Introduction
Immunoglobulin (Ig)A nephropathy (IgAN) is
considered the leading cause of primary glomerulonephritis
worldwide (1). Mesangial
hypercellularity and matrix expansion, alongside glomerular IgA
deposition, which according to renal biopsies is usually
accompanied by complement component 3 and IgG deposition, are all
involved in its pathogenesis (1).
Approximately 40% of patients with IgAN will develop end-stage
renal disease (ESRD) within 20 years of the initial biopsy
(2). IgAN is a complex
multifactorial disease associated with various clinical and
pathological features, and its pathogenic mechanisms remain
unknown.
Non-coding RNAs (ncRNAs) were once considered to
have no significant transcriptional functions; however, they have
been increasingly reported to serve an important role in cell
differentiation and disease progression (3). Long ncRNAs (lncRNAs) are a class of
transcripts >200 nucleotides in length with little or no
protein-coding capacity (4,5).
However, lncRNAs do participate in various biological processes,
including gene expression, recruitment of chromatin modifications,
X chromosome inactivation, chromosome recombination and protein
folding (6,7). Recent studies regarding lncRNAs in
kidney diseases have reported that differentially expressed lncRNAs
are present in ESRD (8) and
IgA-negative mesangial proliferative glomerulonephritis (9). In addition, some lncRNAs may be used
as biomarkers in membrane nephropathy (10) and diabetic nephropathy (11). Although the detailed regulatory
mechanisms and functions of lncRNAs remain uncertain, they form a
basis for subsequent research on the development of renal
diseases.
At present, to the best of our knowledge, the
expression of lncRNAs and their influence on IgAN have yet to be
reported. The present study aimed to determine the altered
expression of lncRNAs and mRNAs profiled in patients with IgAN.
Furthermore, via Gene Ontology (GO), Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway and lncRNA-mRNA co-expression network
analyses, the study aimed to elucidate the functions of these
dysregulated lncRNAs and mRNAs in the pathogenesis and progression
of IgAN.
Subjects and methods
Isolation of peripheral blood
mononuclear cells (PBMCs) from patients with IgAN and controls
A total of 12 patients with IgAN and 12 healthy
controls were recruited to the present study from the First
Affiliated Hospital of China Medical University (Shenyang, China),
between November and December 2015. The patients were diagnosed
with IgAN according to renal immunopathological diagnosis (by
kidney biopsy) and due to the presence of clinical characteristics
without other complications. Patients with secondary IgAN were
excluded from the present study, and the patients studied had never
been treated with immunosuppressive drugs. The present study was
approved by the Ethical Review Committees of the First Affiliated
Hospital of China Medical University, and were conducted in
accordance with the Declaration of Helsinki. Written informed
consent was obtained from all participants, and blood samples were
collected from patients with IgAN and healthy controls. PBMCs were
isolated using Ficoll-Hypaque gradient centrifugation (12).
Microarray analysis
Total RNAs were extracted from PBMCs using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
were purified with the RNeasy kit (Qiagen GmbH, Hilden, Germany)
according to the manufacturers' protocols; individual RNA samples
were stored at −80°C until further use. RNA concentration was
quantified using the NanoDrop ND-2000 (NanoDrop; Thermo Fisher
Scientific, Inc., Wilmington, DE, USA). After passing RNA
measurement quality control using the NanoDrop ND-2000 and
denaturing gel electrophoresis, RNA was used to synthesize
double-stranded cDNA using the SuperScript Double-Stranded cDNA
Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to manufacturer's protocol.
The Agilent Human lncRNA (4*180K, Design ID, 076500;
Agilent Technologies, Inc., Santa Clara, CA, USA) was used to
conduct the microarray analysis according to manufacturer's
protocol. Differentially expressed mRNAs were also identified using
a microarray. Raw data were extracted as pair files by Feature
Extraction software (version 10.7.1.1; Agilent Technologies, Inc.),
and Genespring (version 13.1; Agilent Technologies, Inc.) was used
to analyze the raw data. The threshold set for up- and
downregulated genes was a fold change ≥2.0 and P≤0.05.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) validation
Total RNA was reverse transcribed using Random
Decamers (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
and reverse transcriptase, M-MLV (Invitrogen; Thermo Fisher
Scientific, Inc.): cDNA synthesis was 25°C for 5 min, 42°C for 60
min and 70°C for 5 min. lncRNA expression levels were quantified by
qPCR using a SYBR-Green kit (Thermo Fisher Scientific, Inc.).
Reactions were performed in duplicate and comprised 2X concentrated
Universal Master mix, 1 µl template cDNA, and 100 nM primers in a
final volume of 9 µl. Reactions were analyzed in a 384-well PCR
reaction plate (Axygen; Corning Incorporated, Corning, NY, USA)
using a QuantStudio™ 6 Flex Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
RT-qPCR conditions were as follows: PCR amplification for 40 cycles
at 95°C for 10 sec, at 60°C for 60 sec and at 95°C for 10 sec.
Then, the temperature was allowed to slowly rise from 60 to 99°C.
The relative lncRNA expression levels were calculated using the
ΔΔCq method (13) and
data were normalized with H-actin. Primer sequences were as
follows: Myelin expression factor 2 (MYEF2)-1:1 forward,
5′-CCCAACACAAGCTAGAAGACAACA-3′ and reverse,
5′-GCATCTTGCTACTTTAATTGGTCC-3′; MYEF2-1:3 forward,
5′-TACACACATGCTTACATTTTGAAGG-3′ and reverse,
5′-GCACCAACTAATACCAAGGAACC-3′; MYEF2-1:4 forward,
5′-TTCCCCAATACGTTAATGTTTTGAC-3′ and reverse,
5′-GGAAAAAGGAGGAGTGAATGGGT-3′; arachidonate 15-lipoxygenase
pseudogene 1 (ALOX15P1)-nc forward, 5′-TGTTGACTTTAAGGTTTCGCTGG-3′
and reverse, 5′-CAGAGCGCCTCAGCACCAC-3′; mitogen-activated protein
kinase 8 interacting protein 2 (MAPK8IP2)-1:14 forward,
5′-CAAGCCAGAGCGGGTCAGTT-3′ and reverse,
5′-GGGCACTGGGAGAAGTTAGCAC-3′; folliculogenesis specific BHLH
transcription factor (FIGLA)-1:1 forward,
5′-GCCGACCGGAGATAGCTAAGA-3′ and reverse,
5′-GGCTCCTTCTGTTCACTGCTCA-3′; and H-actin forward, 5-AGC ACA GAG
CCT CGC CTT TG-3′ and reverse, 5′-CTTCTGACCCATGCCCACCA-3′.
GO analysis and KEGG pathway
GO is an ontological classification analysis
ascribing functions of differentially expressed mRNAs to GO
categories. Classification consists of cellular components,
molecular functions and biological processes. The GO categories are
derived from the GO Consortium (http://www.geneontology.org), which comprises three
structured networks of defined terms used to describe the gene
product attributes. KEGG (http://www.genome.jp/kegg/pathway.html/) mapping can
be used to predict which biological pathways differentially
expressed mRNAs are involved in. According to P-value,
significantly enriched GO terms and KEGG pathways were screened.
P≤0.05 was considered to indicate significance.
lncRNA-mRNA co-expression network. The lncRNA-mRNA
co-expression network was based on a correlation between
differentially expressed lncRNAs and mRNAs enriched in the ‘innate
immune response’ term, and was constructed according to normalized
signal intensities of specific expression levels of lncRNAs and
mRNAs. Pearson's correlation coefficients (>0.9) were used to
identify the lncRNAs and associated coding genes. Subsequently, the
lncRNA-mRNA co-expression network was constructed using Cytoscape
software version 2.8.1 (The Cytoscape Consortium, San Diego, CA,
USA).
Statistical analysis
Data are presented as the mean ± standard deviation.
Data obtained from three independent repetitions of RT-qPCR were
analyzed using unpaired Student's t-test. Statistical analyses were
performed using SPSS software version 18.0 (SPSS, Inc., Chicago,
IL, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Differentially expressed lncRNAs and
mRNAs in IgAN
The demographics and baseline clinical
characteristics of the study subjects are summarized in Table I. lncRNA and mRNA expression in
PBMCs from 12 patients with IgAN and 12 healthy controls were
studied using microarray analyses. A total of 167 differentially
expressed lncRNAs (including 55 upregulated lncRNAs and 112
downregulated lncRNAs) and 94 differentially expressed mRNAs
(including 36 upregulated mRNAs and 58 downregulated mRNAs) were
identified. The differentially expressed lncRNAs varied between the
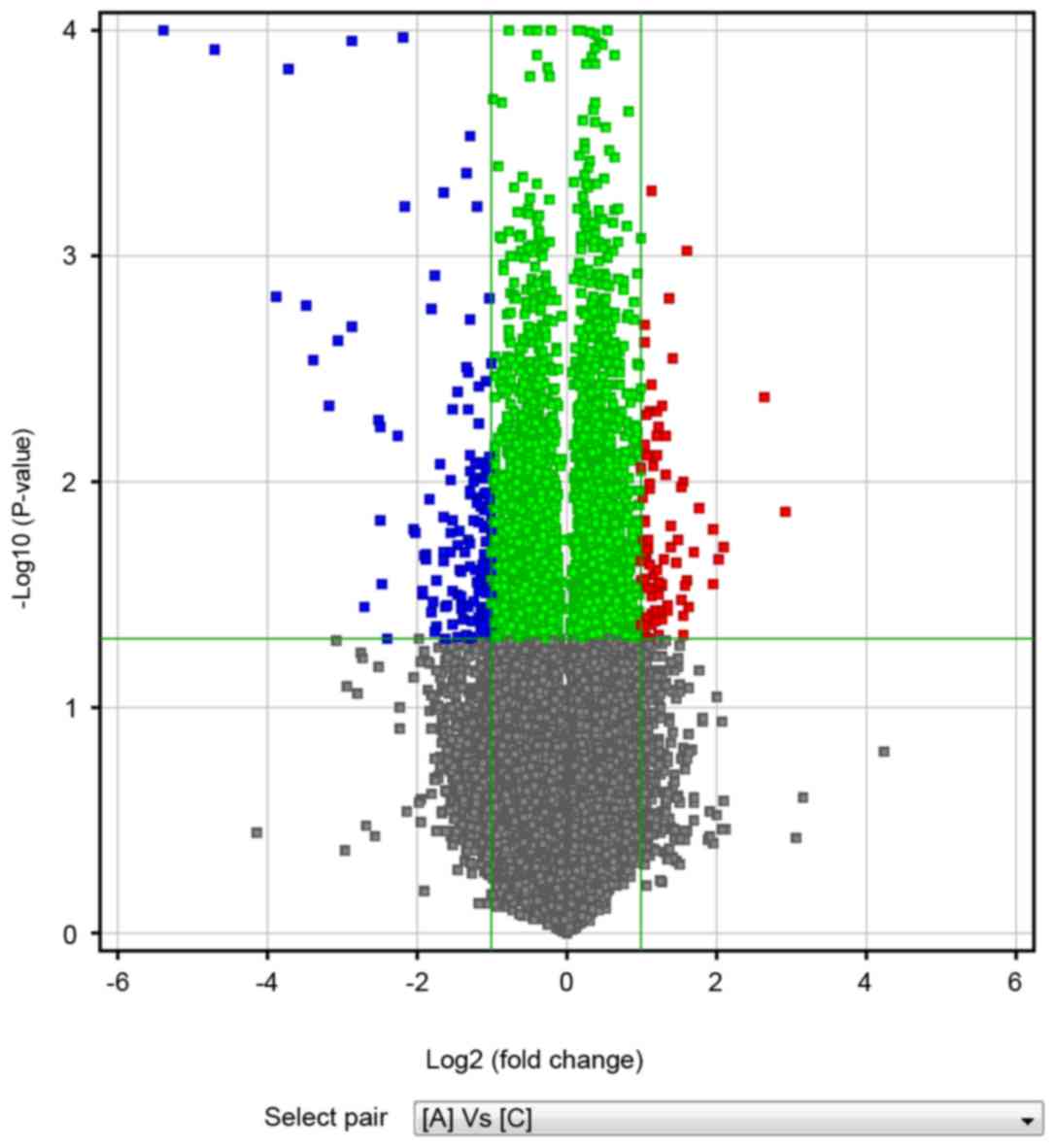
patients with IgAN and the healthy controls, as shown by a volcano
plot (Fig. 1). The red and the
blue spots indicate increased or decreased expression compared with
relative expression, respectively. The top 20 expressed lncRNAs and
mRNAs are listed in Tables II and
III.
 | Table I.Characteristics of patients with IgAN
and healthy controls enrolled in the present study. |
Table I.
Characteristics of patients with IgAN
and healthy controls enrolled in the present study.
| Characteristic | IgAN | Controls |
|---|
| Age, mean ± SD
(years) | 35±12 | 36±10 |
| Sex (M:F) | 7:5 | 1:1 |
| Urine RBCs count
(range, 104/ml) | 115 (40–240) | 0 |
| Proteinuria
(g/day) | 0.87±0.50 | 0.15±0.10 |
| Scr, mean ± SD
(µmol/l) | 151.50±90.20 | 58.50±21.50 |
 | Table II.Top 20 differentially expressed
lncRNAs. |
Table II.
Top 20 differentially expressed
lncRNAs.
| Order | lncRNA |
|---|
| 1 | ENST00000496886 |
| 2 | DNAJC24-3:1 |
| 3 | TMEM63B-1:1 |
| 4 | FAM27L-7:1 |
| 5 | MYEF2-1:1 |
| 6 | TMEM100-3:1 |
| 7 | NRG2-1:6 |
| 8 | DNAJA1-3:1 |
| 9 | PABPC3-3:1 |
| 10 | PARP16-4:1 |
| 11 | ZNF296-2:4 |
| 12 | ENST00000478301 |
| 13 | WHAMM-2:2 |
| 14 | SAP30-2:1 |
| 15 | SOD1-9:1 |
| 16 | ENST00000448680 |
| 17 | CDYL2-2:10 |
| 18 | ENST00000564702 |
| 19 | FBN1-3:2 |
| 20 | CYBRD1-3:1 |
 | Table III.Top 20 differentially expressed
mRNAs. |
Table III.
Top 20 differentially expressed
mRNAs.
| Order | mRNA |
|---|
| 1 | SLC12A1 |
| 2 |
TCONS_l2_00008986 |
| 3 | KIAA1324 |
| 4 | TCONS_00019298 |
| 5 | SLC12A1 |
| 6 | ZFP57 |
| 7 | GUCY1A3 |
| 8 | OLIG2 |
| 9 | PGA3 |
| 10 | IL5RA |
| 11 | IL18RAP |
| 12 | CLEC7A |
| 13 |
TCONS_l2_00008982 |
| 14 | TCONS_00021505 |
| 15 | GOLGA6L6 |
| 16 | ALOX15 |
| 17 | PRSS33 |
| 18 | SIGLEC12 |
| 19 | LOC100653133 |
| 20 | DEFB123 |
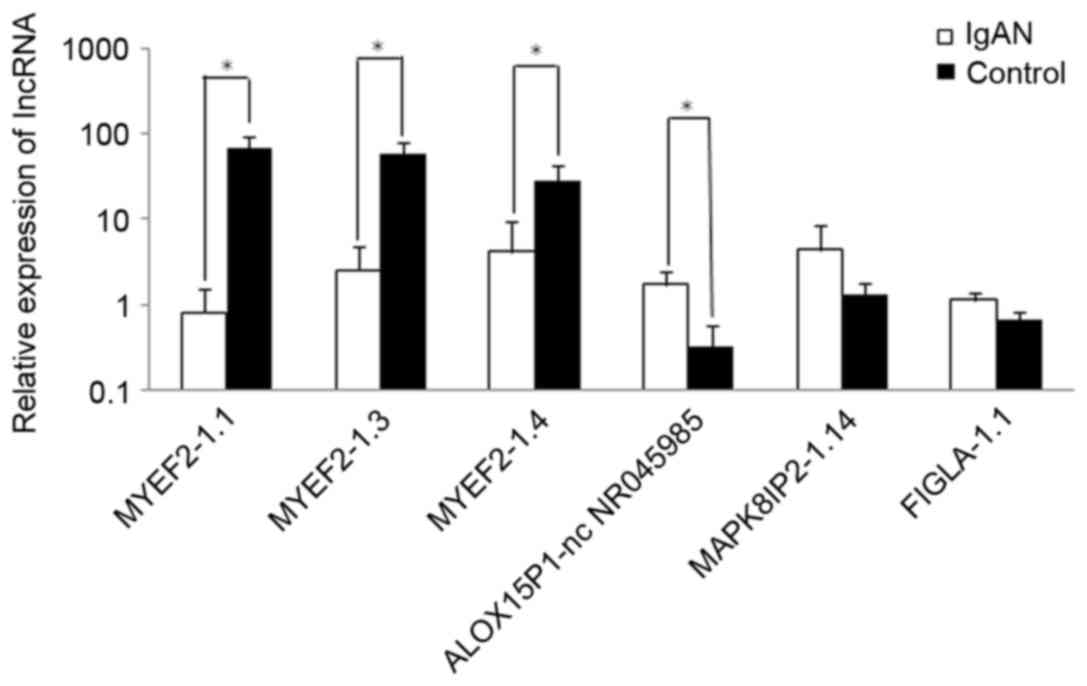
RT-qPCR validation
For the purpose of validating the microarray
analysis results and determining the role of lncRNAs in IgAN, 6
lncRNAs were randomly selected. As presented in Fig. 2, differential expression of these
lncRNAs was detected in patients with IgAN compared with healthy
controls. lncRNA MYEF2-1.1, lncRNA MYEF2-1.3 and lncRNA MYEF2-1.4
exhibited 85-, 22.92- and 6.91-fold lower expression in patients
with IgAN, respectively. In addition, lncRNA ALOX15P1-nc NR045985,
lncRNA MAPK8IP2-1.14 and lncRNA FIGLA-1.1 exhibited 5.15-, 3.38-
and 1.68-fold higher expression in patients with IgAN,
respectively. These results agreed with the findings obtained from
the microarray analysis.
GO and KEGG analyses of differentially
expressed mRNAs
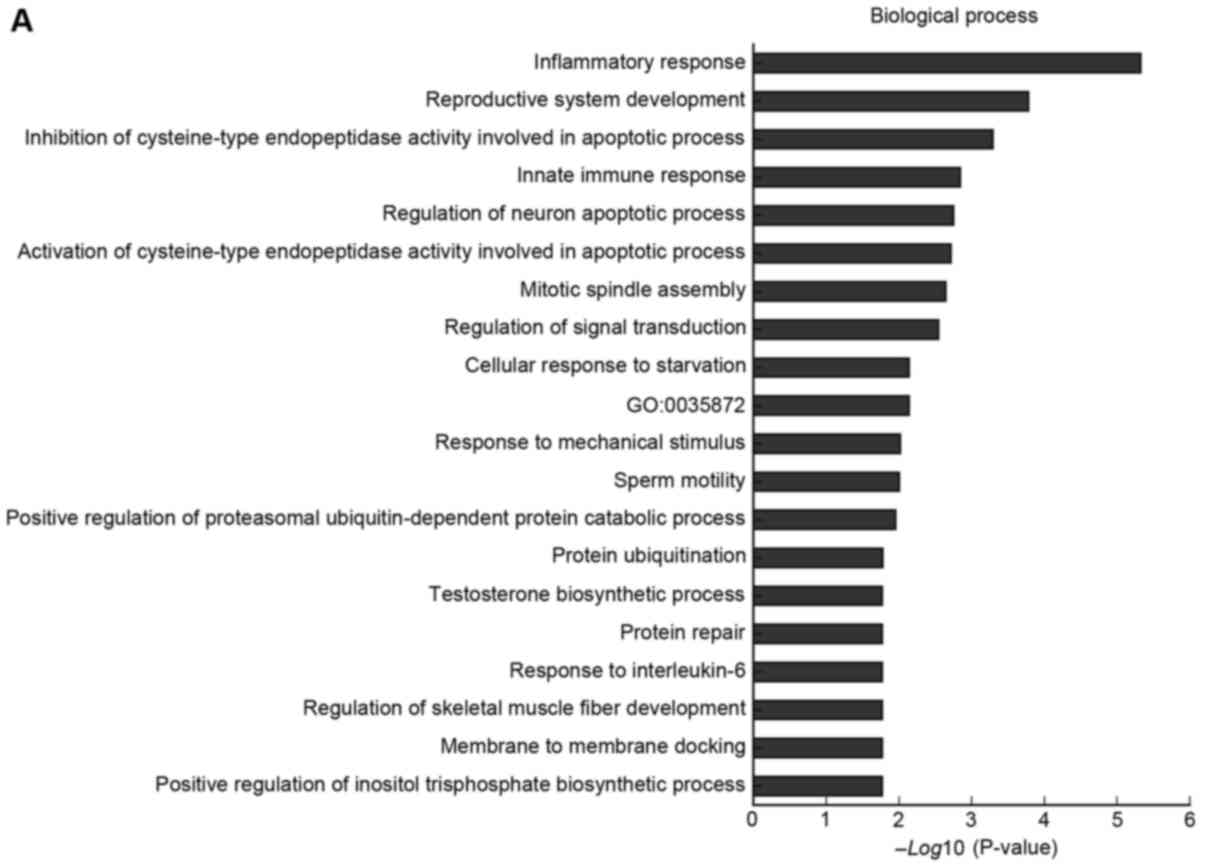
A total of 94 filtered mRNAs were included in the GO
analysis. As shown in Fig. 3. GO
analysis of the differentially expressed mRNAs indicated that they
were enriched in numerous GO terms, including innate immune
response, inflammatory response, IPAF inflammasome complex and
UDP-galactose:β-N-acetylglucosamine β-1, and
3-galactosyltransferase activity.
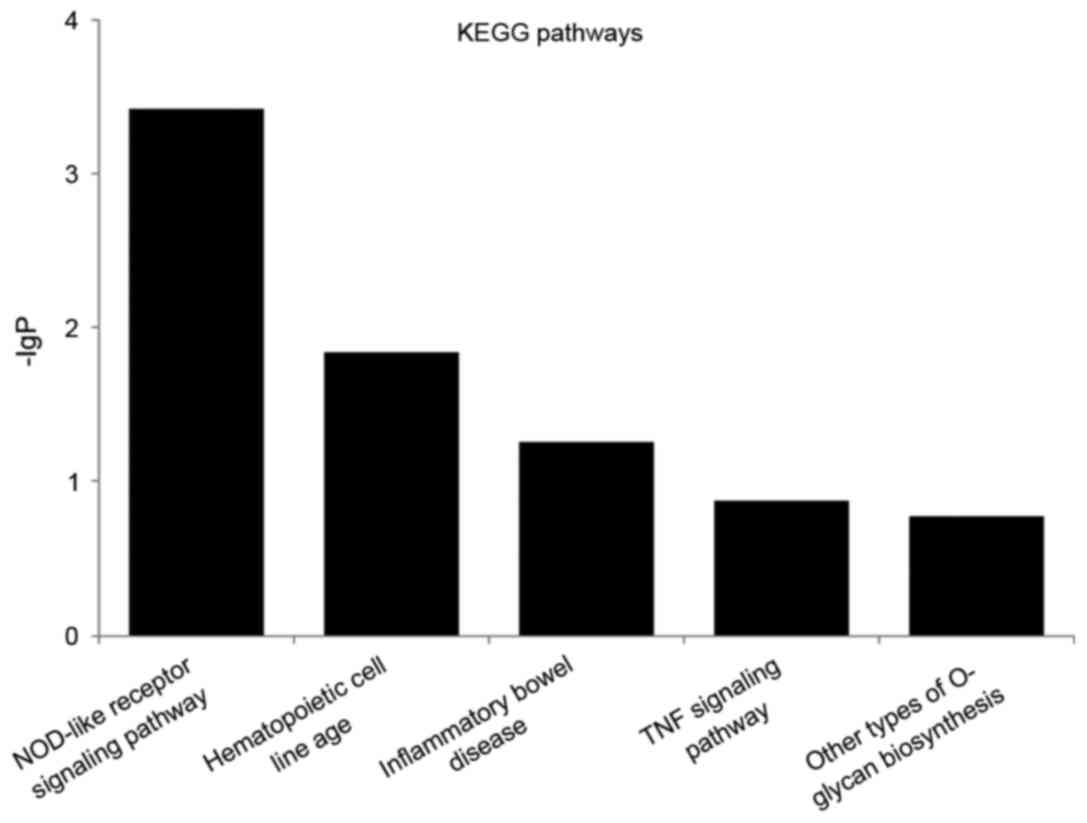
Furthermore, the genes were also mapped to pathways
in a functionally analytical manner; the top five KEGG pathways are
listed in Fig. 4, including
nucleotide-binding oligomerization domain (NOD)-like receptor
signaling pathway, hematopoietic cell lineage, inflammatory bowel
disease (IBD), tumor necrosis factor (TNF) signaling pathway and
other types of O-glycan biosynthesis.
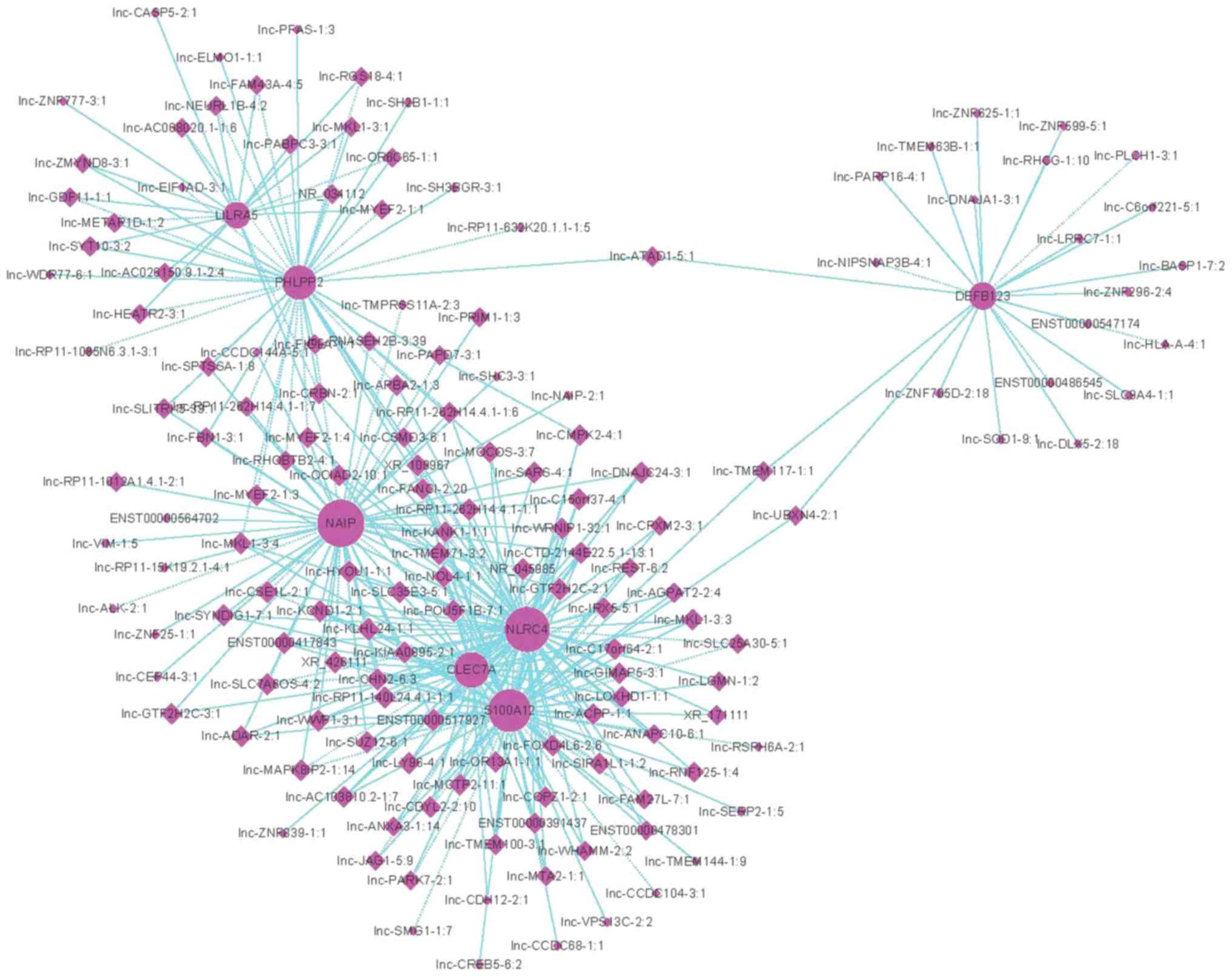
lncRNA-mRNA co-expression network
In the enriched biological processes identified in
the GO analysis, the term ‘innate immune response’ was analyzed
further, in order to generate a lncRNA-mRNA co-expression network.
A total of 167 differentially expressed lncRNAs and 94 mRNAs
comprised the network analysis; 149 lncRNAs were revealed to
interact with 7 mRNAs participating in this term (Fig. 5).
Discussion
IgAN represents a common form of primary
glomerulonephritis, which is characterized by highly heterogeneous
clinicopathological features (1),
and is an important cause of ESRD. Approximately half of patients
with IgAN will eventually depend on dialysis. While the
pathogenesis of IgAN is only partially understood, previous studies
support a multi-hit hypothesis: Specific autoantibodies recognize
galactose-deficient IgA1 molecules, resulting in the
formation of circulating IgA1-IgG immune complexes, which may be
deposited in the glomerular mesangium, thereby inducing renal
injury (1,14).
Regulatory ncRNAs have essential roles in the
regulation of gene expression and mammalian development, and
include microRNAs (miRNAs/miRs) and lncRNAs (15,16).
miRNAs have been reported to contribute to the progression of IgAN
and may potentially serve as biomarkers for diagnosis and disease
monitoring (17); for example,
abnormal expression of miR-148b may explain aberrant glycosylation
of IgA1 (18).
Furthermore, a retrospective international study indicated that
circulating miR-148b and let-7b may be considered serum markers for
detecting primary IgAN (19).
However, research regarding the role of lncRNAs in IgAN is lacking.
The establishment of differentially expressed lncRNA profiles in
human IgAN is important for elucidating an explicit pathogenesis of
this illness.
The present study generated lncRNA and mRNA
expression profiles in PBMCs from patients with IgAN and healthy
controls via microarray analyses. A total of 167 differentially
expressed lncRNAs and 94 differentially expressed mRNAs were
identified, thus suggesting that these mRNAs and lncRNAs may serve
as potential biomarkers for the diagnosis of IgAN. Via GO and KEGG
pathway analyses, detailed information was obtained regarding the
biological functions and potential mechanisms of these mRNAs in
IgAN. Numerous GO terms, including innate immune response and
inflammatory response, were revealed to be significantly enriched
in the identified differentially expressed genes. In addition, 94
differentially expressed mRNAs were associated with numerous KEGG
pathways; the top five pathways included NOD-like receptor
signaling pathway, hematopoietic cell lineage and IBD.
The present study focused on the ‘innate immune
response’ term in GO analysis when evaluating the prominent role of
innate immune dysregulation in IgAN. Toll-like receptors (TLR) are
a family of pathogen recognition molecules, including TLR1, TLR3,
TLR4, TLR9 and TLR10, which have been reported to be associated
with the development of IgAN. A nasal challenge with CpG
oligodeoxynucleotide, which is a ligand of TLR9, was reported to
aggravate renal injury in a murine IgAN model (20), and a single nucleotide polymorphism
in TLR9 (TT genotype in rs352140) was reported to be an important
risk factor for the progression of human IgAN (19). In addition, TLR9 on B cells and
dendritic cells located in the mucosa may serve different roles in
the development of IgAN, via induction of nephritogenic IgA or
IgA-IgG immune complexes, respectively (21). Such findings suggested that TLR9
may be closely associated with the pathogenesis of human IgAN.
Coppo et al demonstrated that the levels of TLR4 in the PBMC
of patients with IgAN were increased, and were correlated with the
degree of proteinuria and gross microscopic hematuria (22). Furthermore, TLR1 (CT and CC
genotype in rs4833095, TT genotype in rs5743557) (23) and TLR10 genes (TA and AA genotype
in rs1004195) (24) may be
associated with susceptibility to IgAN in Korean children.
Recently, He et al (25)
reported that the TLR3-B-cell activating factor axis was involved
in IgA class switch recombination in IgAN.
Activation of the complement system, including the
alternative and lectin pathways, has also been reported to be
closely associated with IgAN, perhaps by contributing to abnormal
IgA1 glycosylation (26,27). Furthermore, local renal
polarization of macrophages has been implicated in the pathological
type of IgAN; activation of M2 macrophages was followed by fibrotic
alterations, whereas M1 polarization induced mesangial
proliferation (21).
Existing evidence has indicated that innate immune
disturbances crucially contribute to the pathogenesis of IgAN;
however, these require further investigation. The present results
obtained from the GO analysis in patients with IgAN supported the
importance of the ‘innate immune response’, which is concordant
with the findings of previous studies. Therefore, the present study
focused on the ‘innate immune response’ term for further
lncRNA-mRNA co-expression network analysis. The co-expression
network was constructed based on the 167 differentially expressed
lncRNAs and the 94 differentially expressed mRNAs obtained from
patients with IgAN compared with healthy controls. A total of 149
lncRNAs were revealed to interact with 7 mRNAs that participated in
the ‘innate immune response’ term. These mRNAs included NAIP,
LILRA5 and CLEC7A. The protein encoded by LILRA5 is a member of the
leukocyte immunoglobulin-like receptor (LIR) family, known to have
activating and inhibitory functions in leukocytes. There are 19
lncRNAs interacting with LILRA5 in the ‘innate immune response’
term. Lnc-CRBN-2:1 is one of these lncRNAs and is also known as
IL5RA. The protein encoded by this gene is an interleukin 5
specific subunit of a heterodimeric cytokine receptor. A potential
interaction of C1GALT1 and IL5RA on the susceptibility of IgAN has
been identified (28). These
findings suggested that the inter-regulation of lncRNAs and mRNAs
may be involved in IgAN-associated innate immune disturbances.
Dysregulation in the lncRNA-mRNA network may be a possible
mechanism underlying IgAN progression.
In conclusion, the present study systematically
screened abnormally expressed lncRNAs and mRNAs between patients
with IgAN and healthy controls, providing novel information
regarding the potential role of lncRNAs in IgAN. GO and KEGG
pathway analyses revealed detailed information regarding the
biological functions and potential mechanisms of these mRNAs. In
addition, the co-expression network generated in the present study
suggested inter-regulation of lncRNAs and mRNAs in patients with
IgAN. These findings form a basis for further studies of lncRNAs in
IgAN, which should focus on exploring the functions and regulatory
mechanisms of identified lncRNAs, thus identifying potential
screening biomarkers and novel treatment targets for this
disease.
References
|
1
|
Wyatt RJ and Julian BA: IgA nephropathy. N
Engl J Med. 368:2402–2414. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schena FP: A retrospective analysis of the
natural history of primary IgA nephropathy worldwide. Am J Med.
89:209–215. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Consortium EP: An integrated encyclopedia
of DNA elements in the human genome. Nature. 489:57–74. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sui W, Yan Q, Li H, Liu J, Chen J, Li L
and Dai Y: Genome-wide analysis of long noncoding RNA expression in
peripheral blood monoclear cells of uremia patients. J Nephrol.
26:731–738. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sui W, Li H, Ou M, Tang D and Dai Y:
Altered long non-coding RNA expression profile in patients with
IgA-negtive mesangial proliferative glomerulonephritis. Int J Mol
Med. 30:173–178. 2012.PubMed/NCBI
|
|
10
|
Huang YS, Hsieh HY, Shih HM, Sytwu HK and
Wu CC: Urinary Xist is a potential biomarker for membranous
nephropathy. Biochem Biophys Res Commun. 452:415–421. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alvarez ML, Khosroheidari M, Eddy E and
Kiefer J: Role of microRNA 1207-5P and its host gene, the long
non-coding RNA Pvt1, as mediators of extracellular matrix
accumulation in the kidney: Implications for diabetic nephropathy.
PLoS One. 8:e774682013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Peng H, Tian J, Ma J, Tang X, Rui
K, Tian X, Wang Y, Chen J, Lu L, et al: Upregulation of long
noncoding RNA TMEVPG1 enhances T helper type 1 cell response in
patients with Sjögren syndrome. Immunol Res. 64:489–496. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Novak J, Renfrow MB, Gharavi AG and Julian
BA: Pathogenesis of immunoglobulin A nephropathy. Curr Opin Nephrol
Hypertens. 22:287–294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zaratiegui M, Irvine DV and Martienssen
RA: Noncoding RNAs and gene silencing. Cell. 128:763–776. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Szeto CC and Li PK: MicroRNAs in IgA
nephropathy. Nat Rev Nephrol. 10:249–256. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Serino G, Sallustio F, Cox SN, Pesce F and
Schena FP: Abnormal miR-148b expression promotes aberrant
glycosylation of IgA1 in IgA nephropathy. J Am Soc Nephrol.
23:814–824. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Serino G, Pesce F, Sallustio F, De Palma
G, Cox SN, Curci C, Zaza G, Lai KN, Leung JC, Tang SC, et al: In a
retrospective international study, circulating miR-148b and let-7b
were found to be serum markers for detecting primary IgA
nephropathy. Kidney Int. 89:683–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suzuki H, Suzuki Y, Narita I, Aizawa M,
Kihara M, Yamanaka T, Kanou T, Tsukaguchi H, Novak J, Horikoshi S
and Tomino Y: Toll-like receptor 9 affects severity of IgA
nephropathy. J Am Soc Nephrol. 19:2384–2395. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kajiyama T, Suzuki Y, Kihara M, Suzuki H,
Horikoshi S and Tomino Y: Different pathological roles of toll-like
receptor 9 on mucosal B cells and dendritic cells in murine IgA
nephropathy. Clin Dev Immunol. 2011:8196462011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Coppo R, Camilla R, Amore A, Peruzzi L,
Daprà V, Loiacono E, Vatrano S, Rollino C, Sepe V, Rampino T and
Dal Canton A: Toll-like receptor 4 expression is increased in
circulating mononuclear cells of patients with immunoglobulin A
nephropathy. Clin Exp Immunol. 159:73–81. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee JS, Park HK, Suh JS, Hahn WH, Kang SW,
Park HJ, Kim MJ, Chung JH and Cho BS: Toll-like receptor 1 gene
polymorphisms in childhood IgA nephropathy: A case-control study in
the Korean population. Int J Immunogenet. 38:133–138. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park HJ, Hahn WH, Suh JS, Kim MJ, Kang SW,
Lee JS, Kim JW, Chung JH and Cho BS: Association between toll like
receptor 10 (TLR10) gene polymorphisms and childhood IgA
nephropathy. Eur J Pediatr. 170:503–509. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He L, Peng X, Wang J, Tang C, Zhou X, Liu
H, Liu F, Sun L and Peng Y: Synthetic double-stranded RNA Poly(I:C)
aggravates IgA nephropathy by triggering IgA class switching
recombination through the TLR3-BAFF axis. Am J Nephrol. 42:185–197.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hashimoto A, Suzuki Y, Suzuki H, Ohsawa I,
Brown R, Hall S, Tanaka Y, Novak J, Ohi H and Tomino Y:
Determination of severity of murine IgA nephropathy by glomerular
complement activation by aberrantly glycosylated IgA and immune
complexes. Am J Pathol. 181:1338–1347. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Karsten CM, Pandey MK, Figge J,
Kilchenstein R, Taylor PR, Rosas M, McDonald JU, Orr SJ, Berger M,
Petzold D, et al: Anti-inflammatory activity of IgG1 mediated by Fc
galactosylation and association of FcgammaRIIB and dectin-1. Nat
Med. 18:1401–1406. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang W, Sun Y, Fu Y, Yu X and Li M:
Interaction of C1GALT1-IL5RA on the susceptibility to IgA
nephropathy in Southern Han Chinese. J Hum Genet. 58:40–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|