Introduction
Esophageal cancer is one of the most common cancers
in the world, whose cases are most prevalent in developing
countries. The North-Central part of China is often referred to as
part of the ‘esophageal cancer belt’, which has the highest
incidence of esophageal cancer in the world. And over 90% of the
newly diagnosed cases were esophageal squamous cell carcinoma
(ESCC) (1). Although recent
development of screening technology contributed to the early
detection of ESCC, the decrease of ESCC morbidity remains slight
(2,3). Surgical resection is considered to be
the main treatment of ESCC, followed with adjuvant chemotherapy and
radiotherapy (4). Although
targeted therapy of ESCC has been widely studied (5,6),
clinical trials were mostly concentrated on relapse or metastasis
cases until now, and only a few agents had entered phase III
clinical trial stage. As the result, much more researches will be
needed to reveal the mechanisms involved in ESCC tumorigenesis, in
order to devise new schemes for therapy.
The Golgi phosphoprotein 3 (GOLPH3), also named
GPP34/GMx33/MIDAS was discovered as a Golgi-associated protein
(7) which was conserved from yeast
to human (8). GOLPH3 binds to
Golgi-localized glycosyltransferase, and contributes to their
steady-state Golgi membrane localization (9,10).
Furthermore, the main functions of GOLPH3 are also achieved via
binding to phosphatidylinositol-4-phosphate [PtdIns(4)P] on the trans-Golgi membranes, which
will simultaneously connect to myosin XVIIIA (MYO18A) to provide
Golgi a tensile force to form tubule and vesicle (11,12).
In sum, GOLPH3 is involved in vesicle budding and retromer cargo
transport (13–18); and all these procedure also
contributes to DNA-damage repairing (19).
GOLPH3 was first considered as a potent oncogene by
Scott et al (20). Further
studies regarding the role of GOLPH3 in several cancers, such as
rhabdomyosarcoma, tongue cancer, breast cancer,
glioblastomamultiforme, gastric cancer and ovarian cancer (21–27),
all illustrated that high expression of GOLPH3 indicated patients'
poor prognosis. Our previous work also showed that GOLPH3 was
overexpressed in ESCC cancer tissues, and correlated with ESCC
patients' poor prognosis (28). In
order to gain deeper insight into the role of GOLPH3 play in ESCC
tumorigenesis, we conducted a series of gain- and loss-of-function
analyses and preliminarily demonstrated the activation of
mechanistic target of rapamycin (mTOR) and Wnt/β-catenin signaling
pathway by GOLPH3 overexpression.
Materials and methods
This research was approved by the
Ethics Committee of Guangdong Science and Technology
Department
All animal work had been conducted according to
relevant national and international guidelines. ESCC cell
suspension was injected subcutaneously into the left flank of
4-week old female BALB/c-nu mice. Mice were sacrificed 30 days
after injection by cervical dislocation method.
Cell lines and culture
ESCC cell line KYSE-140, constructed KYSE-140 cell
lines and the human kidney cell line HEK-293T were cultured under
37°C with 5% CO2 and 95% humidity, in Dulbecco's
modified Eagle's medium (DMEM), supplemented with 10% fetal bovine
serum (FBS) (both from Life Technologies, Carlsbad, CA, USA).
A GOLPH3 stable knockdown KYSE-140 cell line,
labeled KYSE-140-GOLPH3-RNAi, was constructed by infection of
retrovirus expressing shRNA targeted GOLPH3. The negative control
cell line was constructed with scramble sequence and labeled
KYSE-140-scramble. The viruses expressing shRNAs were packaged with
pSuper.retro.puro backbone plasmids.
A GOLPH3 stable overexpressed KYSE-140 cell line was
constructed with retrovirus expressing GOLPH3 cDNA sequence, and an
empty vector packaged retrovirus was used to construct negative
control, which were labeled KYSE-140-GOLPH3 and KYSE-140-vector,
respectively. The viruses expressing cDNA and control were packaged
with pBaBe.retro.puro backbone plasmids.
KYSE-140-GOLPH3-RNAi, KYSE-140-scramble,
KYSE-140-GOLPH3 and KYSE-140-vector cell lines were selected with
puromycin (EMD Millipore, Billerica, MA, USA) and maintained in 10%
FBS DMEM with 0.5 µg/ml puromycin.
Quantitative real-time PCR (qPCR)
Cultured cells were digested with 0.25% trypsin with
EDTA (Life Technologies) and washed with PBS twice. Total RNA of
cells was extracted with TRIzol reagent (Life Technologies). Total
RNA (2 µg) was used as the template of reverse transcription in 2.0
µl system, M-MLV reverse transcriptase (Promega, Madison, WI, USA)
was applied following the manufacturers' instruction. qPCR was
carried out by GoTaq qPCR Master Mix (Promega).
Western blot analysis
Total protein from cells or tissues was extracted
with RIPA lysis buffer. Protein samples were treated with protein
loading buffer, with reducing agent maintained at 100°C for 5 min
after protein quantification with bicinchonininc acid (BCA).
Protein samples were resolved by sodium dodecyl
sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to polyvinylidene fluoride (PVDF) membrane.
Subsequently, Protein samples were incubated with primary antibody
for over 12 h at 4°C, and HRP-conjugated second antibody for 1 h at
room temperature. Pierce ECL Western Blotting Substrate (Thermo
Fisher Scientific, Waltham, MA, USA) was exposed to Kodak X-OMAT BT
XBT-1 Film (Kodak, Rochester, NY, USA).
Cell proliferation assay
Growth curve was drawn based on the cell viability
tested by MTT assay. Cells were digested and washed, and suspended
to 0.5×104/ml with 10% FBS DMEM. 200 µl per well cell
suspension was seeded to 96-well plate. Cell viability of each
group of cells was analyzed by MTT assay, and the OD values of the
first day were used as standards; cell viabilities were measured at
the following 4 days.
Cell cycle assay
Cells were synchronized by 24 h serum starvation and
then cultured with 10% FBS DMEM for 20 h. Cells were digested with
trypsin without EDTA (Life Technologies) and washed twice, and then
were fixed with 75% ethanol at 20°C for 1 h, suspended with PBS.
Prior to flow cytometry analysis, cells were treated with RNase A
at 37°C for 30 min and stained with propidium iodide (PI) on ice
for 30 min.
Plate colony formation assay
Cultured cells were digested and washed with PBS
once. After suspension with 10% FBS DMEM to 0.5×103
cells/ml, 2 ml cell suspension per well was added to 6-well plate
and cultured for 10 days. Cells were fixed with 75% ethanol at room
temperature for 1 hr and stained with crystal violet. Cell colonies
larger than 50 cells were counted.
Dual-luciferase reporter assay
The Dual-Luciferase® Reporter Assay
system (Promega) was used to perform the TOP-Flash and FOP-Flash
assays. The TOP-Luciferase vector and FOP-Luciferase vector were
separately transfected cultured cells which were co-transfected
with a pRL-TK control vector. 48 hr after transfection, the
activity of firefly luciferase and Renilla luciferase were
measured following manufacturers' instructions. The ratio of
firefly and Renilla luciferase activity was recorded as
relative luciferase activity; the relative luciferase activity of
the TOP-Luciferase vector transfected cell was standardized with
data FOP-luciferase vector transfected cells.
Tumorigenesis
Cultured cells were digested and washed, cells were
suspended in 2.5×106/ml with PBS, and 200 µl of cell
suspension were injected subcutaneously into the left flank of
4-week old female BALB/c-nu mice. Tumor volumes were measured every
3 to 4 days and calculated via following: Volume = (length ×
width2)/2. Mice were sacrificed after 30 days, tumor
tissues were excised and photographed. Tumor tissues were preserved
in liquid nitrogen.
Statistic analysis
All experiments were triplicated, and data shown as
mean ± SE. Statistical analyses were performed with PASW Statistics
18 (IBM, Armonk, NY, USA). P-values in graph were depicted as
follows: P<0.05 (significant), P<0.01 (very significant), and
P<0.001 (highly significant).
Results
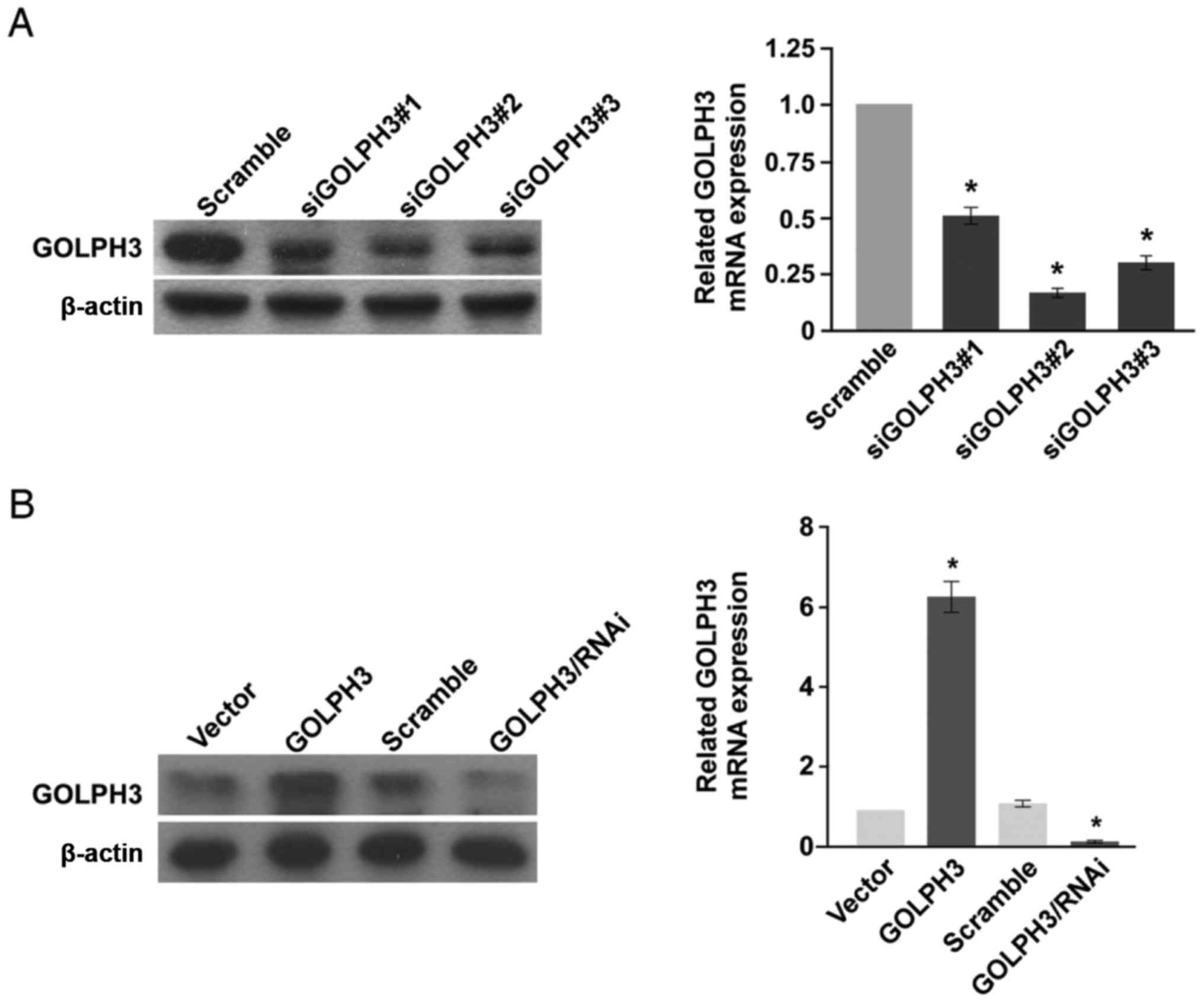
GOLPH3 stable overexpression and
knockdown KYSE-140 cell lines were constructed
GOLPH3 stable overexpression and knockdown ESCC cell
lines were first constructed in order to investigate the function
of GOLPH3 in ESCC (Fig. 1).
KYSE-140 ESCC cell lines were chosen, and stable cell lines were
constructed via retrovirus.
GOLPH3 CDs were cloned and inserted into retrovirus
backbone vector and subsequently packed into retrovirus particles.
KYSE-140 cells were transfected and selected by puromycin. GOLPH3
protein expression of KYSE-140-GOLPH3 was substantively higher than
the negative control KYSE-140-vector cell line (Fig. 1B, left). GOLPH3 mRNA level
was also 5 times higher in KYSE-140-GOLPH3 than in KYSE-140-vector
(Fig. 1B, right).
GOLPH3 knockdown was performed via RNA interference
(RNAi). Three different GOLPH3-targeted siRNAs were transfected to
KYSE-140 cells, following which the protein levels and mRNA levels
of GOLPH3 in transfected cells were analyzed (Fig. 1A). SiGOLPH3#2 demonstrated the best
knockdown efficacy at both the protein and mRNA levels, and the
GOLPH3 mRNA expression level was suppressed by 83.33% (Fig. 1A, right). The sequence of
siGOLPH3#2 was then chosen to construct the retrovirus backbone
vector expressing shRNA. The retrovirus particles were packed and
infected KYSE-140 to construct the GOLPH3 stable knockdown cell
KYSE-140-GOLPH3/RNAi. A negative control cell line was constructed
by scramble sequence labeled KYSE-140-scramble. GOLPH3 protein and
mRNA levels were significantly down regulated in
KYSE-140-GOLPH3/RNAi compared with KYSE-140-scramble, while no
difference was observed between KYSE-140-vector and
KYSE-140-scramble (Fig. 1B).
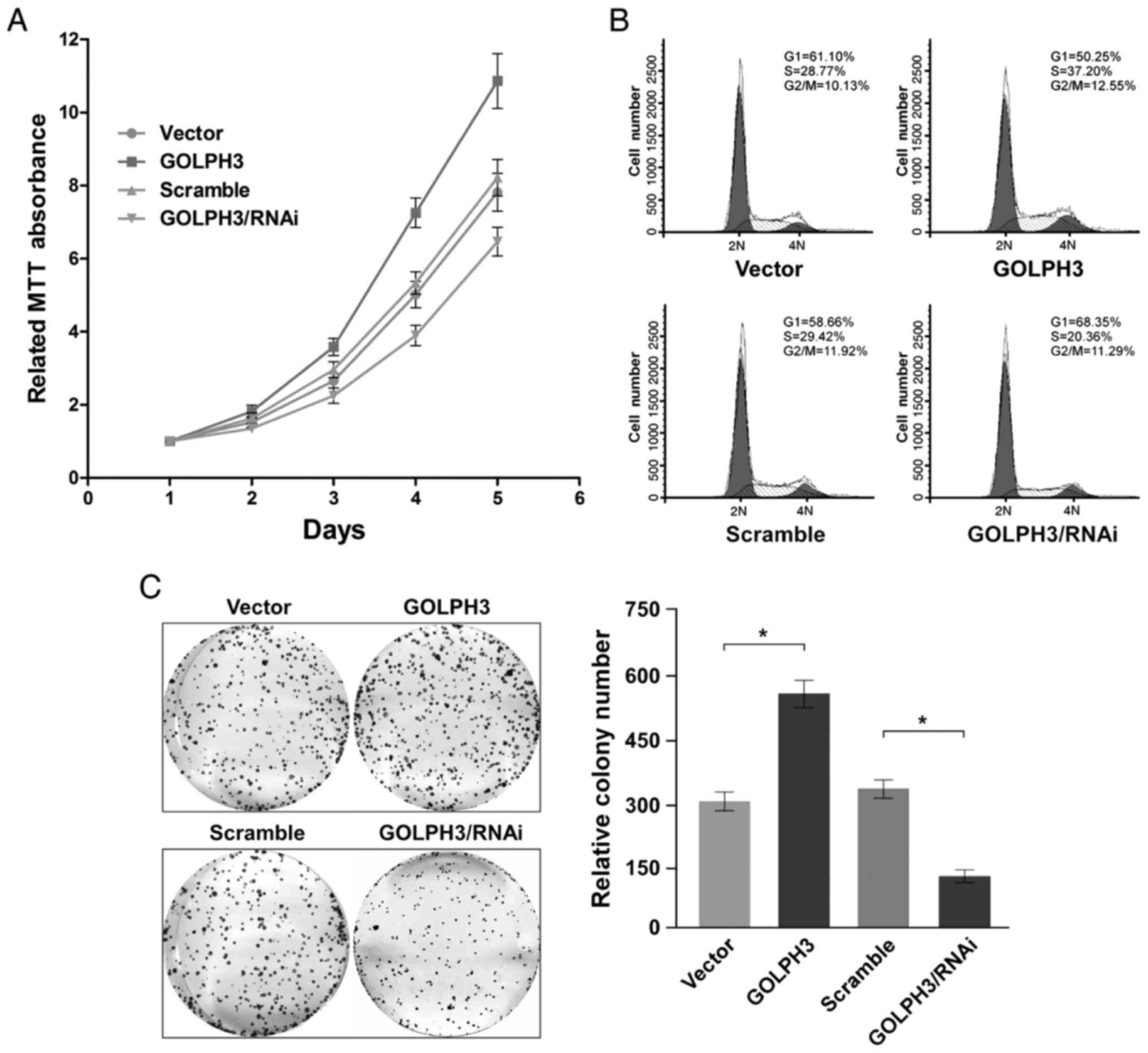
GOLPH3 promoted ESCC cell growth and
proliferation in vitro
The GOLPH3 stable overexpression cell line
KYSE-140-GOLPH3, the negative control KYSE-140-vector; the GOLPH3
stable knockdown cell line KYSE-140-GOLPH3/RNAi, and the negative
control KYSE-140-scramble were all analyzed for cell viabilities
for five days. Growth curves revealed that the proliferation of
GOLPH3 overexpressed cell line KYSE-140-GOLPH3 was accelerated
compared with KYSE-140-vector, cell viability was elevated
significantly from the 3rd day (day 2 P=0.15, day 3 P=0.0002, day 4
P<0.0001, day 5 P<0.0001). Simultaneously, proliferation of
GOLPH3 downregulated cell line KYSE-140-GOLPH3/RNAi was visibly
hindered compared to KYSE-140-scramble, cell viability was
decreased significantly from the 2nd day (day 2 P=0.017, day 3
P=0.0001, day 4 P=0.0010, day 5 P=0.0005). At the same time, cell
curves indicated near-equal proliferation ability between
KYSE-140-scramble and KYSE-140-vector (Fig. 2A).
The cell cycle of each cell line was further
examined to determine the reason for proliferation influence by
GOLPH3. The cell cycle of KYSE-140 with GOLPH3 overexpression was
accelerated compared to the negative control, with the S Phase cell
proportion increased from 28.77 to 37.20%, and the G1 Phase cell
proportion decreased from 61.10 to 50.25%, simultaneously.
Reversely, GOLPH3 knockdown resulted in cell cycle arrest, with S
phase cell decreased from 29.42 to 20.36% and G1 Phase increased
from 58.66 to 68.35% (Fig.
2B).
GOLPH3 could promote cell growth in addition to cell
proliferation; plate colony formation assay demonstrated the colony
of KYSE-140-GOLPH3 to be nearly twice the number of KYSE-140-vector
(P=0.0003). Meanwhile, the colony formed of KYSE-140-GOLPH3/RNAi
was only half the number of KYSE-140-scramble (P=0.0003) (Fig. 2C).
In general, GOLPH3 overexpression promoted ESCC cell
proliferation and growth, while GOLPH3 knockdown inhibited cell
proliferation and growth in vitro.
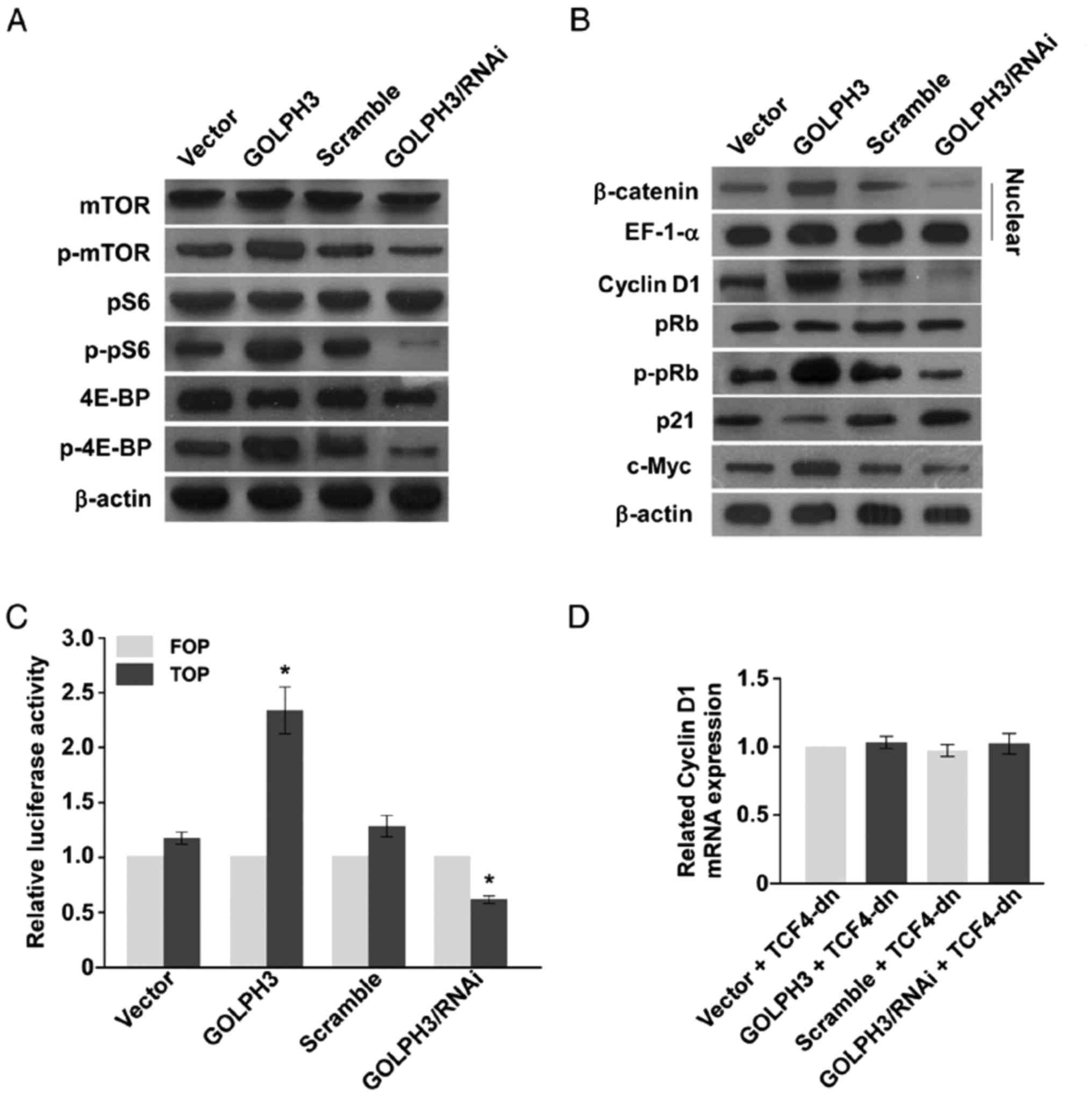
GOLPH3 activated mTOR signaling
pathway
Compared with the KYSE-140-vector cells, the
phosphorylation of the key proteins of mTOR signaling pathway, mTOR
pS6 and 4E-BP, were enhanced in the KYSE-140-GOLPH3 cells. At the
same time, the phosphorylation of the key proteins of mTOR
signaling pathway were suppressed in the KYSE-140-GOLPH3/RNAi cell
compared with KYSE-140-scramble cell (Fig. 3A). As the results determine, GOLPH3
plays a tumorigenesis role in ESCC cells through mTOR signaling
activation.
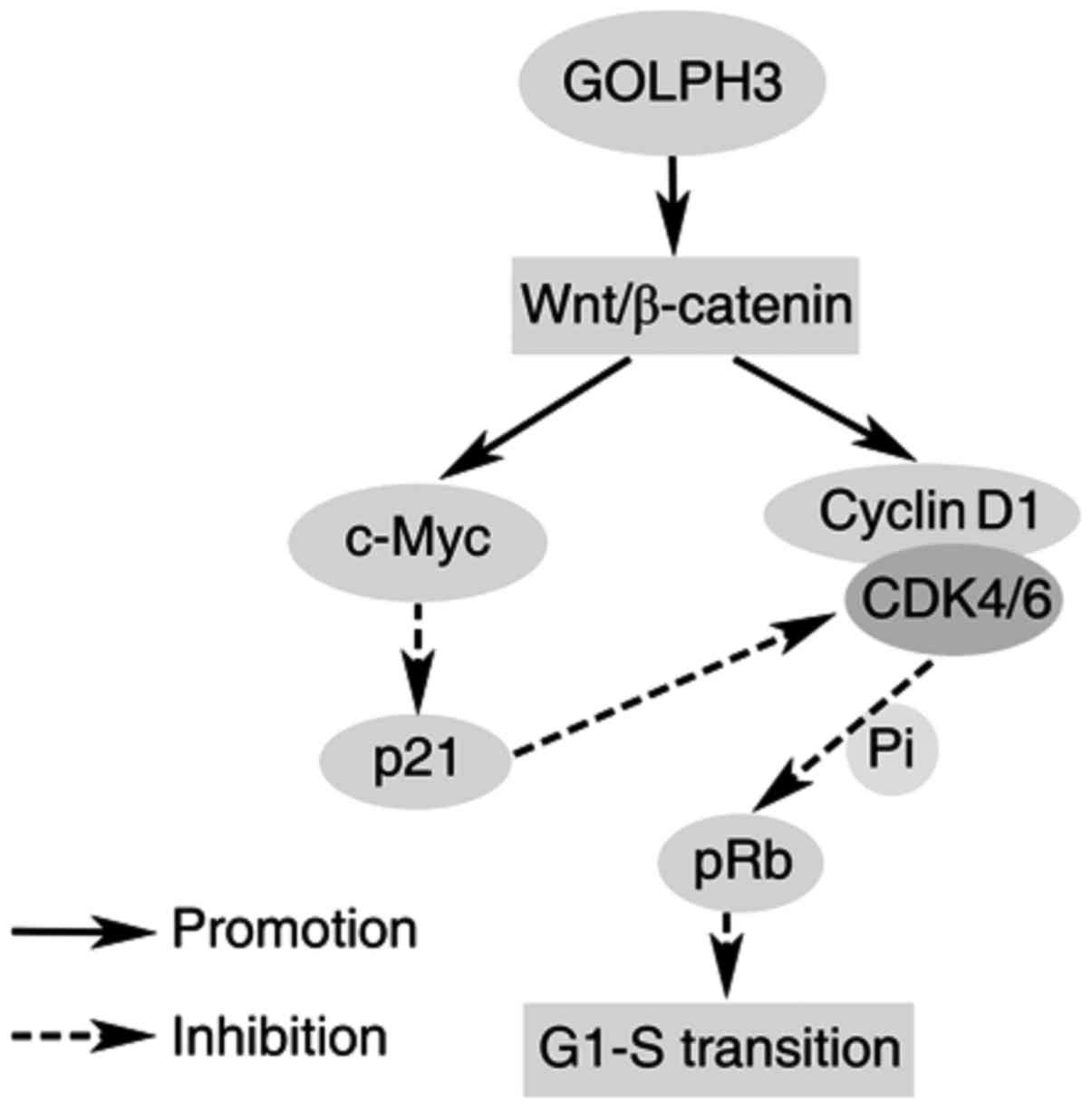
GOLPH3 accelerated cell cycle through
Wnt/β-catenin signaling pathway activation
In order to investigate the mechanisms GOLPH3 played
in cell cycle acceleration when GOLPH3 is overexpressed in the
KYSE-140 cell, the proteins involved in G1-S Phase transition was
analyzed. Cyclin D1 and c-Myc were overexpressed when GOLPH3 was
upregulated, while p21 expression was down regulated. At the same
time, pRb phosphorylation was increased. In KYSE-140 cell with
GOLPH3 knockdown, the trends reversed (Fig. 3B).
The Wnt/β-catenin signaling pathway is the upstream
of all above proteins. Western blot analysis revealed that more
β-catenin progressed to the nucleus of KYSE-140-GOLPH3 compared to
KYSE-140-vector cell, but less β-catenin localized to the nucleus
of KYSE-140-GOLPH3/RNAi than the KYSE-140-scamble cell (Fig. 3B).
TOP-Flash and FOP-Flash assays were performed to
confirm the activity of TCF4 and Wnt/β-catenin signaling activity.
In the KYSE-140-GOLPH3 cell, TCF4 activity was increased compared
to KYSE-140-vector cell. Contrastingly, TCF4 activity was decreased
in KYSE-140-GOLPH3/RNAi cell compared to KYSE-140-scramble cell
(Fig. 3C). When the Wnt/β-catenin
signaling was fully blocked with TCF4-dn, which lacked the
β-catenin binding domain and antagonized Wnt/β-catenin signal, the
GOLPH3 overexpression or knockdown had no effect on cyclin D1
(Fig. 3D).
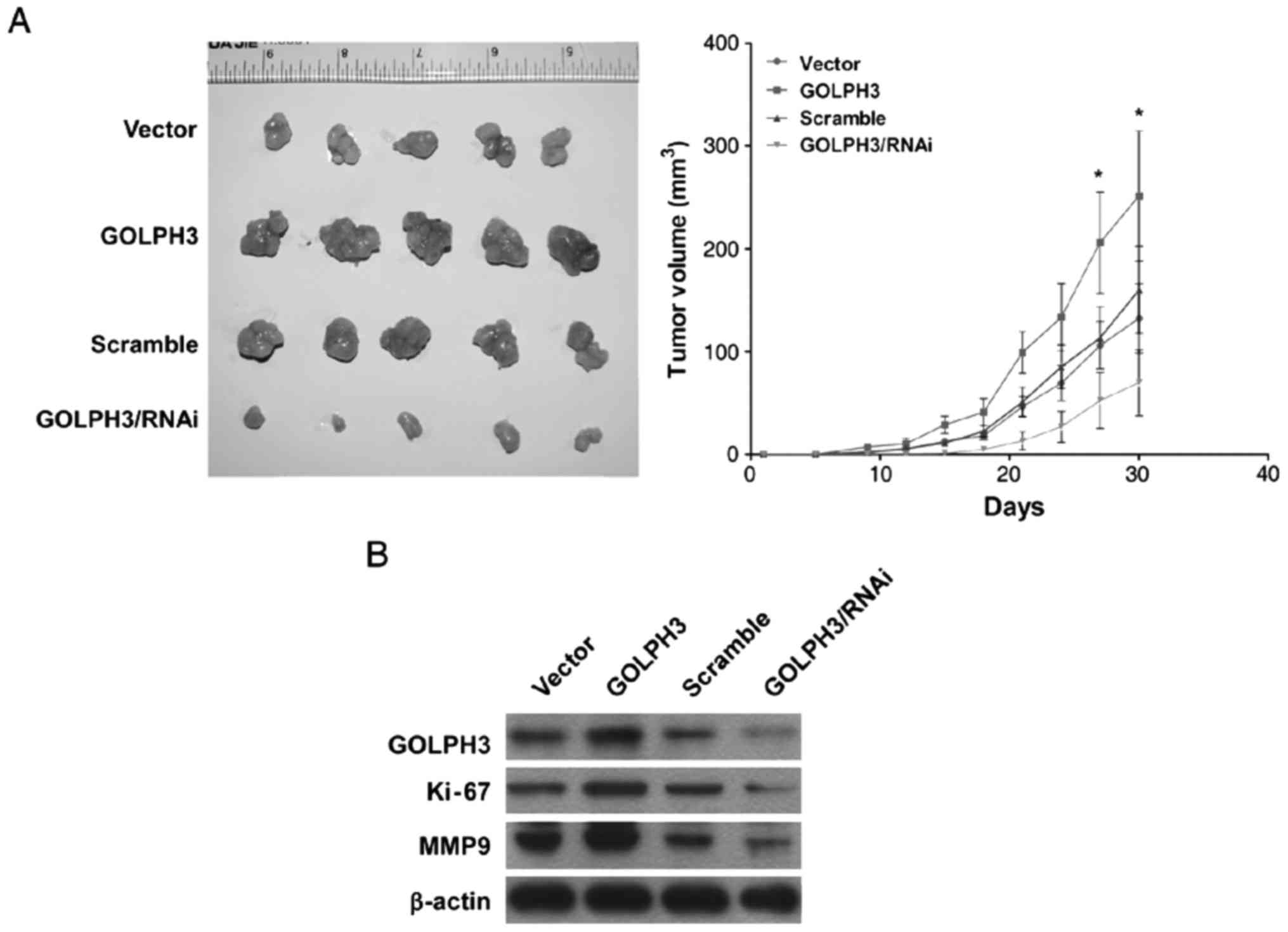
GOLPH3 promoted ESCC tumorigenesis in
vivo
Nude mice tumorigenesis assays demonstratd that the
KYSE-140-GOLPH3 cell-formed tumors were larger than KYSE-140-vector
cells. While KYSE-140-GOLPH3/RNAi cell-formed tumors were smaller
than KYSE-140-scramble cells (Fig.
4A).
Further analysis of protein expression of tumor
tissues formed in vivo by western blot indicated that GOLPH3
in generated tumor tissues remained stable. Cell proliferation
related protein Ki-67 was found upregulated in KYSE-140-GOLPH3
generated tumor tissues, but down regulated in KYSE-140-GOLPH3/RNAi
generated tumor tissues. Interestingly, migration related protein
MMP9 was also upregulated when GOLPH3 was overexpressed, while down
regulated when GOLPH3 was knockdown (Fig. 4B).
In general, GOLPH3 can also play a tumorigenesis
role in vivo. And the tumor progression may due to cell
proliferation and invasion promoting abilities of GOLPH3.
Discussion
Our previous study evidenced that GOLPH3 high
expression in ESCC cancer tissues correlated with patients'
clinical stage, TNM classification, histological differentiation
and shorter overall survival time (28). In this study, gain- and
loss-of-function analysis were performed based on the construction
of the GOLPH3 stable overexpressed cell line KYSE-140-GOLPH3 and
the negative control KYSE-140-vector, and the GOLPH3 stable
knockdown cell line KYSE-140-GOLPH3/RNAi and the negative control
KYSE-140-scramble.
Our both in vitro and in vivo
experiments demonstrated that GOLPH3 promoted cell growth and
proliferation, and in the end triggered tumorigenesis. The
underlying mechanisms may attribute to cell cycle acceleration.
GOLPH3 has already been reported as a potent
oncogene which is able to promote cell growth and activate mTOR
signaling (20). MTOR signaling
pathway was reported to be activated in many other cancers, which
played a role as cell growth accelerator and cell survival
protector. The results of our study corroborated this through
confirmation of the activation of mTOR attributing to GOLPH3
overexpression in ESCC. Aside from mTOR signaling, GOLPH3 was also
reported to be involved in cell autophagy regulation and
coordination of the tumor cell and the fibroblasts in the
surrounding microenvironment, which culminates in the promotion of
tumorigenesis (29). Another study
suggested that GOLPH3 overexpression protected cells from DNA
damage and contributed to tumor growth (24). However, the cell cycle acceleration
function of GOLPH3 could not be explained adequately until now.
Our analysis of the expression level of cell cycle
G1-S phase checkpoint associated protein showed that overexpression
of GOLPH3 led to down regulation of the p21 protein and
upregulation of cyclin D1 at the same time, which also increased
pRb phosphorylation. The total effect was G1-S phase transition and
cell cycle acceleration. Cyclin D1 is one of the most specific
effector of the Wnt/β-catenin signaling pathway (30), thus this pathway became the
prioritie of our following study. We inspected another effecter,
c-Myc of Wnt/β-catenin signal, which also regulates p21 expression
(31–33), and found c-Myc upregulation
followed by GOLPH3 overexpression. TOP-Flash and FOP-Flash assays
and western blot of the nuclear β-catenin protein expression levels
both demonstrated the activation of Wnt/β-catenin signal by GOLPH3
overexpression. Thereafter, elimination of the Wnt/β-catenin signal
by TCF4-dn confirmed that the overexpression of GOLPH3 promoted
tumorigenesis via Wnt/β-catenin signaling activation (33). Oppositely, stable knockdown of
GOLPH3 led to a complete reversed effect. In addition, studies from
the other researchers indicated that the overexpression of cyclin
D1 and p21 also correlated with poor prognosis of ESCC patients
(34,35); our study elucidated the
relationship between GOLPH3, cyclin D1 and p21. Considering the
expression of cyclin D1 and p21 in ESCC influenced by GOLPH3, which
also dedicates to poor prognosis of the patients, the role of
GOLPH3 in ESCC may also be defined to some extent.
In our study, another protein matrix
metallopeptidase 9 (MMP9), which was involved in cancer metastasis
(36,37), was also detected in nude mice
tumorigenesis assays. Tumors originated from KYSE-140-GOLPH3 cell
line also showed a variation of MMP9, indicating a new research
direction of GOLPH3 and metastasis.
In summary, GOLPH3/Wnt/β-catenin signaling plays an
important role in ESCC tumorigenesis (Fig. 5). In this study, mechanisms
relating to the tumorigenesis role of GOLPH3 in ESCC cells were
analyzed, and a new signaling in ESCC is proposed, providing a new
target for ESCC therapy.
Acknowledgements
This study was supported by grants from the Natural
Science Foundation of Guangdong Province (no. S2013010011548), the
National Natural Science Foundation of China (no. 81171948 and
81372275), the Key Program of Natural Science Foundation of
Guangdong Province (no. S2012020011060), and the Project of State
Key Laboratory of Oncology in South China (no. 030041060004), to
M.Z.
Glossary
Abbreviations
Abbreviations:
|
RNAi
|
RNA interference
|
|
siRNA
|
small interfering RNA
|
|
cDNA
|
complementary DNA
|
|
PCR
|
polymerase chain reaction
|
|
qPCR
|
real-time quantitative PCR
|
|
EDTA
|
ethylene diamine tetraacetic acid
|
|
BCA
|
bicinchonininc acid
|
|
SDS-PAGE
|
sodium dodecyl sulfate polyacrylamide
gelelectrophoresis
|
|
PVDF
|
polyvinylidene fluoride
|
|
HRP
|
horseradish peroxidase
|
|
ECL
|
enhanced chemiluminescence
|
|
MTT
|
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium
bromide
|
|
PI
|
propidium Iodide
|
|
pS6
|
ribosomal protein S6
|
|
shRNA
|
short hairpin RNA
|
|
4E-BP1
|
eukaryotic translation initiation
factor 4E binding protein 1
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He YT, Hou J, Chen ZF, Qiao CY, Song GH,
Meng FS, Jin HX and Chen C: Trends in incidence of esophageal and
gastric cardia cancer in high-risk areas in China. Eur J Cancer
Prev. 17:71–76. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y,
Ueda J, Wei W, Inoue M and Tanaka H: Epidemiology of esophageal
cancer in Japan and China. J Epidemiol. 23:233–242. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
D'Journo XB and Thomas PA: Current
management of esophageal cancer. J Thorac Dis. 6 Suppl 2:S253–S264.
2014.PubMed/NCBI
|
|
5
|
Kasper S and Schuler M: Targeted therapies
in gastroesophageal cancer. Eur J Cancer. 50:1247–1258. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mohamed A, El-Rayes B, Khuri FR and Saba
NF: Targeted therapies in metastatic esophageal cancer: Advances
over the past decade. Crit Rev Oncol Hematol. 91:186–196. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu CC, Taylor RS, Lane DR, Ladinsky MS,
Weisz JA and Howell KE: GMx33: A novel family of trans-Golgi
proteins identified by proteomics. Traffic. 1:963–975. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bell AW, Ward MA, Blackstock WP, Freeman
HN, Choudhary JS, Lewis AP, Chotai D, Fazel A, Gushue JN, Paiement
J, et al: Proteomics characterization of abundant Golgi membrane
proteins. J Biol Chem. 276:5152–5165. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tu L, Tai WC, Chen L and Banfield DK:
Signal-mediated dynamic retention of glycosyltransferases in the
Golgi. Science. 321:404–407. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tu L, Chen L and Banfield DK: A conserved
N-terminal arginine-motif in GOLPH3-family proteins mediates
binding to coatomer. Traffic. 13:1496–1507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guzik-Lendrum S, Heissler SM, Billington
N, Takagi Y, Yang Y, Knight PJ, Homsher E and Sellers JR: Mammalian
myosin-18A, a highly divergent myosin. J Biol Chem. 288:9532–9548.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Taft MH, Behrmann E, Munske-Weidemann LC,
Thiel C, Raunser S and Manstein DJ: Functional characterization of
human myosin-18A and its interaction with F-actin and GOLPH3. J
Biol Chem. 288:30029–30041. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dippold HC, Ng MM, Farber-Katz SE, Lee SK,
Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu
S, et al: GOLPH3 bridges phosphatidylinositol-4-phosphate and
actomyosin to stretch and shape the Golgi to promote budding. Cell.
139:337–351. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Graham TR and Burd CG: Coordination of
Golgi functions by phosphatidylinositol 4-kinases. Trends Cell
Biol. 21:113–121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bugarcic A, Zhe Y, Kerr MC, Griffin J,
Collins BM and Teasdale RD: Vps26A and Vps26B subunits define
distinct retromer complexes. Traffic. 12:1759–1773. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bishe B, Syed GH, Field SJ and Siddiqui A:
Role of phosphatidylinositol 4-phosphate (PI4P) and its binding
protein GOLPH3 in hepatitis C virus secretion. J Biol Chem.
287:27637–27647. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lenoir M and Overduin M: PtdIns(4)P
signalling and recognition systems. Adv Exp Med Biol. 991:59–83.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sechi S, Colotti G, Belloni G, Mattei V,
Frappaolo A, Raffa GD, Fuller MT and Giansanti MG: GOLPH3 is
essential for contractile ring formation and Rab11 localization to
the cleavage site during cytokinesis in Drosophila melanogaster.
PLoS Genet. 10:e10043052014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Farber-Katz SE, Dippold HC, Buschman MD,
Peterman MC, Xing M, Noakes CJ, Tat J, Ng MM, Rahajeng J, Cowan DM,
et al: DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3.
Cell. 156:413–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Scott KL, Kabbarah O, Liang MC, Ivanova E,
Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, et al:
GOLPH3 modulates mTOR signalling and rapamycin sensitivity in
cancer. Nature. 459:1085–1090. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kunigou O, Nagao H, Kawabata N, Ishidou Y,
Nagano S, Maeda S, Komiya S and Setoguchi T: Role of GOLPH3 and
GOLPH3L in the proliferation of human rhabdomyosarcoma. Oncol Rep.
26:1337–1342. 2011.PubMed/NCBI
|
|
22
|
Li H, Guo L, Chen SW, Zhao XH, Zhuang SM,
Wang LP, Song LB and Song M: GOLPH3 overexpression correlates with
tumor progression and poor prognosis in patients with clinically N0
oral tongue cancer. J Transl Med. 10:1682012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sotgia F, Whitaker-Menezes D,
Martinez-Outschoorn UE, Salem AF, Tsirigos A, Lamb R, Sneddon S,
Hulit J, Howell A and Lisanti MP: Mitochondria ‘fuel’ breast cancer
metabolism: Fifteen markers of mitochondrial biogenesis label
epithelial cancer cells, but are excluded from adjacent stromal
cells. Cell Cycle. 11:4390–4401. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou J, Xu T, Qin R, Yan Y, Chen C, Chen
Y, Yu H, Xia C, Lu Y, Ding X, et al: Overexpression of Golgi
phosphoprotein-3 (GOLPH3) in glioblastoma multiforme is associated
with worse prognosis. J Neurooncol. 110:195–203. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu BS, Hu H, Zhu CY, Gu YL and Li JP:
Overexpression of GOLPH3 is associated with poor clinical outcome
in gastric cancer. Tumour Biol. 34:515–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou X, Zhan W, Bian W, Hua L, Shi Q, Xie
S, Yang D, Li Y, Zhang X, Liu G and Yu R: GOLPH3 regulates the
migration and invasion of glioma cells though RhoA. Biochem Biophys
Res Commun. 433:338–344. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma Y, Ren Y, Zhang X, Lin L, Liu Y, Rong
F, Wen W and Li F: High GOLPH3 expression is associated with a more
aggressive behavior of epithelial ovarian carcinoma. Virchows Arch.
464:443–452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang JH, Chen XT, Wen ZS, Zheng M, Deng
JM, Wang MZ, Lin HX, Chen K, Li J, Yun JP, et al: High expression
of GOLPH3 in esophageal squamous cell carcinoma correlates with
poor prognosis. PLoS One. 7:e456222012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Salem AF, Whitaker-Menezes D, Lin Z,
Martinez-Outschoorn UE, Tanowitz HB, Al-Zoubi MS, Howell A, Pestell
RG, Sotgia F and Lisanti MP: Two-compartment tumor metabolism:
Autophagy in the tumor microenvironment and oxidative mitochondrial
metabolism (OXPHOS) in cancer cells. Cell Cycle. 11:2545–2556.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Katoh M and Katoh M: WNT signaling pathway
and stem cell signaling network. Clin Cancer Res. 13:4042–4045.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Whitfield JF: Calcium, calcium-sensing
receptor and colon cancer. Cancer Lett. 275:9–16. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Raghu D and Karunagaran D: Plumbagin
downregulates Wnt signaling independent of p53 in human colorectal
cancer cells. J Nat Prod. 77:1130–1134. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
van de Wetering M, Sancho E, Verweij C, de
Lau W, Oving I, Hurlstone A, Van Der Horn K, Batlle E, Coudreuse D,
Haramis AP, et al: The beta-catenin/TCF-4 complex imposes a crypt
progenitor phenotype on colorectal cancer cells. Cell. 111:241–250.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shiozaki A, Nakashima S, Ichikawa D,
Fujiwara H, Konishi H, Komatsu S, Kubota T, Okamoto K, Iitaka D,
Shimizu H, et al: Prognostic significance of p21 expression in
patients with esophageal squamous cell carcinoma. Anticancer Res.
33:4329–4335. 2013.PubMed/NCBI
|
|
35
|
Peng H, Zhong XY, Liu KP and Li SM:
Expression and significance of adenomatous polyposis coli,
beta-catenin, E-cadherin and cyclin D1 in esophageal squamous cell
carcinoma assessed by tissue microarray. Ai Zheng. 28:38–41.
2009.PubMed/NCBI
|
|
36
|
van Kempen LC and Coussens LM: MMP9
potentiates pulmonary metastasis formation. Cancer Cell. 2:251–252.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang S, Liu Z, Wang L and Zhang X:
NF-kappaB signaling pathway, inflammation and colorectal cancer.
Cell Mol Immunol. 6:327–334. 2009. View Article : Google Scholar : PubMed/NCBI
|