Introduction
Laryngeal squamous cell carcinoma (LSCC) is one of
the most common malignancies in head and neck (1). LSCC has been reported to occur more
frequently in middle-aged and elderly men (1). In 2016, 13,430 new cases and 3,620
deaths are estimated of larynx cancer in United States (2). Currently, although marked
developments have been achieved in terms of therapeutic strategies
for LSCC, there are no such enhancements inthe therapeutic outcome
and prognosis of patients with LSCC (2). Therefore, it is extremely urgent to
uncover the molecular mechanisms of LSCC, which can contribute to
improve the clinical therapy of LSCC.
Hsa-miR-145 is one of the most important tumor
suppressor miRNAs, which is lowly expressed in various cancers,
such as colorectal cancer (3),
gallbladder cancer (4), pleural
mesothelioma (5) andprostate
cancer (6). In the researches on
LSCC, miR-145 has also been shown to be downregulated, and
overexpression of miR-145 inhibit the proliferation and metastasis
in Hep-2 cells viainducing cell cycle arrest and apoptosis
(7). A recent study has shown that
miR-145 is able to inhibit stem cell potency of Hep-2 cells, along
with the downregulation of stem cell marks like SOX2,
KLF4, OCT4 and ABCG2 (8). Furthermore, we found that miR-145-5p
was able to inhibit cell proliferation and metastasis in human LSCC
cell line Tu-177, and downregulation of miR-145-5p led to poor
prognosis of patients with LSCC (9). However, currently, the regulatory
effect of miR-145-5p on the gene expression profiling in LSCC
remains unclear.
In the present study, to reveal the variation of
gene expression profiling induced by miR-145-5p overexpression in
LSCC, miR-145-5p mimic was transfected into Tu-177 cells to
generate miR-145-5p-overexpressed LSCC cells. Then, based on gene
microarrays, differentially expressed genes (DEGs) between
miR-145-5p-overexpressed Tu-177 cells and negative control (NC)
cells were identified. The target genes of miR-145-5p among the
DEGs were identified and their potential functions and
protein-protein interactions (PPIs) were analyzed. These results
were expected to be helpful for the better understanding of the
effects of miR-145-5p on gene expressions in LSCC.
Materials and methods
Cell culture and establishment of
hsa-miR-145-5p-overexpressed cell model
Human LSCC cell line Tu-177 was purchased from
Shanghai Bioleaf Biotech Company (Shanghai, China). Cells were
maintained in Roswell Park Memorial Institute-1640 (HyClone, Logan,
UT, USA) containing 10% fetal bovine serum (Biological Industries,
Cromwell, CT, USA) at 37°C in a humidified chamber supplemented
with 5% CO2. For miRNA mimic transfection, cells were
plated in 6-well dishes (2.0×105 cells per well).
MiR-145-5p mimic and NC (GenePharma Co., Ltd, Shanghai, China) were
transfected at a final concentration of 50 nM by using
Lipofectamine® 2000 (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's instructions. After 48 h of
transfection, cells were harvested for the following
procedures.
RNA extraction and array
procedures
Total RNA was extracted from transfected cells using
TRIzol reagent (Invitrogen) following the manufacturer's protocol.
Double-strand cDNA (ds-cDNA) was synthesized from 5 µg of total RNA
using anSuperScript ds-cDNA synthesis kit (Invitrogen) in the
presence of 100 pmol oligo dT primers. Subsequently, ds-cDNA was
cleaned and labeled in accordance with the NimbleGen Gene
Expression Analysis protocol (NimbleGen Systems, Inc., Madison, WI,
USA). Briefly, ds-cDNA was incubated with 4 µg RNase A at 37°C for
10 min and cleaned using phenol:chloroform:isoamyl alcohol,
followed by ice-cold absolute ethanol precipitation. After
purification, the generated cDNA was quantified using NanoDrop
ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA).
Subsequently, the cDNA was labeled by a monochrome DNA labeling kit
(NimbleGen Systems, Inc., Madison, WI, USA) following the
manufacturer's protocol, and the labeled cDNA was hybridized
withNimbleGen Human 12×135 k expression microarrays at 42°C for
16–20 h. Finally, slides were washed using NimbleGen Wash Buffer
kit (NimbleGen Systems, Inc.), and scanned by Axon GenePix 4000B
microarray scanner (Axon Instruments Inc., Foster City, CA, USA).
The microarray data were submitted to Gene Expression Omnibus (GEO)
with an accession number GSE92678.
Real-time quantitative PCR (qPCR)
First-strand cDNA was synthesized using All-in-One
miRNA First-Strand cDNA Synthesis kit (GeneCopoeia, Inc.,
Rockville, MD, USA). The real-time quantitative PCR (RT-qPCR) was
performed using ChamQ SYBR qPCR Master Mix (Vazyme, Piscataway, NJ,
USA) on a ABI 7500 FAST real time PCR system (Applied Biosystems,
Foster City, CA, USA). The procedures for the qPCR were as follows:
95°C for 30 sec, followed by 40 cycles of 95°C for 10 sec and 60°C
for 30 sec. The specificity of the primer amplicons was examined by
the analysis of a melting curve. For miRNA-145-5p, the comparative
Ct method was employed for quantification of target mRNA expression
that was normalized to β-actin expression and relative to the
calibrator. For ACACB, FGFR1, PPP3CA and
SYK, the comparative Ct method was used for quantifying
target mRNA expression normalized to that of 18S rRNA. The forward
primers used in qPCR are as follows:
miR-145-5p-5′-GTCCAGTTTTCCCAGGAATCC-3′;
RNU6-5′-TCGCTTCGGCAGCACATAT-3′, universal reverse primers were
provided in the cDNA synthesis kit; ACACB-F: CTG GAG AAG GGC GTC
ATA TC, ACACB-R: CTG GAT ATG CAC GTG ACT CAG; FGFR1-F: TTA ATA CCA
CCG ACA AAG AG, FGFR1-R: GTA GAC GAT GAC CGA CCC; PPP3CA-F: TGG TCC
CTT CCA TTT GTT, PPP3CA-R: AAG CCT TTC AGC GTC AGC; SYK-F: GTT AGA
GAA AGG AGA GCG GAT G; SYK-R: TTC CTG TGA TTG CTC CTG TG; 18S-F:
GTA ACC CGT TGA ACC CCA TT; 18S-R: CCA TCC AAT CGG TAG TAG CG.
Identification of DEGs
Expression data were normalized by quantile
normalization and the Robust Multichip Average (RMA) algorithm. The
probe level was generated after normalization. The unpaired
Student's t-test was used to identify genes that were
differentially expressed between the miR-145-5p-overexpression
group and the NC group. The fold-change (FC) value of gene
expression between two groups was calculated, and genes with FC
value ≥1.5 and P-value <0.05 were selected as DEGs.
Enrichment analysis of DEGs
The online tool DAVID (10) (version: 6.8, Database for
Annotation, Visualization and Integrated Discovery) was used to
perform the Gene Ontology (GO) functional and pathway enrichment
analyses of DEGs based on GO database (11) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) PATHWAY database (12). The P-value of each term calculated
by Fisher's exact test (13) was
adjusted by the Benjamini-Hochberg method (14). Only function terms with adjusted
P-value <0.05 were considered significant.
Prediction of hsa-miR-145-5p target
genes
Target genes of hsa-miR-145-5p were predicted from
the DEGs identified above based on the miRNA-targeted interaction
information in the miRWalk2.0 (15), miRDB (16), RNA22 (17), miRanda (18), RNAhybrid (19) and TargetScan (20) databases.
Functional analysis of hsa-miR-145-5p
targets
The GO functions in the category of biological
process (BP) and KEGG pathway enrichment analyses of the DEGs
targeting by hsa-miR-145-5p were conducted using DAVID, and only
function terms with adjusted P-value <0.05 were considered
significant. Subsequently, the network consisting of GO and KEGG
terms was visualized using the plugin ClueGO in Cytoscape (version
2.2.6) (21). Additionally, the
network of significant pathways was visualized using the plugin
CyKEGGParser (version 1.2.7) (22).
Construction of PPI network
The PPIs of the DEGs targeting by hsa-miR-145-5p
were extracted from STRING database (version 10.0) (23) with default parameters. The PPIs
meeting the combined score >0.4 were used to construct PPI
network, which was then visualized by Cytoscape (version 3.4.0)
(24). In PPI network, each node
represents a protein and each edge between two nodes represents an
interaction between these two proteins.
Furthermore, network topological features of nodes
in the PPI network, including degree, betweenness and closeness,
were analyzed by the plugin CytoNCA (version 2.1.6) (25). The hub nodes were revealed
according to the scores of degree, betweenness and closeness.
Results
DEGs in hsa-miR-145-5p-overexpressed
Tu-177 cells
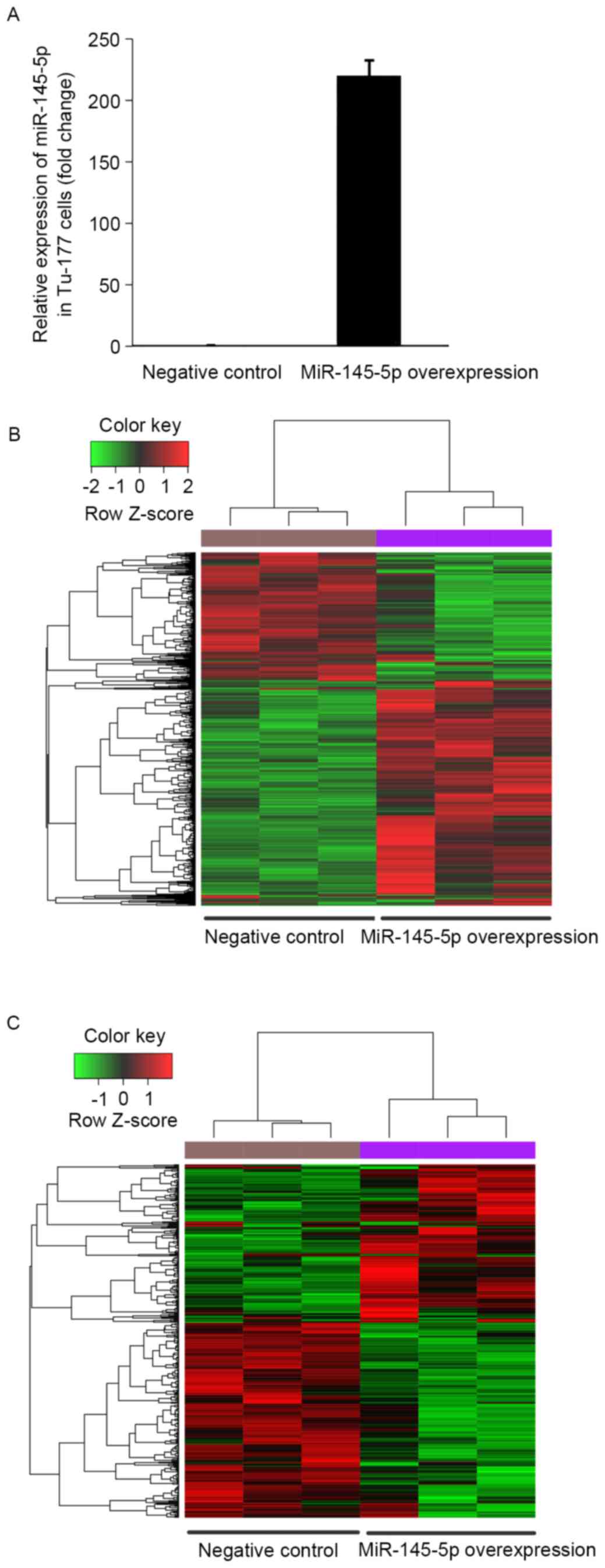
To validate the miR-145-5p expression change between
the miR-145-5p mimic and the NC transfected Tu-177 cells, the
miR-145-5p expression level was determined by qPCR. The results
showed that miR-145-5p was significantly upregulated in miR-145-5p
mimic transfected cells than that in NC cells (Fig. 1A).
Based on the analysis of mRNA microarray data, a
total of 1501 upregulated and 887 downregulated genes were
identified in the hsa-miR-145-5p-overexpressed Tu-177 cells,
compared with the NC cells. The heat map of these DEGs showed that
the identified DEGs were able to distinguish the two group samples
(Fig. 1B).
Potential functions of the DEGs
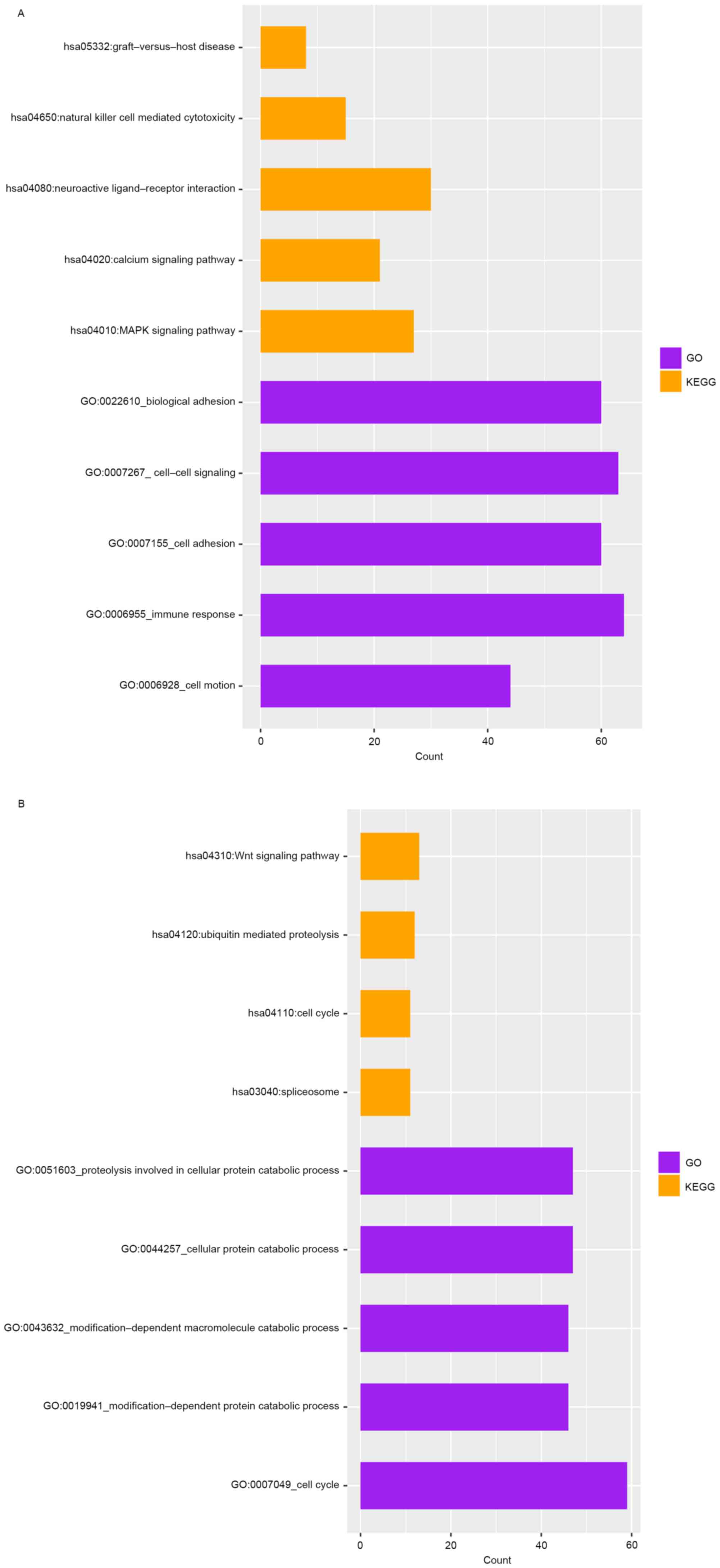
The upregulated DEGs were significantly enriched in
185 GO terms and 7 KEGG pathways, such as immune response, cell
adhesion, and the pathway of neuroactive ligand−receptor
interaction (Fig. 2A). Meanwhile,
the downregulated DEGs were significantly enriched in 97 GO terms
and 4 pathways, such as cell cycle, cellular protein catabolic
process, and Wnt signaling pathway (Fig. 2B).
The predicted target genes of
hsa-miR-145-5p
Among the identified DEGs, 164 upregulated and 221
downregulated genes were predicted to be targeted by
hsa-miR-145-5p. These target genes were able to distinguish the
hsa-miR-145-5p-overexpressedcells from the NC cells (Fig. 1C).
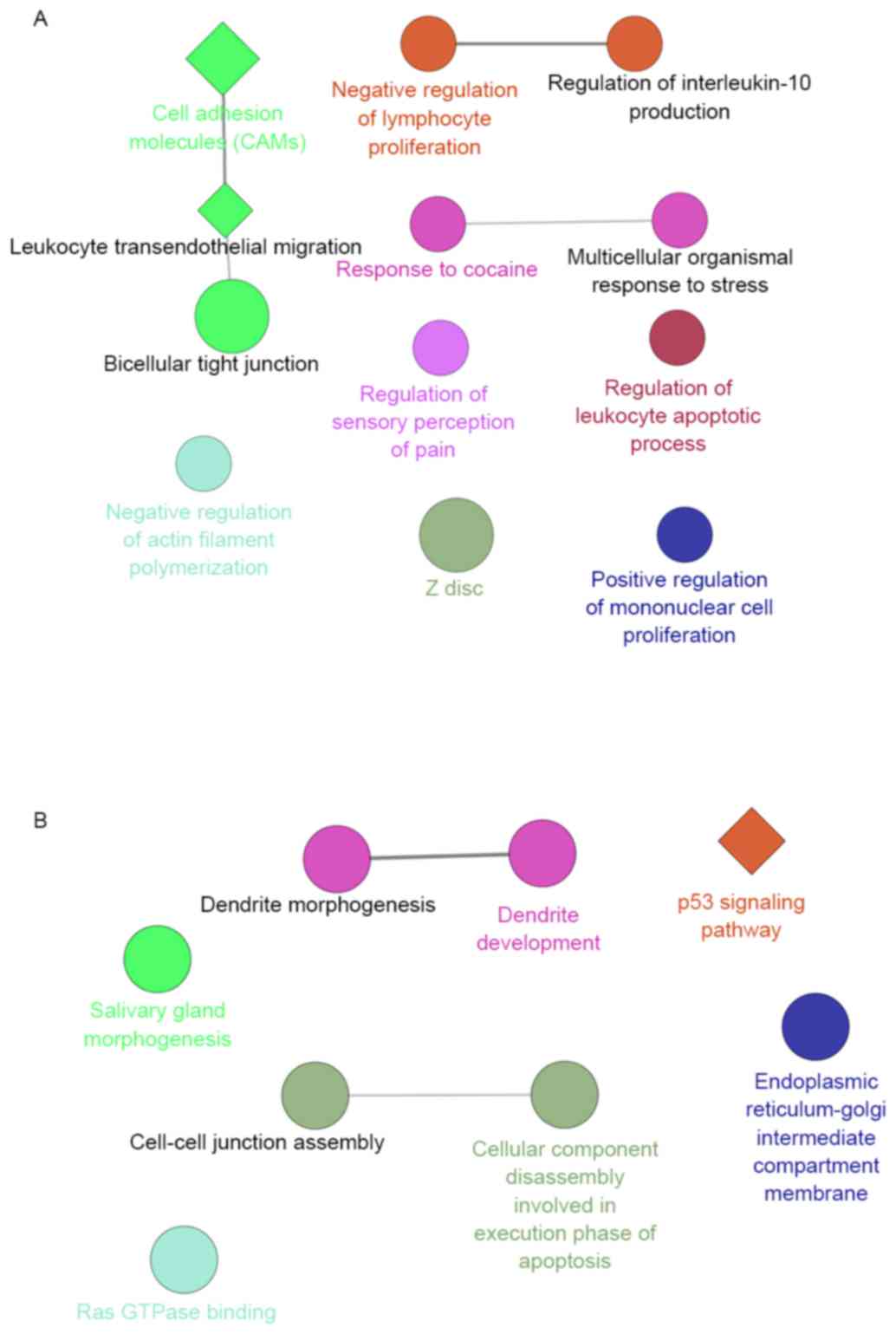
Furthermore, the potential functions of the DEGs
targeting by hsa-miR-145-5p were analyzed to reveal the potential
biological functions of hsa-miR-145-5p in LSCC. The upregulated
target genes were significantly enriched in functions of immunity,
such as the GO terms of negative regulation of lymphocyte
proliferation, which interacted with the GO terms of regulation of
interleukin-10 production. Furthermore, the pathway of cell
adhesion molecules (CAMs) enriched by the upregulated target genes
interacted with the pathway of leukocyte transendothelial
migration, which was predicted to have a correlation with
bicellular tight junction (Fig.
3A). Meanwhile, the downregulated target genes were
significantly enriched in the p53 signaling pathway, and the GO
term of cell-cell junction assembly which interacted with the GO
term of cellular component disassembly involved in execution phase
of apoptosis (Fig. 3B).
The PPI analysis of hsa-miR-145-5p
target genes
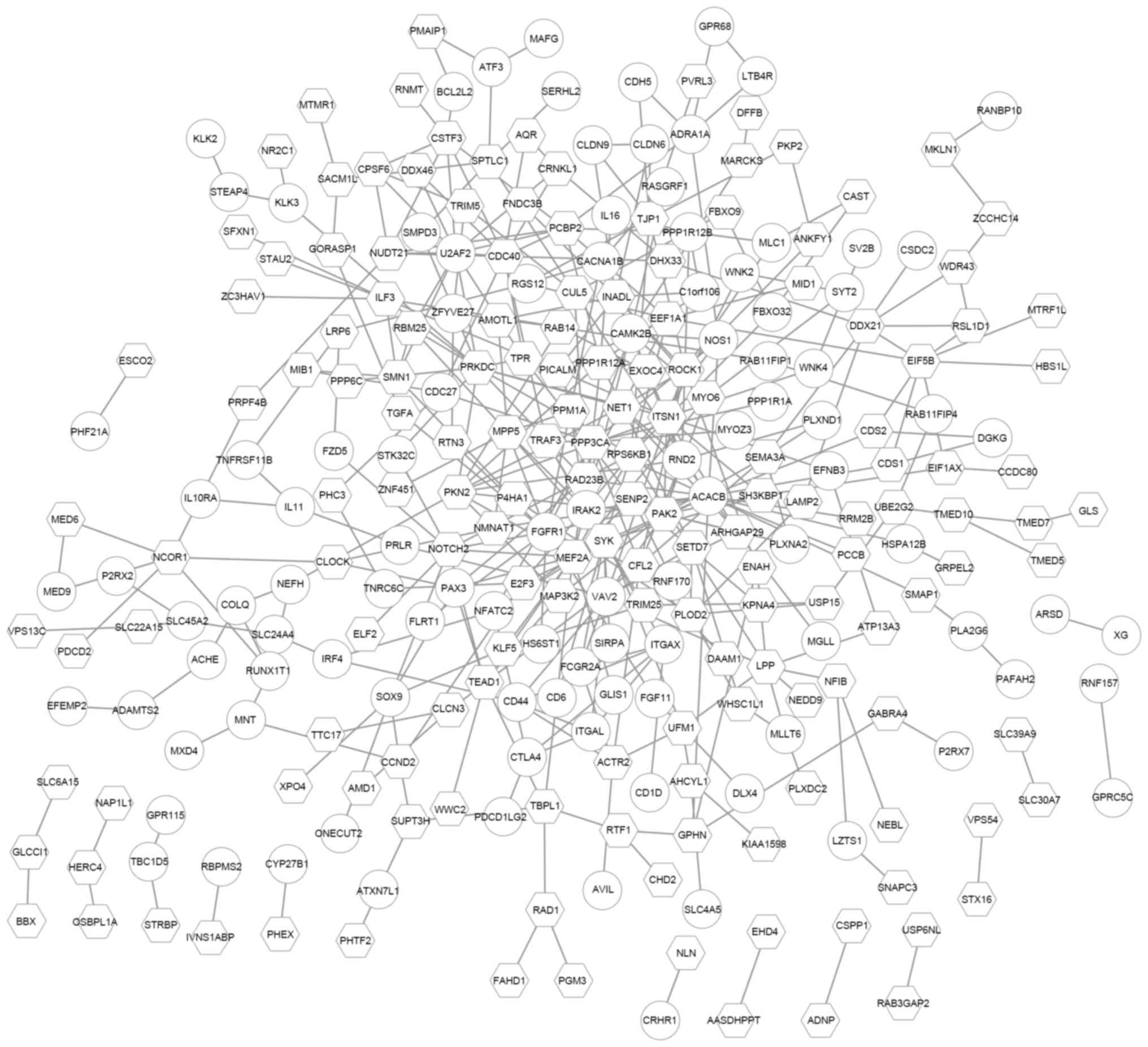
To further investigate the potential PPIs of the
DEGs targeted by hsa-miR-145-5p, a PPI network was constructed.
This network consisted of 267 nodes and 416 PPI pairs. The
upregulated ACACB had the highest degree and interacted with
genes like the downregulated PPP3CA and SYK, as well
as the upregulated FGFR1. PPP3CA interacted with 15
genes, such as MEF2A and FGFR1 (Fig. 4).
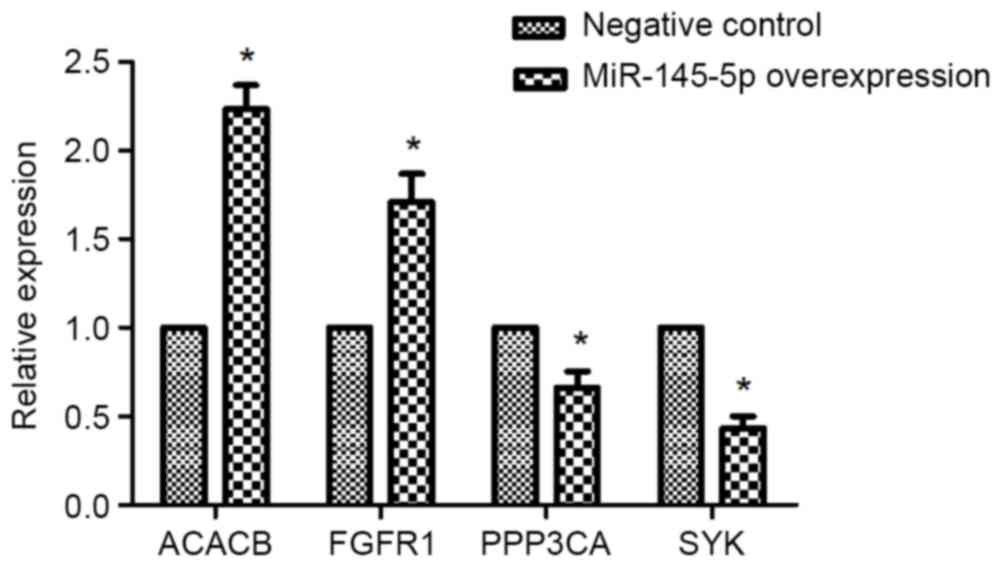
The expression of ACACB, FGFR1,
PPP3CA, and SYK in hsa-miR-145-5p-overexpressed Tu-177 cells
RT-PCR results revealed that the mRNA expressions of
ACACB and FGFR1 were obviously enhanced in
miR-145-5p-overexpressed Tu-177 cells, while overexpressing
miR-145-5p significantly reduced mRNA expression of PPP3CA
and SYK (Fig. 5).
Discussion
In the present study, a total of 1,501 upregulated
and 887 downregulated genes were identified in the
hsa-miR-145-5p-overexpressed laryngeal squamous carcinoma Tu-177
cells, compared with the NC cells. Among these DEGs, 164
upregulated and 221 downregulated genes were predicted to be
targeted by hsa-miR-145-5p. The upregulated target genes were
mainly related to functions of immunity, whereas the downregulated
target genes were significantly enriched in the p53 signaling
pathway. In the PPI network, 267 target genes were included, and
the upregulated ACACB had the highest degree. Besides, the
mRNA expressions of ACACB and FGFR1 were obviously
enhanced in miR-145-5p-overexpressed Tu-177 cells, while
overexpressing miR-145-5p significantly reduced mRNA expression of
PPP3CA and SYK.
ACACB encodes acetyl-CoA carboxylase (ACC)
beta. ACC is a biotin-containing enzyme which catalyzes the
carboxylation of acetyl-CoA to malonyl-CoA in fatty acid synthesis,
and regulates fatty acid oxidation (26). In human colorectal cancer cells,
fatty acid synthase suppression mediates the anticancer substance
oridonin-induced apoptosis (27).
Mak et al also found that ACC involved in lipid metabolism
may be a potential anticancer target protein (28). Although there is no any evidence to
prove the association of ACACB with LSCC or miR-145-5p, the
results above indicate that miR-145-5p overexpression may induce
ACACB expression in LSCC cells, which results in abnormity
of fatty acid metabolism in LSCC.
In the PPI network, a set of DEGs were predicted to
interact with ACACB, such as the downregulated PPP3CA
and SYK, as well as the upregulated FGFR1.
PPP3CA encodes protein phosphatase 3 catalytic subunit
alpha, also named calcineurin A alpha (29). Ostenfeld et al have reported
that miR-145 induces cell death in human urothelial cancer cells by
targeting PPP3CA, clathrin interactor 1 and core-binding
factor β subunit (30).
Furthermore, PPP3CA has been also reported to be involved in
the invasiveness of other cancers, such as breast cancer (31), pancreatic cancer (32), and small cell lung cancer (33). In the present study, PPP3CA
was predicted to be correlated with the functions of cell cycle,
which is commonly deregulated in cancers, including in LSCC.
Collectively, miR-145-5p overexpression might induce the
downregulation of PPP3CA, which likely affected the cell
cycle and invasiveness of LSCC cells. SYK encodes spleen
associated tyrosine kinase, which can couple activated
immunoreceptors to downstream signaling pathways that regulate
multiple cell progresses, such as cell proliferation,
differentiation and phagocytosis (34). Currently, SYK has not been
confirmed to be associated with LSCC or miR-145-5p. We speculated
that miR-145-5p overexpression resulted in downregulation of
SYK, thus suppressing the cell proliferation of LSCC cells.
FGFR1 (fibroblast growth factor receptor 1) has been
previously found to be frequently amplified in head and neck
squamous cell carcinoma (HNSCC), and it has been identified as a
candidate prognostic biomarker of HNSCC (35,36).
Furthermore, fibroblast growth factor receptor family member
proteins (FGFR1-4) were recently reported to be also
frequently highly expressed in oral cavity and oropharyngeal
squamous cell carcinoma (37). In
the PPI network, FGFR1 interacted with PPP3CA, and
the mRNA expressions of FGFR1 and PPP3CA were
obviously enhanced and reduced by overexpressing of miR-145-5p,
respectively. Therefore, miR-145-5p overexpression may also change
the expression of FGFR1 in LSCC cells via downregulating
PPP3CA.
Additionally, in this study, we also found that the
upregulated target genes were mainly enriched in functions of
immunity, such as regulation of lymphocyte proliferation and
interleukin-10 production. The downregulated was significantly
related to the p53 signaling pathway. In LSCC, lymphocytes are
proliferated and lymphokines are overexpressed (38,39).
Disruption of p53 function has been independently shown to occur in
the majority of HNSCC (40).
Recently, p53 has been discovered to mediate the amplification of
the transcription of pro-apoptotic miRNAs in LSCC (41). Therefore, miR-145-5p overexpression
may also changes immune responses and p53 signaling pathway to
exert its anticancer efficiency in LSCC.
Despite the aforementioned results, this study still
had several limitations. For example, the expression levels of the
DEGs are needed to be confirmed, and the regulatory correlations
between miR-145-5p and its predicted targets should also be
validated by experiments. In our further study, we will confirm the
predicted results in animal models and clinical samples.
In conclusion, based on microarray technology, we
investigated expression changes of downstream genes of the
miR-145-5p in human LSCC. A total of 1501 upregulated and 887
downregulated genes were identified in the
hsa-miR-145-5p-overexpressed Tu-177 cells, compared with the NC
cells. A series of genes were predicted to be targeted by
hsa-miR-145-5p, and they were predicted to be related to immunity
or p53 signaling pathway. Furthermore, some target genes were
predicted to have interactions in the PPI network, such as
ACACB that was associated with fatty acid metabolism,
PPP3CA that was related to cell cycle and invasiveness,
SYK that was correlated with cell proliferation. Therefore,
hsa-miR-145-5p may function in LSCC via regulating multiple cell
processes, such as cell proliferation and invasiveness, fatty acid
metabolism, immunity and p53 signaling pathway. These findings
provide novel information for the investigation of miR-145-5p
functions and mechanisms in LSCC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81602394, 81572670,
81402256), Scientific and Technological Innovation Programs of
Higher Education Institutions in Shanxi (STIP, 2016-92), the
Research Project of Shanxi Province Health and Family Planning
Commission (201301073, 2014028, 201601038, 201601037), the
Excellent Talent Science and Technology Innovation Project of
Shanxi Province (201605D211029), and China Postdoctoral Science
Foundation (2016M591412), and the Shanxi Scholarship Council of
China (2016–118).
Glossary
Abbreviations
Abbreviations:
|
LSCC
|
laryngeal squamous cell carcinoma
|
|
NC
|
negative control
|
|
DEGs
|
differentially expressed genes
|
|
PPIs
|
protein-protein interactions
|
|
GEO
|
Gene Expression Omnibus
|
|
RMA
|
Robust Multichip Average
|
|
GO
|
Gene Ontology
|
|
FC
|
fold-change
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
CAMs
|
cell adhesion molecules
|
|
ACC
|
acetyl-CoA carboxylase
|
|
FGFR1
|
fibroblast growth factor receptor
1
|
|
BP
|
biological process
|
References
|
1
|
Zhang SY, Lu ZM, Luo XN, Chen LS, Ge PJ,
Song XH, Chen SH and Wu YL: Retrospective analysis of prognostic
factors in 205 patients with laryngeal squamous cell carcinoma who
underwent surgical treatment. PloS One. 8:e601572013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Slattery ML, Herrick JS, Mullany LE,
Valeri N, Stevens J, Caan BJ, Samowitz W and Wolff RK: An
evaluation and replication of miRNAs with disease stage and
colorectal cancer-specific mortality. Int J Cancer. 137:428–438.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Letelier P, García P, Leal P, Álvarez H,
Ili C, López J, Castillo J, Brebi P and Roa JC: miR-1 and miR-145
act as tumor suppressor microRNAs in gallbladder cancer. Int J Clin
Exp Pathol. 7:1849–1867. 2014.PubMed/NCBI
|
|
5
|
Andersen M, Grauslund M, Ravn J, Sørensen
JB, Andersen CB and Santoni-Rugiu E: Diagnostic potential of
miR-126, miR-143, miR-145, and miR-652 in malignant pleural
mesothelioma. J Mol Diagn. 16:418–430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ozen M, Karatas OF, Gulluoglu S, Bayrak
OF, Sevli S, Guzel E, Ekici ID, Caskurlu T, Solak M, Creighton CJ
and Ittmann M: Overexpression of miR-145-5p inhibits proliferation
of prostate cancer cells and reduces SOX2 expression. Cancer
Invest. 33:251–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karatas OF, Yuceturk B, Suer I, Yilmaz M,
Cansiz H, Solak M, Ittmann M and Ozen M: Role of miR-145 in human
laryngeal squamous cell carcinoma. Head Neck. 38:260–266. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karatas OF, Suer I, Yuceturk B, Yilmaz M,
Hajiyev Y, Creighton CJ, Ittmann M and Ozen M: The role of miR-145
in stem cell characteristics of human laryngeal squamous cell
carcinoma Hep-2 cells. Tumor Biol. 37:4183–4192. 2016. View Article : Google Scholar
|
|
9
|
WeiGao CZ, Chen G, Li Y, Wen S, Huang F
and Wang B: Novel mechanism and clinical significance of
Hsa-miR-145-5P/FSCN1 regulatory axis in laryngeal squamous cell
carcinoma. Tumor academic conference across the Taiwan Straits.
2014;
|
|
10
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
11
|
Gene Ontology Consortium: Gene ontology
consortium: Going forward. Nucleic Acids Res. 43(Database issue):
D1049–D1056. 2015.PubMed/NCBI
|
|
12
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Preacher KJ and Briggs NE: Calculation for
Fisher's Exact Test. 2015.
|
|
14
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Stat Soci Series B (Methodological).
57:289–300. 1995.
|
|
15
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43(Database issue): D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of microRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36(Database issue): D149–D153. 2008.PubMed/NCBI
|
|
19
|
Kruger J and Rehmsmeier M: RNAhybrid:
microRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34(Web Server issue): W451–W454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nersisyan L, Samsonyan R and Arakelyan A:
CyKEGGParser: Tailoring KEGG pathways to fit into systems biology
analysis workflows. F1000Res. 3:1452014.PubMed/NCBI
|
|
23
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(Database issue): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saddik M, Gamble J, Witters LA and
Lopaschuk GD: Acetyl-CoA carboxylase regulation of fatty acid
oxidation in the heart. J Biol Chem. 268:25836–25845.
1993.PubMed/NCBI
|
|
27
|
Kwan HY, Yang Z, Fong WF, Hu YM, Yu ZL and
Hsiao WL: The anticancer effect of oridonin is mediated by fatty
acid synthase suppression in human colorectal cancer cells. Journal
of Gastroenterology. 48:182–192. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mak L, Liggi S, Tan L, Kusonmano K,
Rollinger JM, Koutsoukas A, Glen RC and Kirchmair J: Anti-cancer
drug development: Computational strategies to identify and target
proteins involved in cancer metabolism. Curr Pharm Des. 19:532–577.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang MG, Yi H, Guerini D, Klee CB and
Mcbride OW: Calcineurin A alpha (PPP3CA), calcineurin A beta
(PPP3CB) and calcineurin B (PPP3R1) are located on human
chromosomes 4, 10q21→q22 and 2p16→p15 respectively. Cytogenet Cell
Genet. 72:236–241. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ostenfeld MS, Bramsen JB, Lamy P,
Villadsen SB, Fristrup N, Sørensen KD, Ulhøi B, Borre M, Kjems J,
Dyrskjøt L and Orntoft TF: miR-145 induces caspase-dependent and
-independent cell death in urothelial cancer cell lines with
targeting of an expression signature present in Ta bladder tumors.
Oncogene. 29:1073–1084. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gabrovska PN, Smith RA, Haupt LM and
Griffiths LR: Investigation of two Wnt signalling pathway single
nucleotide polymorphisms in a breast cancer-affected Australian
population. Twin Res Hum Genet. 14:562–567. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Buchholz M, Schatz A, Wagner M, Michl P,
Linhart T, Adler G, Gress TM and Ellenrieder V: Overexpression of
c-myc in pancreatic cancer caused by ectopic activation of NFATc1
and the Ca2+/calcineurin signaling pathway. Embo J. 25:3714–3724.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Y, Zhang Y, Min J, Liu LL, Ma NQ, Feng
YM, Liu D, Wang PZ, Huang DD, Zhuang Y and Zhang HL: Calcineurin
promotes proliferation, migration, and invasion of small cell lung
cancer. Tumor Biol. 31:199–207. 2010. View Article : Google Scholar
|
|
34
|
Buchner M, Fuchs S, Prinz G, Pfeifer D,
Bartholomé K, Burger M, Chevalier N, Vallat L, Timmer J, Gribben
JG, et al: Spleen tyrosine kinase is overexpressed and represents a
potential therapeutic target in chronic lymphocytic leukemia.
Cancer Res. 69:5424–5432. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Göke F, Bode M, Franzen A, Kirsten R,
Goltz D, Göke A, Sharma R, Boehm D, Vogel W, Wagner P, et al:
Fibroblast growth factor receptor 1 amplification is a common event
in squamous cell carcinoma of the head and neck. Mod Pathol.
26:1298–1306. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Koole K, Brunen D, van Kempen PM, Noorlag
R, de Bree R, Lieftink C, van Es RJ, Bernards R and Willems SM:
FGFR1 Is a potential prognostic biomarker and therapeutic target in
head and neck squamous cell carcinoma. Clin Cancer Res.
22:3884–3893. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Koole K, Clausen MJ, van Es RJ, van Kempen
PM, Melchers LJ, Koole R, Langendijk JA, van Diest PJ, Roodenburg
JL, Schuuring E and Willems SM: FGFR family members protein
expression as prognostic markers in oral cavity and oropharyngeal
squamous cell carcinoma. Mol Diagn Ther. 20:363–374. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bergmann C, Strauss L, Zeidler R, Lang S
and Whiteside TL: Expansion of human T regulatory type 1 cells in
the microenvironment of cyclooxygenase 2 overexpressing head and
neck squamous cell carcinoma. Cancer Res. 67:8865–8873. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jebreel A, Mistry D, Loke D, Dunn G, Hough
V, Oliver K, Stafford N and Greenman J: Investigation of
interleukin 10, 12 and 18 levels in patients with head and neck
cancer. J Laryngol Otol. 121:246–252. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Allen CT, Ricker JL, Chen Z and Van Waes
C: Role of activated nuclear factor-kappaB in the pathogenesis and
therapy of squamous cell carcinoma of the head and neck. Head Neck.
29:959–971. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kan X, Sun Y, Lu J, Li M, Wang Y, Li Q,
Liu Y, Liu M and Tian L: Co-inhibition of miRNA-21 and miRNA-221
induces apoptosis by enhancing the p53-mediated expression of
pro-apoptotic miRNAs in laryngeal squamous cell carcinoma. Mol Med
Rep. 13:4315–4320. 2016. View Article : Google Scholar : PubMed/NCBI
|