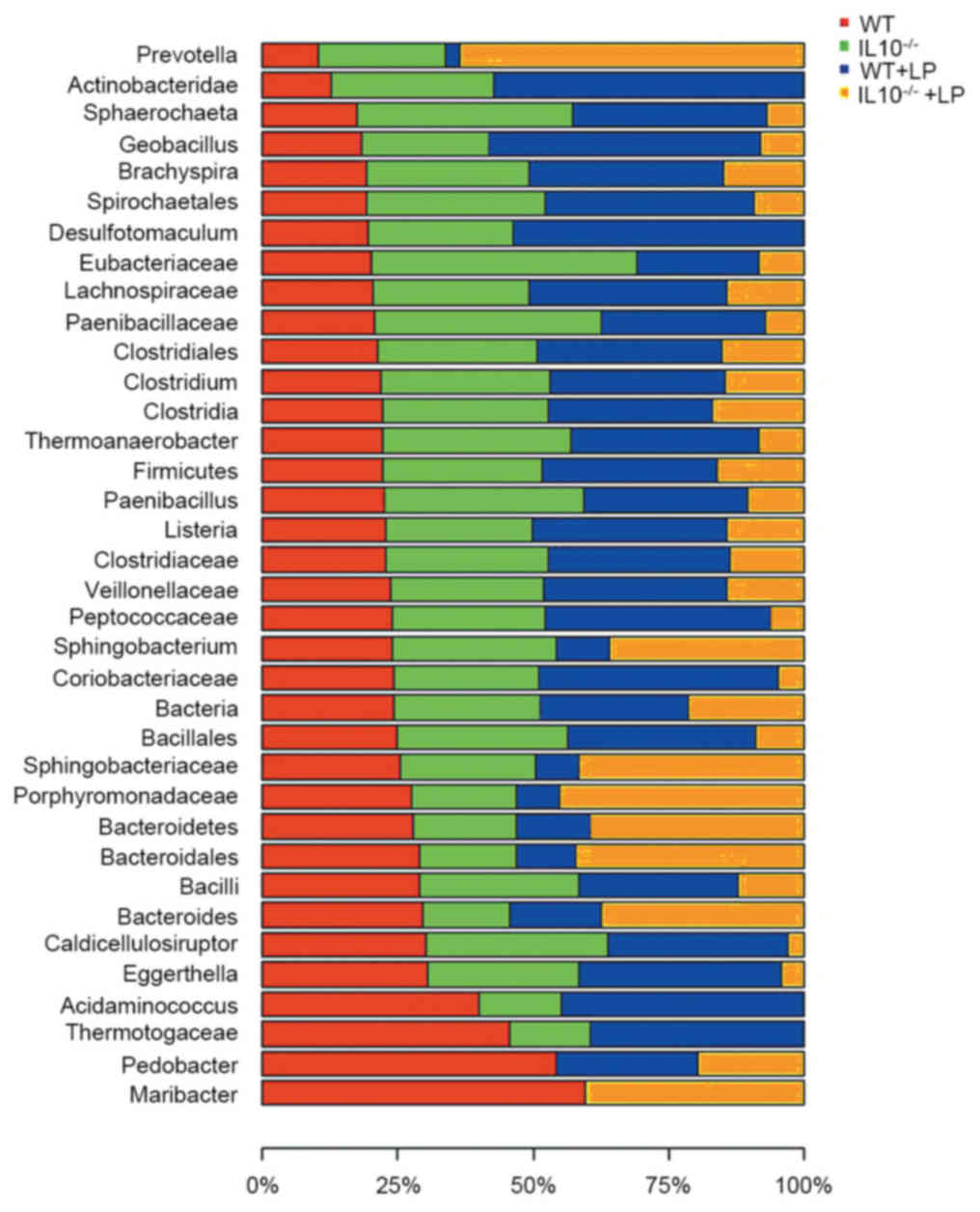
also observed in IBD in the current study. Furthermore, a total of
36 genera/phylum were affected by the inflammation and LP
administration. The variation of the abundance of microbiomes in
the four groups is presented in Fig.
5.
Functional activities of the mice gut
microbiota
Gut microbial activities are associated with host
physiological function, may influence metabolism and are a key
factor in the inflammation state of IBD. Therefore, to characterize
the functional activities of the mice gut microbiota, the predicted
genes were functionally annotated to the eggCOG database and the
relative abundance of COGs was assessed. The marked variation of
COG abundance was observed between the three groups of mice gut
microbiotas (Fig. 6). Twenty COG
classes demonstrated significant differences in at least two groups
of microbiomes. Numerous categories of COG were significantly
decreased in the IL-10−/− mice compared with the WT + LP
groups, which included ‘Cell cycle control, cell division’, ‘Amino
acid transport and metabolism’, ‘Carbohydrate transport and
metabolism’, ‘Transcription’, ‘Replication, recombination and
repair’, ‘Cell wall/membrane/envelope biogenesis’ and ‘General
function prediction only’, while the abundance of these classes was
increased in the LP administration groups of IL-10−/−
mice. Furthermore, the abundance of categories ‘Intracellular
trafficking, secretion and vesicular transport’ was significantly
enriched in the IL10−/− group.
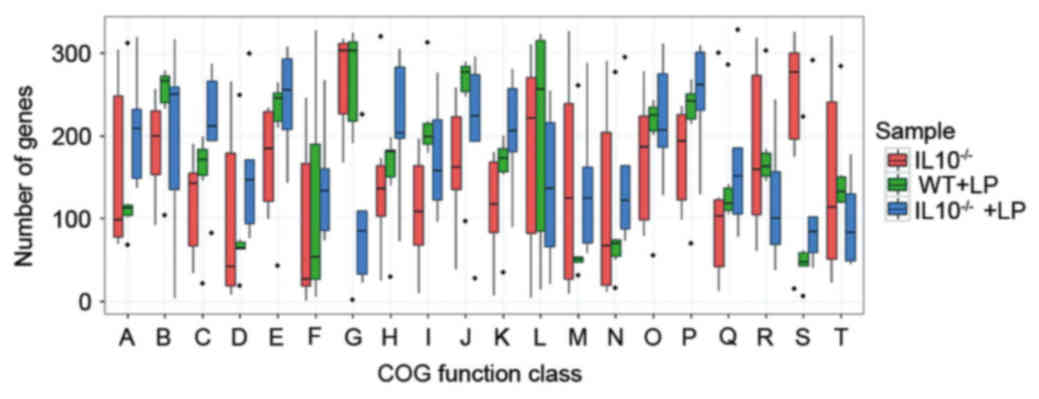 | Figure 6.COGs of proteins affected by the
IL-10−/− and LP treatments. Occurrence of COGs of
proteins according to the following COG functional categories: (A)
Energy production and conversion; (B) Cell cycle control cell
division; (C) Amino acid transport and metabolism; (D) Nucleotide
transport and metabolism; (E) Carbohydrate transport and
metabolism; (F) Coenzyme transport and metabolism; (G) Lipid
transport and metabolism; (H) Translation, ribosomal structure and
biogenesis; (I) Transcription; (J) Replication, recombination and
repair; (K) Cell wall/membrane/envelope biogenesis; (L) Cell
motility; (M) Posttranslational modification, protein turnover,
chaperones; (N) Inorganic ion transport and metabolism; (O)
Secondary metabolites biosynthesis, transport and catabolism; (P)
General function prediction only; (Q) Function unknown; (R) Signal
transduction mechanisms; (S) Intracellular trafficking, secretion
and vesicular transport; (T) Defense mechanisms. COGs, Clusters of
Orthologous Groups; WT, wild type; LP, Lactobacillus
plantarum LP-Onlly; IL, interleukin; IL-10−/−, IL-10
knock out. |
Discussion
Probiotics have an important role in maintaining gut
microbiota homeostasis, the imbalances in microbial communities may
contribute to the gut disease of the host (14). Therefore, probiotics have been
utilized as an effective treatment for gut disease. The onset and
progression of IBD have been attributed to alteration of microbiota
composition and the interaction between immune system and
microbiota. Our previous work and other studies have demonstrated
that probiotic treatment attenuates inflammation of colitis
(19). In the current study, a
colitis model was constructed using IL-10−/− mice. A
metagenome sequence approach was conducted to investigate the
effect of LP in colitis in IL-10−/− mice. Weight loss
was observed in the IL-10−/−mice compared with the
control group and the inflammation score was particularly high.
However, the inflammation score and weight loss were improved
following LP treatment in IL-10−/−-deficient mice. This
indicated that the colitis mice model had successfully been
constructed, and LP administration is effective in the treatment of
colitis.
During the metagenome analysis, clean reads were
mapped to microbial reference genomes. The principal component
analysis clearly differentiated the IL10−/− + LP group
and WT + LP group; however, the samples of IL-10−/− are
scattered. This may be because inflammation of colitis influences
the abundance and diversity of microbiota. The abundance of A.
muciniphila and P. distasonis was significantly
increased in IL-10−/−+ LP group compared with the WT +
LP group. The abundance of B. uniformis and P.
distasonis was decreased in the IL-10−/− group. A
previous study revealed that the increase of A. muciniphila
was harmful to IBD and that B. uniformis has been identified
as an essential member of gut microbiota (34). However, a previous study
demonstrated that P. distasonis may be beneficial to IBD as
it prevents intestinal inflammation in murine models (31). In addition, taxonomic analysis
revealed that Firmicutes and Bacteroidetes are
dominated in mice guts, which is consistent with previous studies.
The Firmicutes/Bacteroidetes ratio increases in
IL-10−/−, and following LP administration this ratio
decreased to a normal level when compared with the ratio of WT.
However, this ratio increased markedly in the WT + LP group. The
results demonstrate that alterations to the microbiota composition
may serve an important role in IBD, and the administration of LP
may regulate the abundance and diversity of gut microbiota.
In the analysis of functional activities, the
relative abundance of COGs was observed to change in
IL-10−/−mice. The function categories of COG, including
‘Cell cycle control, cell division’, ‘Amino acid transport and
metabolism’, ‘Carbohydrate transport and metabolism’,
‘Transcription’ and ‘Replication, recombination and repair’ were
significantly influenced. The ‘Carbohydrate and nucleotide
metabolism’, ‘Lipid and amino acid metabolism’ and ‘Amino acid
transport and metabolism’ were found to be associated with IBD in a
recent study (5).
In conclusion, the present study further
demonstrated the effectiveness of LP in the treatment of colitis.
The current study provided an overview of gut microbiota components
of colitis, and revealed the ability of LP to regulate the gut
microbiota, which may be important in attenuating the inflammation
of colitis. However, further investigations regarding LP and gut
microbiota in colitis are required to reveal how metabolic changes
to LP attenuates the colitis, and how the host immune system
interacts with LP.
Acknowledgements
The present study was supported by National Natural
Science Foundation of China (grant no. 81200264).
References
|
1
|
Guarner F and Malagelada JR: Gut flora in
health and disease. Lancet. 361:512–519. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ussar S, Griffin NW, Bezy O, Fujisaka S,
Vienberg S, Softic S, Deng L, Bry L, Gordon JI and Kahn CR:
Interactions between gut microbiota, host genetics and diet
modulate the predisposition to obesity and metabolic syndrome. Cell
Metab. 22:516–530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abraham C and Cho JH: Inflammatory bowel
disease. N Engl J Med. 361:2066–2078. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scaldaferri F, Gerardi V, Lopetuso LR, Del
Zompo F, Mangiola F, Boškoski I, Bruno G, Petito V, Laterza L,
Cammarota G, et al: Gut microbial flora, prebiotics, and probiotics
in IBD: Their current usage and utility. Biomed Res Int.
2013:4352682013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hansen JJ: Immune responses to intestinal
microbes in inflammatory bowel diseases. Curr Allergy Asthma Rep.
15:612015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rup L: The human microbiome project.
Indian J Microbiol. 52:3152012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gevers D, Knight R, Petrosino JF, Huang K,
McGuire AL, Birren BW, Nelson KE, White O, Methé BA and Huttenhower
C: The human microbiome project: A community resource for the
healthy human microbiome. PLoS Biol. 10:e10013772012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ott SJ, Musfeldt M, Wenderoth DF, Hampe J,
Brant O, Fölsch UR, Timmis KN and Schreiber S: Reduction in
diversity of the colonic mucosa associated bacterial microflora in
patients with active inflammatory bowel disease. Gut. 53:685–693.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Morgan XC, Tickle TL, Sokol H, Gevers D,
Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et
al: Dysfunction of the intestinal microbiome in inflammatory bowel
disease and treatment. Genome Biol. 13:R792012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sokol H, Lay C, Seksik P and Tannock GW:
Analysis of bacterial bowel communities of IBD patients: What has
it revealed? Inflamm Bowel Dis. 14:858–867. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Swidsinski A, Ladhoff A, Pernthaler A,
Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U,
Schreiber S, Dietel M and Lochs H: Mucosal flora in inflammatory
bowel disease. Gastroenterology. 122:44–54. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Petschow B, Doré J, Hibberd P, Dinan T,
Reid G, Blaser M, Cani PD, Degnan FH, Foster J, Gibson G, et al:
Probiotics, prebiotics, and the host microbiome: The science of
translation. Ann N Y Acad Sci. 1306:1–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rembacken BJ, Snelling AM, Hawkey PM,
Chalmers DM and Axon AT: Non-pathogenic Escherichia coli versus
mesalazine for the treatment of ulcerative colitis: A randomised
trial. Lancet. 354:635–639. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Prantera C and Scribano ML: Probiotics and
Crohn's disease. Dig Liver Dis. 34 Suppl 2:S66–S67. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hedin C, Whelan K and Lindsay JO: Evidence
for the use of probiotics and prebiotics in inflammatory bowel
disease: A review of clinical trials. Proc Nutr Soc. 66:307–315.
2007; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gibson GR: Dietary modulation of the human
gut microflora using the prebiotics oligofructose and inulin. J
Nutr. 129 7 Suppl:1438S–1441S. 1999.PubMed/NCBI
|
|
17
|
Sartor RB: Therapeutic manipulation of the
enteric microflora in inflammatory bowel diseases: Antibiotics,
probiotics, and prebiotics. Gastroenterology. 126:1620–1633. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Everard A, Lazarevic V, Gaïa N, Johansson
M, Ståhlman M, Backhed F, Delzenne NM, Schrenzel J, François P and
Cani PD: Microbiome of prebiotic-treated mice reveals novel targets
involved in host response during obesity. ISME J. 8:2116–2130.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xia Y, Chen HQ, Zhang M, Jiang YQ, Hang XM
and Qin HL: Effect of Lactobacillus plantarum LP-Onlly on gut flora
and colitis in interleukin-10 knockout mice. J Gastroenterol
Hepatol. 26:405–411. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Human Microbiome Project Consortium: A
framework for human microbiome research. Nature. 486:215–221. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Human Microbiome Project Consortium:
Structure, function and diversity of the healthy human microbiome.
Nature. 486:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zerbino DR and Birney E: Velvet:
Algorithms for de novo short read assembly using de Bruijn graphs.
Genome Res. 18:821–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu W, Lomsadze A and Borodovsky M: Ab
initio gene identification in metagenomic sequences. Nucleic Acids
Res. 38:e1322010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huerta-Cepas J, Szklarczyk D, Forslund K,
Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn
M, et al: eggNOG 4.5: A hierarchical orthology framework with
improved functional annotations for eukaryotic, prokaryotic and
viral sequences. Nucleic Acids Res. 44:D286–D293. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Edgar RC: MUSCLE: Multiple sequence
alignment with high accuracy and high throughput. Nucleic Acids
Res. 32:1792–1797. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sinha S and Lynn AM: HMM-ModE:
Implementation, benchmarking and validation with HMMER3. BMC Res
Notes. 7:4832014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dray S and Dufour AB: The ade4 package:
Implementing the duality diagram for ecologists. J Stat Softw.
22:202007. View Article : Google Scholar
|
|
28
|
Wixon J and Kell D: The Kyoto encyclopedia
of genes and genomes-KEGG. Yeast. 17:48–55. 2000.PubMed/NCBI
|
|
29
|
Markowitz VM: Microbial genome data
resources. Curr Opin Biotechnol. 18:267–272. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ganesh BP, Klopfleisch R, Loh G and Blaut
M: Commensal Akkermansia muciniphila exacerbates gut inflammation
in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One.
8:e749632013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kverka M, Zakostelska Z, Klimesova K,
Sokol D, Hudcovic T, Hrncir T, Rossmann P, Mrazek J, Kopecny J,
Verdu EF and Tlaskalova-Hogenova H: Oral administration of
Parabacteroides distasonis antigens attenuates experimental murine
colitis through modulation of immunity and microbiota composition.
Clin Exp Immunol. 163:250–259. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qin J, Li R, Raes J, Arumugam M, Burgdorf
KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al: A
human gut microbial gene catalogue established by metagenomic
sequencing. Nature. 464:59–65. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fenner L, Roux V, Mallet MN and Raoult D:
Bacteroides massiliensis sp. nov., isolated from blood culture of a
newborn. Int J Syst Evol Microbiol. 55:1335–1337. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Png CW, Linden SK, Gilshenan KS, Zoetendal
EG, McSweeney CS, Sly LI, McGuckin MA and Florin TH: Mucolytic
bacteria with increased prevalence in IBD mucosa augment in vitro
utilization of mucin by other bacteria. Am J Gastroenterol.
105:2420–2428. 2010. View Article : Google Scholar : PubMed/NCBI
|