Introduction
Pancreatic cancer is one of the most
life-threatening malignancies, accounting for more than 200,000
annual mortalities worldwide (1).
It is one of the most hard-to-diagnose and aggressive malignancies,
despite the increased understanding of its etiopathogenesis
(2–4). Due to its fatal and asymptomatic
nature, pancreatic cancer has become one of the top causes of
cancer-associated mortality (5,6).
Among the different classes of pancreatic cancers, based on its
histology, pancreatic ductal adenocarcinoma is the most predominant
type, accounting for ~85% of exocrine pancreatic cancers; the
remainder are accounted for by papillary carcinoma and
cystadenocarcinoma (7). The
appropriate prediction markers are essential for clinical
application; however, the performance of a single independent
factor is limited in terms of accurate prediction, which, has
promoted the use of combinatorial markers in clinical practice.
Recent advancement in gene expression profiling technology, such as
the high throughput detection method, enables researchers to
identify gene-based biomarkers for PAAD diagnosis and prognosis
(8). However, there are still no
specific targets for the diagnosis and prognosis prediction of PAAD
(9). To improve strategies applied
for improving the survival of patients with PAAD, it is of great
importance to identify more effective biomarkers for the diagnosis,
prognosis prediction and therapeutic response of PAAD.
To date, microRNA (miRNAs) have been proposed as
promising prognostic predictors for different types of cancers.
miRNAs are small size RNAs, with 18–25 nucleotides, that belong to
the family of non-coding RNAs (10). miRNAs have been reported to be
involved in cell differentiation, apoptosis, proliferation and
metabolism by binding to the 3′-untranslated region of their target
mRNAs, resulting in mRNA degradation and/or inhibition of mRNA
translation (11). Furthermore,
the associations between miRNAs and malignancies have been
investigated in depth in recent years and it has been suggested
that miRNAs may serve potential roles as oncogenes or tumor
suppressive genes in various cancer pathogeneses (12), which, has inspired further research
in to their clinical value. Previous studies have demonstrated that
miRNAs are extremely stable in plasma (13), formalin-fixed paraffin-embedded
tissues, and in fresh and frozen tissues (14,15).
Thus, miRNAs have the potential to serve as novel biomarkers for
cancer diagnosis and prognosis prediction, as they remain largely
intact. Several miRNAs have been reported to serve vital roles in
the tumorigenesis and prognosis of PAAD (16–21).
However, the associations between other miRNAs and the survival of
patients with PAAD remains unclear. In addition, an miRNA profile
based on The Cancer Genome Atlas (TCGA) data has not been
published. Therefore, the present study analyzed the associated
TCGA data, evaluating a large number of miRNA sequencing
information for patients with PAAD in order to systematically
assess the predictive value of a specific miRNA signature for
overall survival in patients with PAAD (22).
Materials and methods
Patient characteristics and miRNA
dataset in TCGA
The level 3 data of miRNA sequencing and the
corresponding clinical data for 175 patients with PAAD were
obtained from TCGA (tcga-data.nci.nih.gov/doc/publications/tcga/?) in
July 2016 (23,24). As all data are publicly available
and open-access, ethical approval was not necessary for the present
study. Clinical characteristics of PAAD patients are presented in
Table I.
 | Table I.Clinical characteristics of patients
with pancreatic cancer based on the The Cancer Genome Atlas. |
Table I.
Clinical characteristics of patients
with pancreatic cancer based on the The Cancer Genome Atlas.
|
Characteristics | Number (%) |
|---|
| Age (years) |
|
≥60 | 119 (68.0) |
|
<60 | 56
(32.0) |
| Sex |
|
Female | 79
(45.1) |
|
Male | 96
(54.9) |
| Stage |
|
I+II | 164 (95.9) |
|
III+IV | 7
(4.1) |
| Grade |
|
G1+G2 | 125 (71.4) |
|
G3+G4 | 50
(28.6) |
| Neoplasm
status |
| With
tumor | 100 (64.1) |
| Tumor
free | 56
(35.9) |
| Chronic
pancreatitis |
|
Yes | 14
(10.1) |
| No | 125 (89.9) |
| Diabetes |
|
Yes | 37
(25.7) |
| No | 107 (74.3) |
| Alcohol
history |
|
Yes | 99
(61.1) |
| No | 63
(38.9) |
Survival analysis
A multivariate model was constructed with miRNA
expression, sex, age, grade and Tumor-Nodes-Metastasis stage.
Multivariate Cox regression was performed to evaluate the
associations between the expression of all miRNAs and overall
survival (OS). Hazard ratio (HR) and 95% confidence intervals (CI)
were also assessed. The top 10 miRNAs, those that exhibited the
most significant prognostic values, were selected for further
analysis. The median expression of each miRNA was set as cutoff
value. Once miRNAs were grouped into those with high or low
expression according to its median value (Table II), Kaplan-Meier (K-M) survival
curves were also generated. A permutation and combination method
was adopted for the present study; it is considered overexpressed
when all selected miRNAs presented overexpression simultaneously.
Multivariate cox regression and Kaplan-Meier survival curves were
conducted with SPSS version 22.0 software (IBM Corp., Armonk, NY,
USA). The survival time of each miRNA were presented as the mean ±
standard error. P<0.05 was considered to indicate a
statistically significant difference.
 | Table II.Prognostic role of the 10 most
valuable microRNAs in pancreatic cancer based on The Cancer Genome
Atlas. |
Table II.
Prognostic role of the 10 most
valuable microRNAs in pancreatic cancer based on The Cancer Genome
Atlas.
|
| Kaplan-meier | Multivariate cox
regression |
|---|
|
|
|
|
|---|
| miRNA | Cutoff value | High expression
(months) | Low expression
(months) | Log rank
P-value | HR | 95% CI | P-value |
|---|
| miR-1301 | 4.80 |
45.62±4.96 |
24.27±3.63 | <0.001 | 0.450 | 0.293–0.692 | <0.001 |
| miR-125a | 650.29 |
45.97±5.00 |
21.82±1.94 |
0.001 | 0.484 | 0.314–0.747 |
0.001 |
| miR-376c | 0.78 |
40.67±4.01 |
28.54±3.97 |
0.001 | 0.499 | 0.327–0.761 |
0.001 |
| miR-328 | 22.85 |
46.01±5.08 |
24.21±3.16 |
0.002 | 0.517 | 0.339–0.788 |
0.002 |
| miR-376b | 0.76 |
39.96±3.97 |
29.35±4.10 |
0.002 | 0.521 | 0.343–0.794 |
0.002 |
| miR-29b | 23.32 |
41.83±4.56 |
24.36±2.86 |
0.002 | 0.522 | 0.340–0.800 |
0.003 |
| miR-454 | 3.97 |
45.93±5.10 |
22.12±1.83 |
0.003 | 0.523 | 0.339–0.806 |
0.003 |
| miR-664a | 2.21 |
46.77±5.30 |
26.51±3.14 |
0.004 | 0.540 | 0.353–0.825 |
0.004 |
| miR-3613 | 5.59 |
43.54±4.88 |
22.46±1.96 |
0.011 | 0.576 | 0.376–0.884 |
0.012 |
| miR-126 | 1,372.53 |
37.39±3.81 |
30.19±4.39 |
0.030 | 0.634 | 0.418–0.960 |
0.031 |
| miRNA pool |
|
65.74±5.30 |
30.17±3.32 | <0.001 | 0.139 | 0.043–0.443 | <0.001 |
Target prediction for miRNAs and
functional enrichment analysis
The prospective target genes of miRNAs were
predicted using 12 different programs including miRWalk2.0,
DIANA-microTv4.0, miRanda-rel2010, mirBridge, miRDB4.0, miRmap,
miRNAMap, doRiNA i.e., PicTar2, PITA, RNA22v2, RNAhybrid2.1 and
Targetscan6.2 (25).
The overlapping genes were processed using the
functional enrichment analyses, Gene Ontology (GO) enrichment,
Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein-protein
interaction (PPI) (26). Among
these approaches, GO is an international standardized gene
functional classification system, offering a dynamic-updated
controlled vocabulary and a strictly defined concept to
comprehensively define properties of genes and their products in
any organism. GO analysis reveals the cataloging of biological
process (BP), cellular component (CC) and molecular function (MF).
The GO enrichment study was performed using OmicShare tools, a free
online platform for data analysis (www.omicshare.com/). Firstly, all target genes were
mapped to GO terms in the Gene Ontology database (www.geneontology.org/) and gene numbers were
calculated for each term. Significantly enriched GO terms in target
genes, as compared with the genome background, were defined by a
hypergeometric test. GO terms that met these criteria were defined
as significantly enriched GO terms in target genes. This
examination identified the key biological functions associated with
the target genes.
KEGG is the major public pathway-associated
database, which identifies significantly enriched metabolic
pathways or signal transduction pathways in target genes when
compared with the whole genome background. Pathway enrichment
analysis was also performed through the OmicShare tools.
Significantly enriched pathways in targets, when compared with the
genome background, were defined by a hypergeometric test. Pathways
meeting this criterion were regarded as significantly enriched
pathways in target genes. GO and KEGG pathways were analyzed by
calculating the False Discovery Rate Correction; 0.05 was applied
as a threshold.
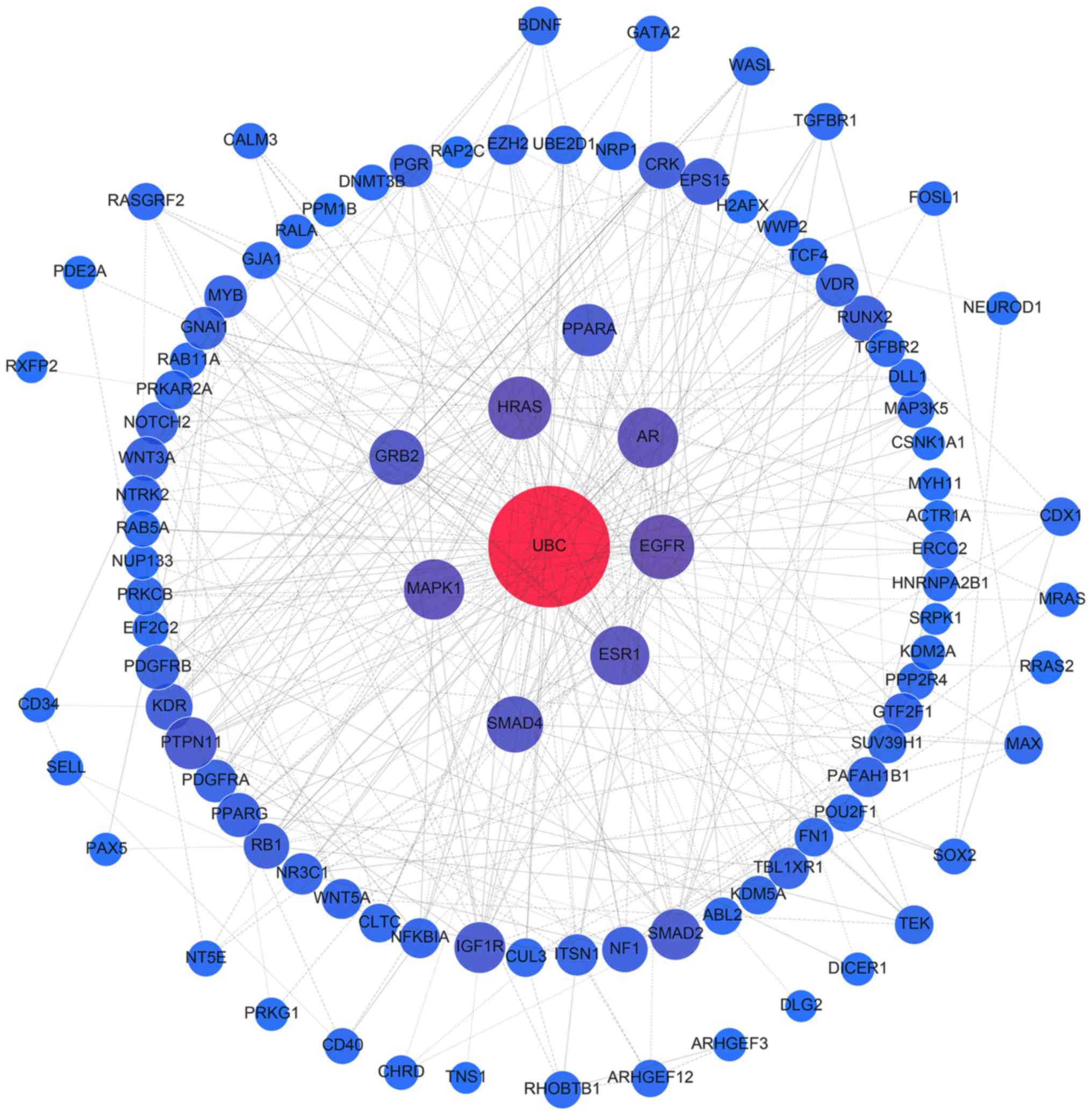
The interactions of all target genes were observed
using the STRING online database (string-db.org), with the cut-off criterion of a
combined score >0.9, and Cytoscape software version 3.4.0
(www.cytoscape.org/). Network nodes
represent proteins, while edges are the protein-protein
associations. The topological centralities, including the local
scale (degree) and the global scale (betweenness, closeness and
stress), were extensively utilized in the network analysis. Of
these, degree is a simple and evident measure that defines the
number of links between a node and its neighboring nodes (27). In the present study, nodes with a
certain degree were proposed to be hub genes (28,29).
A sub-network composed of hub genes and their corresponding
interactions were extracted from the target network.
Immunohistochemistry of The Human
Protein Atlas (THPA)
Immunohistochemistry images were downloaded from the
publicly available database, THPA (www.proteinatlas.org/) (30,31).
THPA, a database that includes >5 million images of
immunohistochemically stained tissues and cells, is based on 6,122
antibodies, representing 5,011 human proteins encoded by ~25% of
the human genome. Data regarding the expression levels of UBC,
SMAD4, MAPK1, AR, ESR1, HRAS and EGFR in PAAD tissues was obtained
from THPA. The protein expression level of each gene was ranked
according to whether it presented low, medium or high antibody
staining.
Results
Survival analysis
In total, the present study successfully assessed
494 miRNAs associated with survival. The prognostic value of all
miRNAs was analyzed by multivariate Cox regression (data not
shown), and then the miRNAs were ranked according to the HR and
P-values. The top 10 candidates (miR-1301, miR-125a, miR-376c,
miR-328, miR-376b, miR-29b, miR-454, miR-664a, miR-3613 and
miR-126) presented the most significant prognostic value (Fig. 1; Table II). Among these 10 miRNAs, the
miRNA with the lowest HR was miR-126, which, had a HR of 0.634 (95%
CI: 0.418–0.96, P=0.031), and the miRNA with the greatest
prognostic value was miR-1301 (HR=0.45; 95% CI, 0.293–0.692;
P<0.001). The high and low expression groups of each miRNA were
formed based on the cutoff median level, and the K-M method was
performed. The patients in the high expression group containing the
top 10 miRNAs had markedly better survival than those of the low
expression group (Table II). For
example, the average survival of the high expression miR-1301 group
was 45.62±4.96 months, a significantly longer survival period than
the 24.27±3.63 months exhibited by the low expression group
(P<0.001). In addition, miR-1301 also had the most significant
prognostic value based on K-M analysis. To produce a more effective
prognostic tool for clinical practice, the present study combined
the top 5 miRNAs into a pool. It was demonstrated that the miRNA
pool comprised of miR-1301, miR-125a, miR-376c, miR-328 and
miR-376b effectively enhanced the efficiency of prognostic
prediction for survival (Fig. 2).
The multivariate Cox regression produced consistent results,
revealing that all 10 miRNA candidates were able to act as
protective factors as their HRs were <1, ranging from 0.634
(miR-126) to 0.450 (miR-1301; Table
II). Notably, the 5-miRNA-pool exhibited an ideal prognostic
value, with a HR value of 0.139 (95% CI, 0.043–0.443; P<0.001),
which, supported the notion that this 5-miRNA-pool may have a
greater predictive capacity for prognosis than individual miRNA
(Table II).
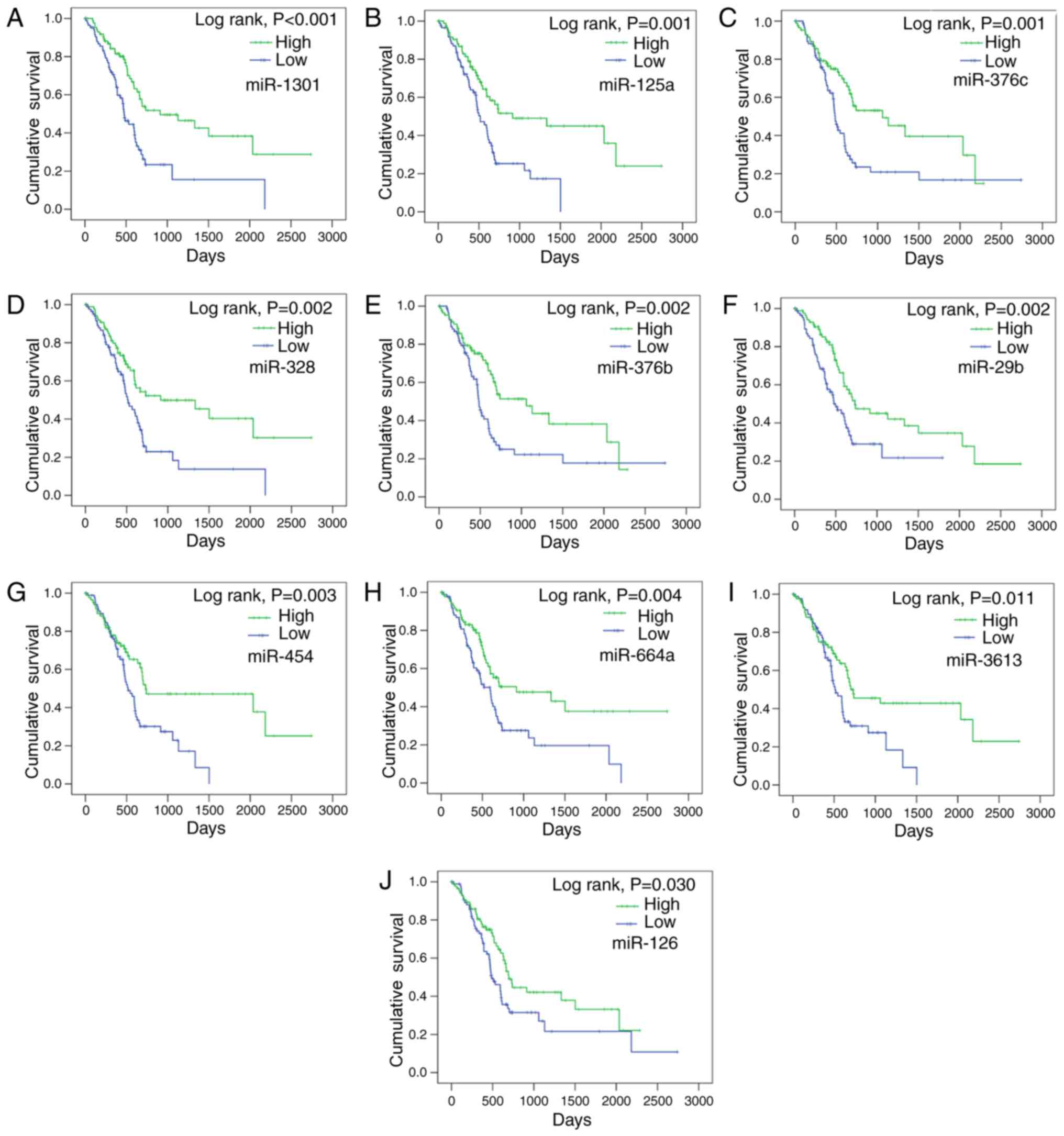 | Figure 1.Associations between the levels of
candidate miRNAs and survival in pancreatic cancer. The
associations between the levels of the top 10 candidate miRNAs, (A)
miR-1301, (B) miR-125a, (C) miR-376c, (D) miR-328, (E) miR-376b,
(F) miR-29b, (G) miR-454, (H) miR-664a, (I) miR-3613 and (J)
miR-126. miR, and survival in pancreatic cancer were analyzed using
the Kaplan-Meier method. miRNA/miR, microRNA. |
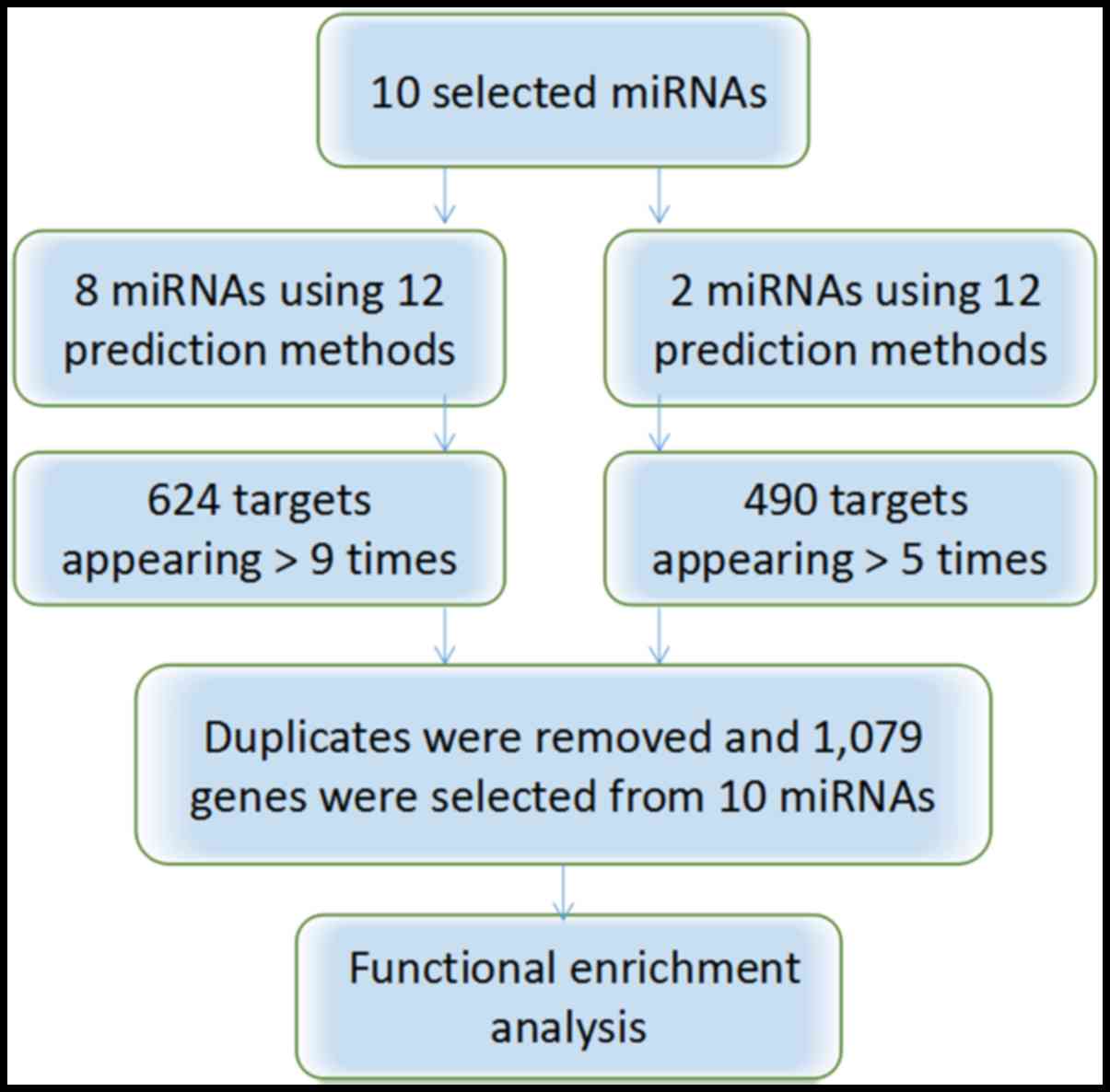
Target prediction
The target genes (n=490) of miR-376b and miR-376c
were selected when they appeared in >5 of the 8 programs
available. The 624 target genes from the remaining 8 miRNAs
(miR-1301, miR-125a, miR-328, miR-29b, miR-454, miR-664a, miR-3613
and miR-126) were selected when they appeared >9 times in the 12
software. The 35 overlapping genes were omitted, leaving a total of
1,079 target genes for further functional enrichment analysis
(Fig. 3).
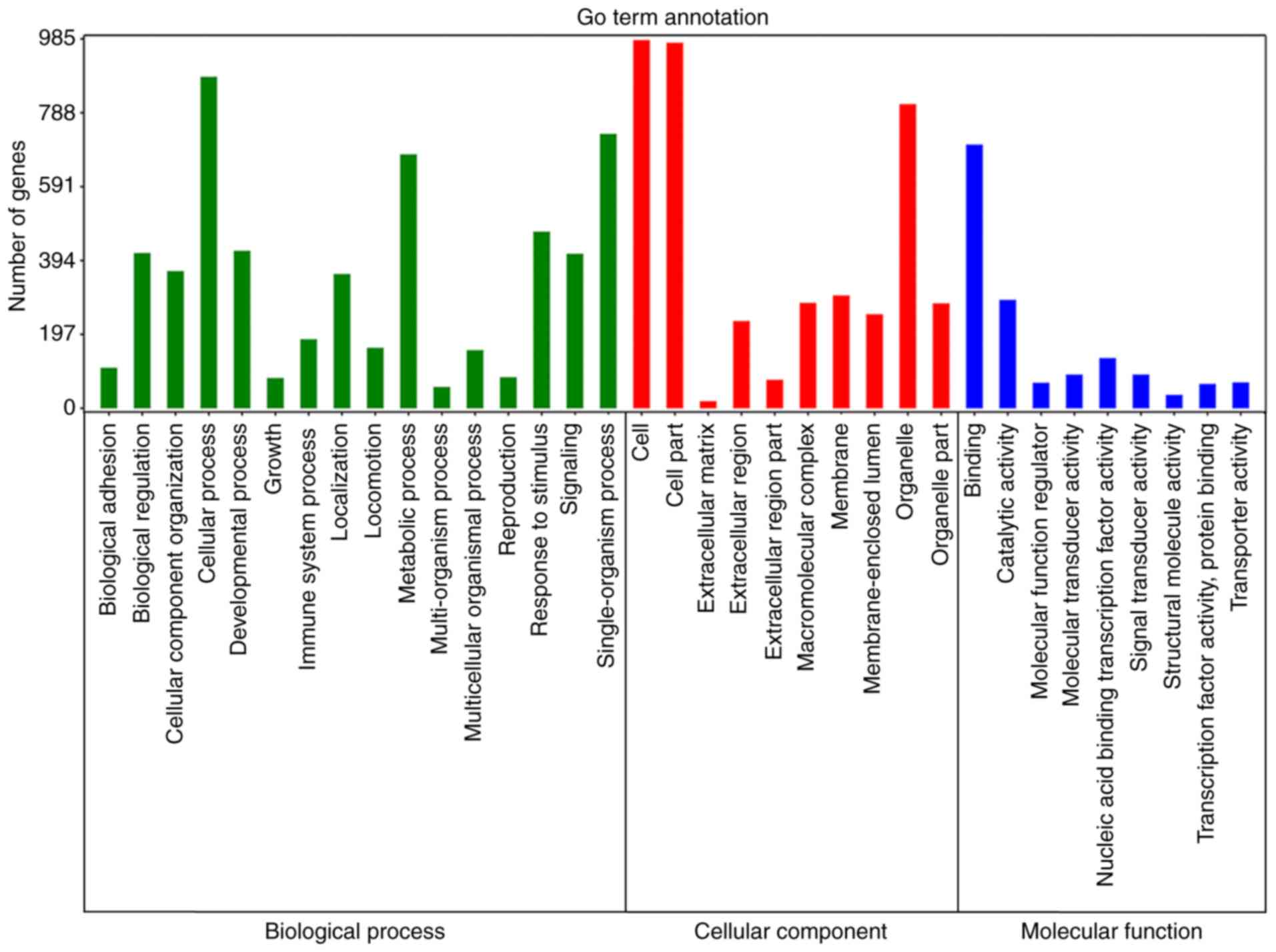
Functional enrichment analysis
Enrichment analyses were then performed to elucidate
the biological function of the target genes of 10 miRNAs. GO
analysis revealed that 52 signaling pathways were significantly
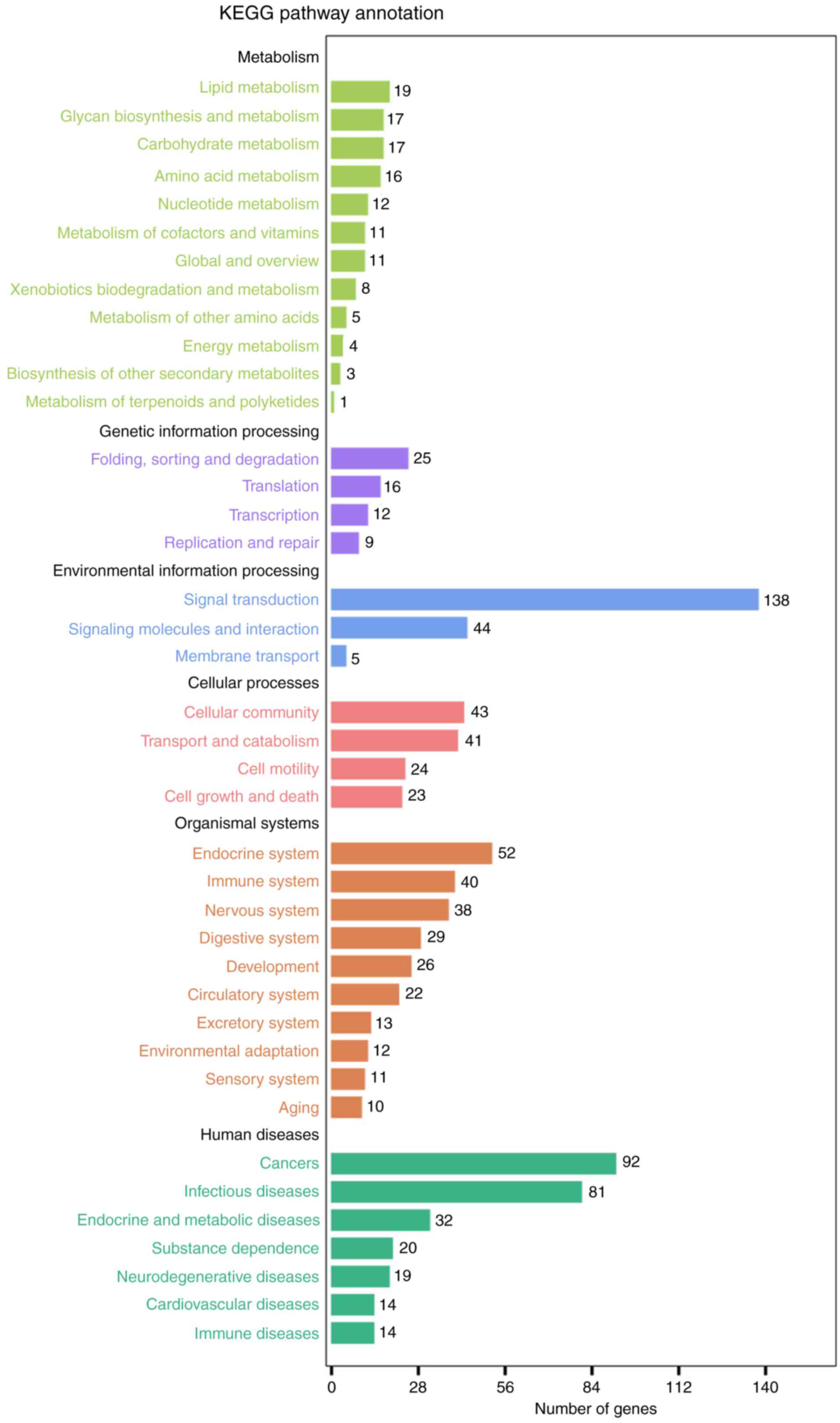
enriched for BP, 20 for CC and 22 for MF (Fig. 4; Table III). KEGG analysis demonstrated
that the top pathway in the KEGG_A_class was ‘Human Diseases’, and
in KEGG_B_class it was ‘Cancers’, of which, several pathways were
closely associated, including ‘Pathways in cancer’, ‘Chronic
myeloid leukemia’, ‘Glioma’ and ‘microRNAs in cancer’ (Fig. 5; Table IV).
 | Table III.Enriched Gene Ontology terms for the
target genes of the top 10 microRNAs. |
Table III.
Enriched Gene Ontology terms for the
target genes of the top 10 microRNAs.
| # | Pathway ID | Pathway
description | P-value | FDR |
|---|
|
| Biological
process |
|
|
|
| 1 | GO:0032502 | Developmental
process |
7.40×10−13 |
6.95×10−11 |
| 2 | GO:0048856 | Anatomical
structure development |
8.33×10−13 |
6.95×10−11 |
| 3 | GO:0009653 | Anatomical
structure morphogenesis |
1.96×10−11 |
1.09×10−9 |
| 4 | GO:0044767 | Single-organism
developmental process |
2.17×10−9 |
9.07×10−8 |
| 5 | GO:0030154 | Cell
differentiation |
3.52×10−9 |
1.17×10−7 |
|
| Cellular
component |
|
|
|
| 1 | GO:0005622 | Intracellular |
2.43×10−12 |
9.20×10−11 |
| 2 | GO:0044424 | Intracellular
part |
2.92×10−12 |
9.20×10−11 |
| 3 | GO:0005634 | Nucleus |
7.69×10−11 |
1.61×10−9 |
| 4 | GO:0043227 | Membrane-bounded
organelle |
4.37×10−9 |
5.51×10−8 |
| 5 | GO:0043231 | Intracellular
membrane-bounded organelle |
4.37×10−9 |
5.51×10−8 |
|
| Molecular
function |
|
|
|
| 1 | GO:0005488 | Binding |
6.53×10−25 |
4.11×10−23 |
| 2 | GO:0001071 | Nucleic acid
binding transcription factor activity |
1.85×10−16 |
5.82×10−15 |
| 3 | GO:0043167 | Ion binding |
1.25×10−15 |
2.62×10−14 |
| 4 | GO:0003677 | DNA binding |
1.75×10−15 |
2.75×10−14 |
| 5 | GO:0003676 | Nucleic acid
binding |
1.02×10−10 |
9.16×10−10 |
 | Table IV.Kyoto Encyclopedia of Genes and
Genomes pathway terms for the target genes of the top 10
microRNAs. |
Table IV.
Kyoto Encyclopedia of Genes and
Genomes pathway terms for the target genes of the top 10
microRNAs.
| Pathway ID | KEGG_A_class | KEGG_B_class | Pathway | P-value | Q-value |
|---|
| ko05200 | Human diseases | Cancers | Pathways in
cancer |
4.79×10−6 |
5.40×10−4 |
| ko05220 | Human diseases | Cancers | Chronic myeloid
leukemia |
6.50×10−6 |
5.40×10−4 |
| ko05214 | Human diseases | Cancers | Glioma |
9.60×10−6 |
5.40×10−4 |
| ko05206 | Human diseases | Cancers | MicroRNAs in
cancer |
1.04×10−5 |
5.40×10−4 |
| ko04010 | Environmental
information processing | Signal
transduction | MAPK signaling
pathway |
1.04×10−5 |
5.40×10−4 |
| ko05212 | Human diseases | Cancers | Pancreatic
cancer |
3.90×10−5 |
1.47×10−3 |
| ko05205 | Human diseases | Cancers | Proteoglycans in
cancer |
3.97×10−5 |
1.47×10−3 |
| ko05215 | Human diseases | Cancers | Prostate
cancer |
7.69×10−5 |
2.49×10−3 |
| ko04540 | Cellular
processes | Cellular
commiunity | Gap junction |
1.11×10−4 |
3.20×10−3 |
| ko04810 | Cellular
processes | Cell motility | Regulation of actin
cytoskeleton |
3.36×10−4 |
8.70×10−3 |
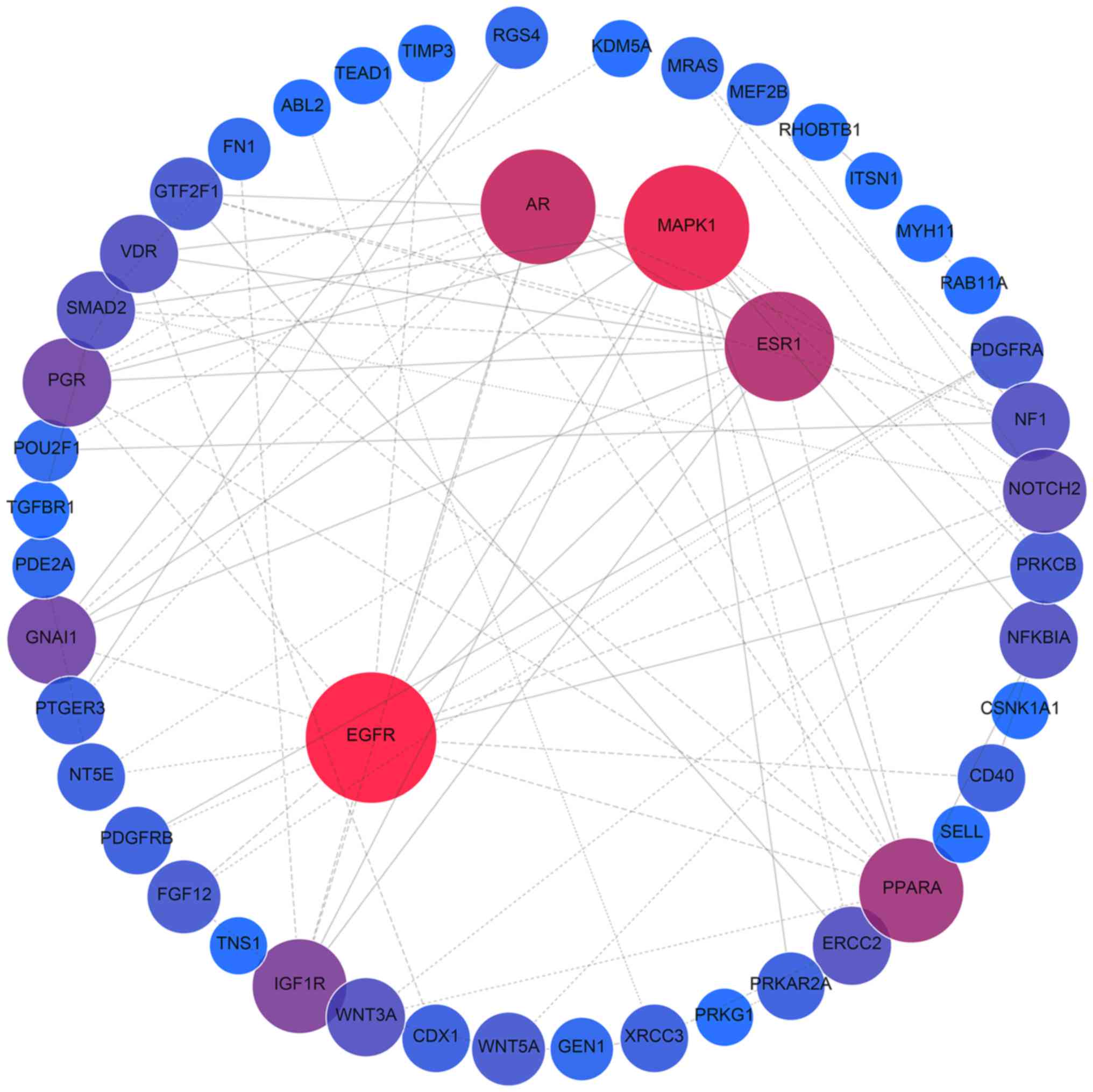
PPI network analysis of all target genes using the
STRING database revealed that 854 interactions were involved, with
combined scores of no less than 0.9. Networks were also constructed
for miR-376b and miR-376c (Fig.
6), and the remaining 8 miRNAs (Fig. 7), as different predicting programs
were utilized. Nodes with ≥20 degrees were defined as hub genes,
including ubiquitin C (UBC; n=132), mothers against decapentaplegic
homolog 4 (SMAD4; n=25), mitogen-activated protein kinase 1 (MAPK1;
n=24), androgen receptor (AR; n=23), estrogen receptor 1 (ESR1;
n=21), HRas proto-oncogene (HRAS; n=20) and epidermal growth factor
receptor (EGFR; n=20). Sub-networks comprised of hub genes and
their corresponding interactions were extracted from the target
networks, as depicted in Fig.
8.
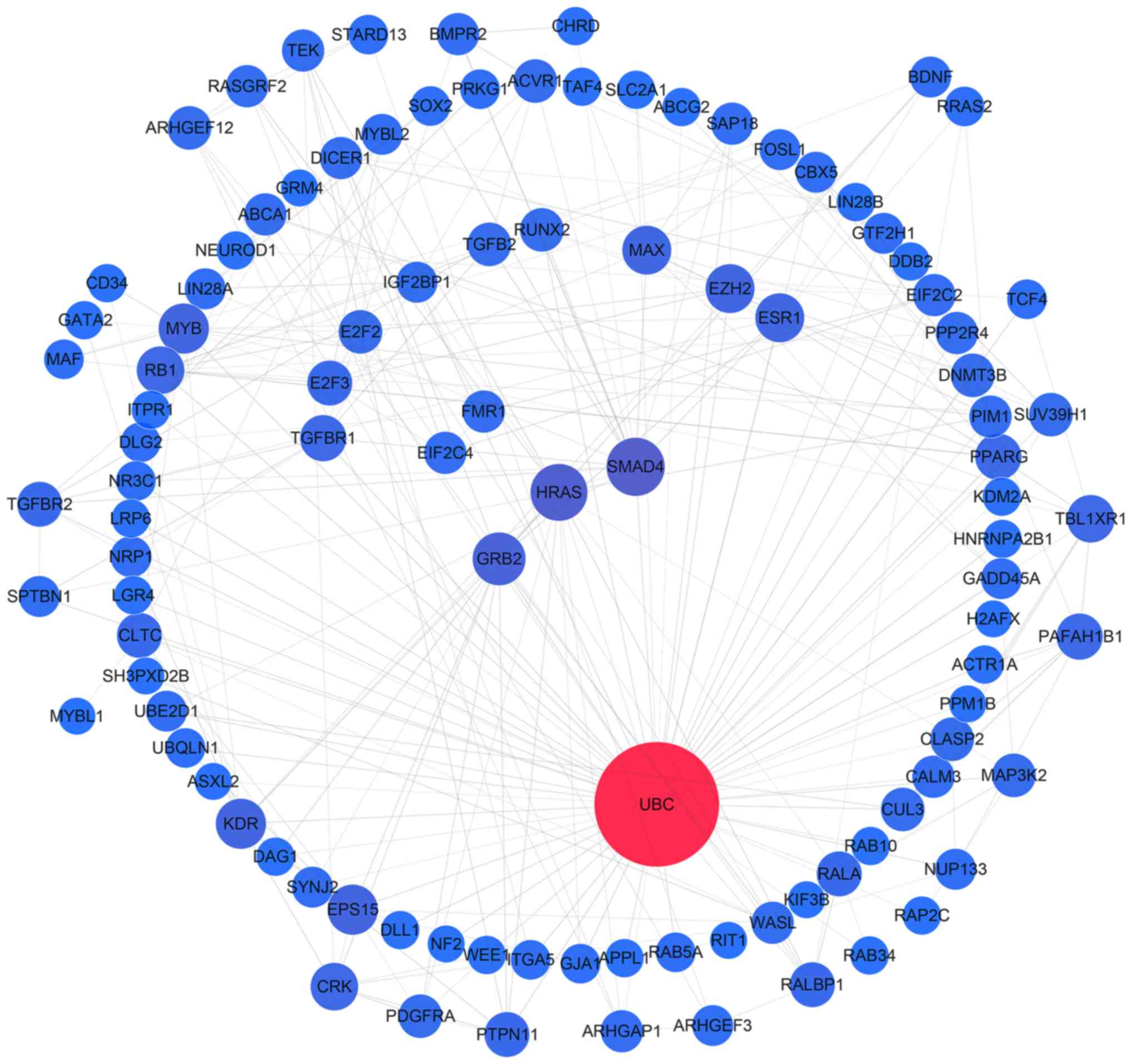 | Figure 7.Protein-protein interaction network
analysis of the target genes of miR-1301, miR-125a, miR-328,
miR-29b, miR-454, miR-664a, miR-3613 and miR-126. Gene interactions
were constructed using the STRING online database (string-db.org) and Cytoscape (version 3.4.0;
www.cytoscape.org/). Network nodes
represent proteins and edges represent protein-protein
associations. Predicted target genes (n=624) of the remaining 8
miRNAs (miR-1301, miR-125a, miR-328, miR-29b, miR-454, miR-664a,
miR-3613 and miR-126) were analyzed. miRNA/miR, microRNA. |
Validation of hub gene expression by
immunochemistry
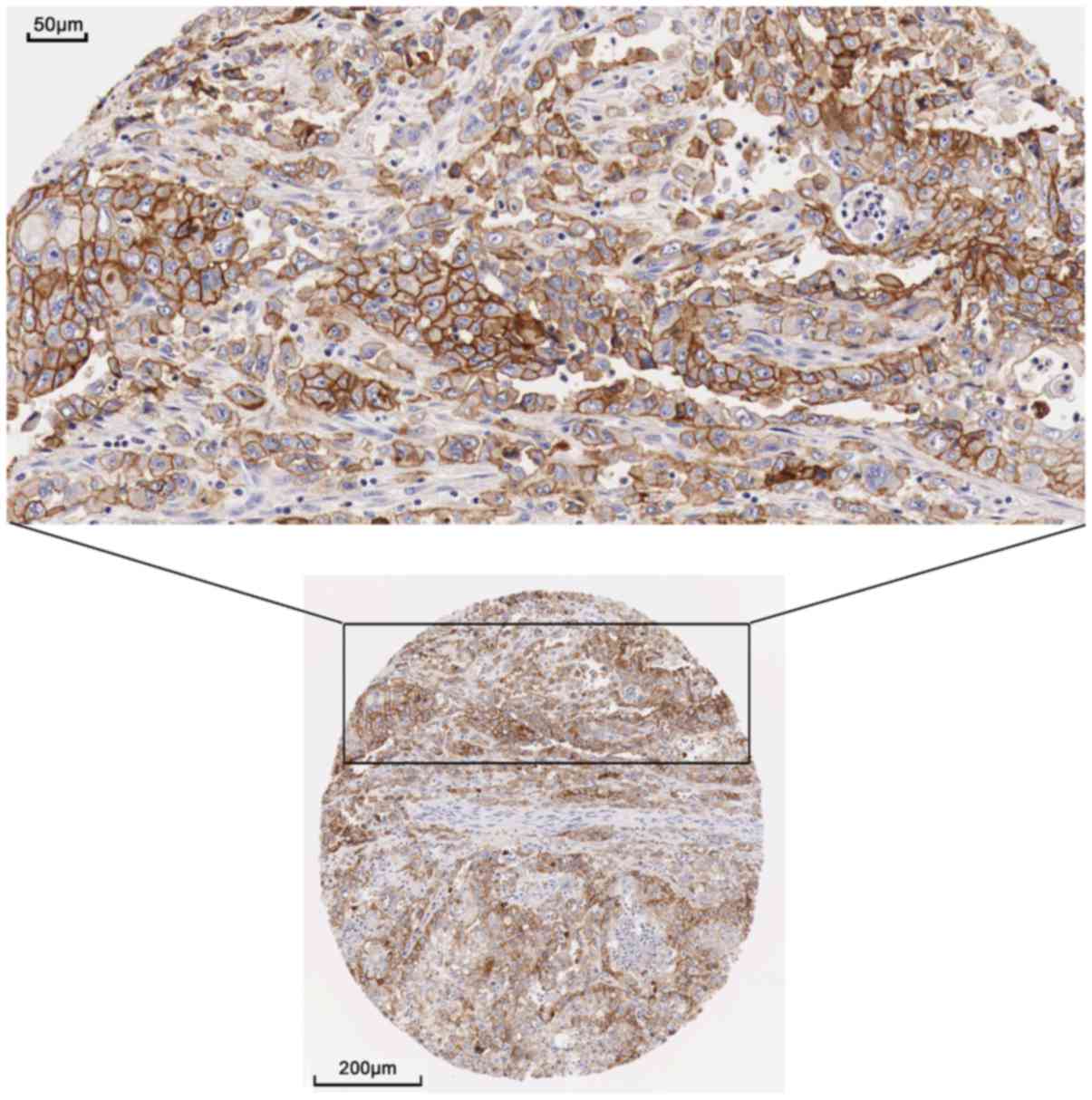
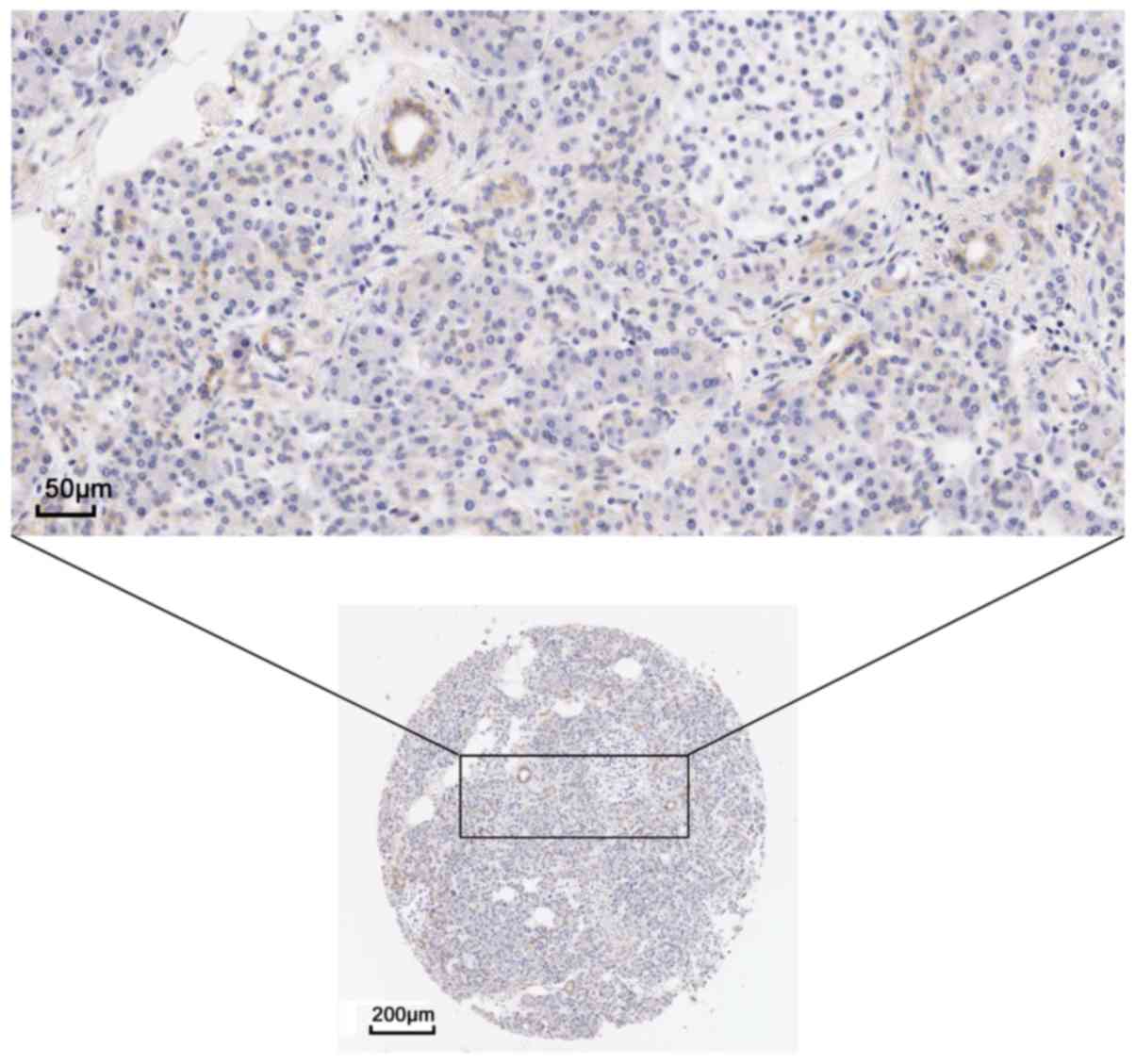
The present study examined the protein expression of
the hub genes. THPA images revealed a markedly high expression of
EGFR could be observed using the CAB000035 antibody in pancreatic
cancer. In 12 patients, 2 presented high levels of staining, 7 had
medium levels of staining and 3 exhibited low levels of staining;
however, no EGFR expression was detected in the 3 normal
counterparts (Figs. 9 and 10). The remaining protein expression
data were not markedly different in the protein atlas.
Discussion
In the present study, the prognostic significance of
each miRNA was calculated by multivariate cox regression analysis,
and was adjusted to the influence of clinical features and factors
including tumor grade, age and sex. The results identified the top
10 miRNAs with the highest prognostic value which, were selected
for subsequent analysis in PAAD based on the TCGA data. These 10
miRNAs, which, may be able to serve as significant independent
predictors for survival, included miR-1301, miR-125a, miR-376c,
miR-328, miR-376b, miR-29b-2, miR-454, miR-664a, miR-3613 and
miR-126. The miRNA expression data was then separated into high and
low expression groups. The 5-miRNA-pool, which, included the top 5
miRNAs in a combined signature (miR-1301, miR-125a, miR-376c,
miR-328 and miR-376b), had the greatest ability to predict the
prognosis of patients with pancreatic cancer. This 5-miRNA-pool
presented an ideal prognostic value with a HR of 0.139 (95% CI,
0.043–0.443; P<0.001), and may provide novel biomarkers for
clinical application. The potential target genes of the
aforementioned miRNAs were enriched in ‘Pathways in cancer’, the
signaling pathways of ‘Chronic myeloid leukemia’ and ‘Glioma’.
Consequently, among the predicted targets of the relevant miRNAs
for PAAD, UBC, EGFR, MAPK1, ESR1, SMAD4 and AR were considered to
be hub genes in the present study.
miRNAs are gaining more attention in current
research on account of their close association with tumor
progression, particularly in PAAD. Previously, Ma et al
(32) published an evaluated
meta-review based on miRNA expression profiling studies from the
Scopus database, Gene Expression Omnibus, ArrayExpress and PubMed,
using the vote-counting strategy and robust rank aggregation
method. The group revealed that the most significantly upregulated
miRNAs included miR-155, miR-100, miR-21, miR-221, miR-31, miR-143
and miR-23a, and the frequently downregulated miRNAs from the
different approaches were miR-217, miR-148a and miR-375. Notably,
when evaluating the prognostic role of these aberrantly expressed
miRNAs by multivariate analyses, miR-21, miR-31 and miR-375 were
able to act as prognostic biomarkers for PAAD (32). Frampton et al (33) also performed a meta-analysis in
2015 in order to examine the effect of miRNAs on OS and
disease-free survival in PAAD, based on 20 eligible studies with
1,525 patients and assessed via microarrays, reverse
transcription-quantitative polymerase chain reaction or in
situ hybridization. The group confirmed that OS was markedly
shortened in patients with high tumoral miR-21. In addition, the
significant miRNAs, miR-155, miR-203, miR-34a, miR-222 and miR-10b,
were identified, and were thought to be potential prognostic
biomarkers for PAAD (33). Thus,
different miRNAs have previously been shown to have prognostic
value in PAAD.
However, to the best of our knowledge, no report has
been published for the prognostic role of miRNAs based on the
public data from TCGA. TCGA has provided clinicians and researchers
unprecedented access to information regarding various malignancies
through numerous platforms, including exome sequencing, comparative
genomic hybridization arrays, DNA methylation arrays, RNA
sequencing and reverse protein phase arrays, as well as
clinicopathological characteristics (34). The miRNA-seq TCGA data of PAAD has
not been utilized for the investigation of the prognostic role of
miRNAs. In the present study, the prognostic values of all miRNAs
for PAAD available in TCGA were calculated, and the 10 most
efficient miRNAs for prognosis prediction were revealed to be
miR-1301, miR-125a, miR-376c, miR-328, miR-376b, miR-29b, miR-454,
miR-664a, miR-3613, and miR-126. This appears to be a novel finding
as these 10 miRNAs were not reported in the meta-analyses performed
either by Ma et al (32) or
Frampton et al (33). All
10 miRNAs were confirmed as protective factors, with the HR values
ranging from 0.450 to 0.634. Furthermore, among the 10 miRNAs, the
5-miRNA signature, consisting of miR-1301, miR-125a, miR-376c,
miR-328 and miR-376b, were pooled as a combined predictor with a HR
of 0.139. The combination of these 5 miRNAs may provide a novel
tool for clinical application in predicting the prognosis of
patients with PAAD. However, this hypothesis requires further
studies involving larger cohorts and multiple detection methods to
confirm.
Some of the functions and molecular mechanisms
associated with the 10 miRNAs selected in the present study have
been reported previously. For example, miR-1301-3p has been
extensively studied in some cancers. Fang et al (35) concluded that miR-1301-3p may
function as an inhibitor of tumorigenesis, regulating cell
apoptosis through B-cell lymphoma-2 (Bcl-2) and Bcl-xL in
hepatocellular carcinoma HepG2 cells. Lin et al (36) indicated that miR-1301-3p-induced
downregulation of Ran GTPase activating protein 1 may activate the
p73-dependent apoptosis pathway by mediating breakpoint cluster
region-Abelson tyrosine-protein kinase nuclear entrapment, which,
may be a novel strategy for improving the current Imatinib
treatment for chronic myeloid leukemia. Another study demonstrated
that miR-1301-3p was upregulated in prostate cancer cells and
tissues (37). miR-1301-3p
accelerated the G1/S transition to advance cell cycle progression
by impeding p27 expression and increasing Cyclin D1 expression
(37). This study also revealed
that miR-1301-3p targeted protein phosphatase 2 regulatory subunit
Bg, further suppressing tumor proliferation. However, a study
investigating miR-1301-3p in PAAD is currently not available.
Aberrant activation of the EGFR pathway may suppress
tumor progression in retinoblastoma. Zhang et al (38) reported that miR-125a-5p was
involved in the EGFR signaling pathway by targeting tafazzin in
retinoblastoma. However, there is currently no research involving
miR-125a-5p in PAAD. Li et al (39) demonstrated that miR-376b-5p
suppressed angiogenesis in cerebral ischemia and halted
angiogenesis by targeting hypoxia-inducible factor 1α, thereby
mediating the vascular endothelial growth factor A/Notch1 signaling
pathway. It was identified that miR-328-3p was downregulated in
endometrioid endometrial carcinoma, which, was considered an
inhibition factor that affected cancer growth and metastasis
(40). Leidinger et al
(41) also demonstrated that
miR-328-5p was upregulated and confirmed to be relevant in lung
cancer. Mishra et al (42)
identified that miR-454-3p was upregulated in breast cancer, that
it may be closely linked to the ErbB signaling pathway and the
group also delineated the importance of miR-454-3p as a prospective
circulating surrogate molecular signature for the early diagnosis
of breast cancer. Recently, Shao et al (43) demonstrated that the survival of
those with a high expression of miR-454-3p was poorer when compared
with that of those with low miR-454-3p. Wu et al (44) also indicated that BTG
anti-proliferation factor 1 was a direct target gene of miR-454-3p.
The overexpression of miR-454-3p rendered tumor cells sensitive to
radiation (44). In addition,
miR-644a-5p has been shown to promote tumor aggressiveness, at
least partially, via the downregulation of the Akt/glycogen
synthase kinase-3β/β-catenin signaling pathway by targeting paired
like homeodomain 2 in esophageal squamous cell carcinoma (45). Chen et al (46) demonstrated that miR-126-3p was
associated with advanced pathological stage and lymph node
metastasis in patients with lung adenocarcinoma. miR-126-3p is the
most endothelial-specific miRNA that alters vascular integrity and
angiogenesis, and it served as a tumor inhibitor targeting
phophoinositide-3-kinase regulatory subunit 2 in Kaposi's sarcoma
cells (47). Its expression
appeared to be upregulated in colon cancer, as demonstrated by Ji
et al (48). Furthermore,
Suresh et al (49) observed
miR-3613-5p enrichment in an extracellular vesicle subtype.
Extracellular vesicles were involved in the mechanism associated
with the secretion of miRNAs; this enrichment was marked (49). In addition, the miR-29 family,
including miR-29a and miR-29b, is well characterized for their
ability to regulate extracellular matrix proteins in pancreatic β
cells (50). Taken together,
analysis of TCGA data identified certain miRNAs that were connected
with the prognosis of PAAD. As the efficacy of a single biomarker
is limited, the pooled signature may be employed instead. Notably,
the results of the present study demonstrated that the pooled
signature, containing 5 miRNAs, had a greater ability to predict
survival. However, the associations between the aforementioned 10
miRNAs and PAAD have not been elucidated. Therefore, the clinical
values, as well as the underlying molecular mechanisms, require
validation with further studies investigating PAAD.
The present study also attempted to assess the
prospective molecular mechanisms of these miRNAs in PAAD. The
progression of PAAD tumors is a multi-step process induced by a
succession of genetic and epigenetic alterations, that result in
the progression of normal cells to cancer cells. miRNAs are
involved in this progression by combining the target genes, and
then suppressing or decreasing gene expression (51). In the present study, by performing
enrichment and function analyses of GO, the target genes of 10
miRNAs were revealed to be involved in a number of crucial
biological processes, which, were consistent with developmental
processes. From the KEGG pathway analysis, it was concluded that
the ‘Pathways in cancer’ of ‘Human Diseases’ and ‘MAPK signaling
pathway’ of ‘Environmental Information Processing’ were the most
important signaling pathways in PAAD. The hub genes obtained from
the interaction network were assessed in order to comprehensively
understand the key role of tumor procession. UBC, SMAD4, MAPK1, AR,
ESR1, HRAS and EGFR were identified as hub genes, all of which were
shown to participate in ‘Signal transduction’ of ‘Environmental
Information Processing’. However, the exact molecular mechanism
associated with miRNAs in PAAD requires further study, with in
vitro and in vivo experiments.
Recently, Zhou et al (52) reported that a miRNA signature with
13 miRNAs (miR-103-2, miR-125a, miR-126, miR-328, miR-340, miR-361,
miR-374b, miR-454, miR-627, miR-664, miR-193b, miR-21 and miR-584)
had the potential to be an independent marker for predicting
survival in patients with PAAD, which, was also based on TCGA data.
Consistent with the results of the present study, miR-125a,
miR-328, miR-126 and miR-454 were also identified by Zhou et
al (52). However, unlike the
present study, Zhou et al (52) did not assess the potential
signaling pathways and network of the target genes. The present
study conducted bioinformatics analysis of the 10 miRNAs, predicted
their target genes and evaluated their roles in the progression of
pancreatic cancer. Furthermore, the prognostic information of the
patients with pancreatic cancer involved in the present study was
updated. In addition, the 5-miRNA signature of the present study
had high prognostic significance in clinical practice, and it is
more convenient to measure 5 indicators in clinical operations.
Also, Zhou et al (52)
performed univariate regression analysis in order to investigate
their prognostic significance. The 5 miRNAs in the present study
differ from those identified by Zhou et al (52) as the prognostic value of each miRNA
in the present study was calculated using a multivariate COX
regression model which, was adjusted by tumor grade, age and
sex.
The present study did have some limitations.
Firstly, the miRNA expression data published on TCGA was only
collected from pancreatic tumor tissues which, may not represent
the expression levels in serum or plasma. Furthermore, the sample
size of the PAAD TCGA dataset was quite small. In addition, the
10-miRNA signature investigated has not been validated by other
detection methods. Although the clinical studies involving the
miRNA signature did have significant prognostic value based on
TCGA, the functions of the 10 miRNAs are still poorly understood.
Thus, well-designed studies are required to investigate the
molecular function of these miRNAs in PAAD.
In conclusion, the results of the present study
indicated that the 10 miRNAs assessed may be effective markers for
predicting the prognosis of PAAD, based on TCGA data. A 5-miRNA
signature was also identified which, may be an important predictor
of poor survival in patients with PAAD as it exhibited a HR of
0.139. However, these results require further verification.
Acknowledgements
The present study was supported by the Fund of
Guangxi Natural Science Foundation (grant nos. 2014GXNSFBA118167
and 2016GXNSFBA380039), the Promoting Project of Basic Capacity for
University Young and Middle-aged Teachers in Guangxi (grant no.
KY2016LX031), the Fund of the Innovation Project of Guangxi
Graduate Education (2016) and Natural Science Foundation of China
(NSFC 81560448). The authors would also like to acknowledge TCGA
Research Network for generating the TCGA datasets (cancergenome.nih.gov/).
References
|
1
|
Yu J, Ohuchida K, Mizumoto K, Fujita H,
Nakata K and Tanaka M: MicroRNA miR-17-5p is overexpressed in
pancreatic cancer, associated with a poor prognosis, and involved
in cancer cell proliferation and invasion. Cancer Biol Ther.
10:748–757. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Z, Li J, Chen X, Duan W, Ma Q and Li
X: Disrupting the balance between tumor epithelia and stroma is a
possible therapeutic approach for pancreatic cancer. Med Sci Monit.
20:2002–2006. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang S, Yang Z, Li D, Miao X, Yang L, Zou
Q and Yuan Y: The clinical and pathological significance of
Nectin-2 and DDX3 expression in pancreatic ductal adenocarcinomas.
Dis Markers. 2015:3795682015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee SH, Kang CM, Kim H, Hwang HK, Song SY,
Seong J, Kim MJ and Lee WJ: Pathological complete remission of
pancreatic cancer following neoadjuvant chemoradiation therapy; not
the end of battles. Medicine (Baltimore). 94:e21682015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen Y, Wang X, Ke N, Mai G and Liu X:
Inferior mesenteric vein serves as an alternative guide for
transection of the pancreatic body during pancreaticoduodenectomy
with concomitant vascular resection: A comparative study evaluating
perioperative outcomes. Eur J Med Res. 19:422014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang YF, Cao XH, Bao CE and Wan X:
Concurrent radiotherapy with oral fluoropyrimidine versus
gemcitabine in locally advanced pancreatic cancer: A systematic
review and meta-analysis. Onco Targets Ther. 8:3315–3322.
2015.PubMed/NCBI
|
|
7
|
Hezel AF, Kimmelman AC, Stanger BZ,
Bardeesy N and Depinho RA: Genetics and biology of pancreatic
ductal adenocarcinoma. Genes Dev. 20:1218–1249. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ali S, Dubaybo H, Brand RE and Sarkar FH:
Differential expression of MicroRNAs in tissues and plasma
co-exists as a biomarker for pancreatic cancer. J Cancer Sci Ther.
7:336–346. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Loosen SH, Neumann UP, Trautwein C,
Roderburg C and Luedde T: Current and future biomarkers for
pancreatic adenocarcinoma. Tumour Biol. 39:10104283176922312017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pillai RS: MicroRNA function: Multiple
mechanisms for a tiny RNA? RNA. 11:1753–1761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiao J, Peng F, Yu C, Wang M, Li X, Li Z,
Jiang J and Sun C: microRNA-137 modulates pancreatic cancer cells
tumor growth, invasion and sensitivity to chemotherapy. Int J Clin
Exp Pathol. 7:7442–7450. 2014.PubMed/NCBI
|
|
13
|
Lin MS, Chen WC, Huang JX, Gao HJ and
Sheng HH: Aberrant expression of microRNAs in serum may identify
individuals with pancreatic cancer. Int J Clin Exp Med.
7:5226–5234. 2014.PubMed/NCBI
|
|
14
|
Kai Y, Qiang C, Xinxin P, Miaomiao Z and
Kuailu L: Decreased miR-154 expression and its clinical
significance in human colorectal cancer. World J Surg Oncol.
13:1952015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hou LJ and Zhai JJ: Aberrant expression
profile of translationally controlled tumor protein and
tumor-suppressive microRNAs in cervical cancer. J BUON.
20:1504–1509. 2015.PubMed/NCBI
|
|
16
|
Lu Y, Hu J, Sun W, Li S, Deng S and Li M:
MiR-29c inhibits cell growth, invasion, and migration of pancreatic
cancer by targeting ITGB1. Onco Targets Ther. 9:99–109.
2016.PubMed/NCBI
|
|
17
|
Dong Q, Li C, Che X, Qu J, Fan Y, Li X, Li
Y, Wang Q, Liu Y, Yang X and Qu X: MicroRNA-891b is an independent
prognostic factor of pancreatic cancer by targeting Cbl-b to
suppress the growth of pancreatic cancer cells. Oncotarget.
7:82338–82353. 2016.PubMed/NCBI
|
|
18
|
Li BS, Liu H and Yang WL: Reduced
miRNA-218 expression in pancreatic cancer patients as a predictor
of poor prognosis. Genet Mol Res. 14:16372–16378. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brunetti O, Russo A, Scarpa A, Santini D,
Reni M, Bittoni A, Azzariti A, Aprile G, Delcuratolo S, Signorile
M, et al: MicroRNA in pancreatic adenocarcinoma:
Predictive/prognostic biomarkers or therapeutic targets?
Oncotarget. 6:23323–23341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Sun Y, Wu H, Yu S, Zhang L, Meng
Y, Liu M, Yang H, Liu P, Mao X, et al: Elevated miR-483-3p
expression is an early event and indicates poor prognosis in
pancreatic ductal adenocarcinoma. Tumour Biol. 36:9447–9456. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bai Z, Sun J, Wang X, Wang H, Pei H and
Zhang Z: MicroRNA-153 is a prognostic marker and inhibits cell
migration and invasion by targeting SNAI1 in human pancreatic
ductal adenocarcinoma. Oncol Rep. 34:595–602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi H, Chen J, Li Y, Li G, Zhong R, Du D,
Meng R, Kong W and Lu M: Identification of a six microRNA signature
as a novel potential prognostic biomarker in patients with head and
neck squamous cell carcinoma. Oncotarget. 7:21579–21590. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang M, Wen TF, He LH, Li C, Zhu WJ and
Trishul NM: A six-microRNA set as prognostic indicators for bile
duct cancer. Int J Clin Exp Med. 8:17261–17270. 2015.PubMed/NCBI
|
|
24
|
Zheng H, Guo X, Tian Q, Li H and Zhu Y:
Distinct role of Tim-3 in systemic lupus erythematosus and clear
cell renal cell carcinoma. Int J Clin Exp Med. 8:7029–7038.
2015.PubMed/NCBI
|
|
25
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang SW, Wang J and Pan J: Identification
of altered pathways in hypertrophic cardiomyopathy based on
combined data of protein-protein interactions and molecular
pathways. Genet Mol Res. 15:2016.
|
|
28
|
Jiang M, Zeng Q, Dai S, Liang H, Dai F,
Xie X, Lu K and Gao C: Comparative analysis of hepatocellular
carcinoma and cirrhosis gene expression profiles. Mol Med Rep.
15:380–386. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gu Y, Lu L, Wu L, Chen H, Zhu W and He Y:
Identification of prognostic genes in kidney renal clear cell
carcinoma by RNA-seq data analysis. Mol Med Rep. 15:1661–1667.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Uhlen M, Oksvold P, Fagerberg L, Lundberg
E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S,
et al: Towards a knowledge-based human protein atlas. Nat
Biotechnol. 28:1248–1250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma MZ, Kong X, Weng MZ, Cheng K, Gong W,
Quan ZW and Peng CH: Candidate microRNA biomarkers of pancreatic
ductal adenocarcinoma: Meta-analysis, experimental validation and
clinical significance. J Exp Clin Cancer Res. 32:712013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Frampton AE, Krell J, Jamieson NB, Gall
TM, Giovannetti E, Funel N, Mato Prado M, Krell D, Habib NA,
Castellano L, et al: microRNAs with prognostic significance in
pancreatic ductal adenocarcinoma: A meta-analysis. Eur J Cancer.
51:1389–1404. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng PF, Dummer R and Levesque MP: Data
mining The Cancer Genome Atlas in the era of precision cancer
medicine. Swiss Med Wkly. 145:w141832015.PubMed/NCBI
|
|
35
|
Fang L, Yang N, Ma J, Fu Y and Yang GS:
microRNA-1301-mediated inhibition of tumorigenesis. Oncol Rep.
27:929–934. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lin TY, Chen KC, Liu HJ, Liu AJ, Wang KL
and Shih CM: MicroRNA-1301-Mediated RanGAP1 Downregulation Induces
BCR-ABL nuclear entrapment to enhance imatinib efficacy in chronic
myeloid leukemia cells. PLoS One. 11:e01562602016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bi D, Ning H, Liu S, Que X and Ding K:
miR-1301 promotes prostate cancer proliferation through directly
targeting PPP2R2C. Biomed Pharmacother. 81:25–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Xue C, Zhu X, Zhu X, Xian H and
Huang Z: Suppression of microRNA-125a-5p upregulates the TAZ-EGFR
signaling pathway and promotes retinoblastoma proliferation. Cell
Signal. 28:850–860. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li LJ, Huang Q, Zhang N, Wang GB and Liu
YH: miR-376b-5p regulates angiogenesis in cerebral ischemia. Mol
Med Rep. 10:527–535. 2014.PubMed/NCBI
|
|
40
|
Xiong H, Li Q, Liu S, Wang F, Xiong Z,
Chen J, Chen H, Yang Y, Tan X, Luo Q, et al: Integrated microRNA
and mRNA transcriptome sequencing reveals the potential roles of
miRNAs in stage I endometrioid endometrial carcinoma. PLoS One.
9:e1101632014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Leidinger P, Brefort T, Backes C, Krapp M,
Galata V, Beier M, Kohlhaas J, Huwer H, Meese E and Keller A:
High-throughput qRT-PCR validation of blood microRNAs in non-small
cell lung cancer. Oncotarget. 7:4611–4623. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mishra S, Srivastava AK, Suman S, Kumar V
and Shukla Y: Circulating miRNAs revealed as surrogate molecular
signatures for the early detection of breast cancer. Cancer Lett.
369:67–75. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shao N, Wang L, Xue L, Wang R and Lan Q:
Plasma miR-454-3p as a potential prognostic indicator in human
glioma. Neurol Sci. 36:309–313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu X, Ding N, Hu W, He J, Xu S, Pei H, Hua
J, Zhou G and Wang J: Down-regulation of BTG1 by miR-454-3p
enhances cellular radiosensitivity in renal carcinoma cells. Radiat
Oncol. 9:1792014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang JX, Chen ZH, Xu Y, Chen JW, Weng HW,
Yun M, Zheng ZS, Chen C, Wu BL, Li EM, et al: Downregulation of
MicroRNA-644a promotes esophageal squamous cell carcinoma
aggressiveness and stem cell-like phenotype via dysregulation of
PITX2. Clin Cancer Res. 23:298–310. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen Q, Hu H, Jiao D, Yan J, Xu W, Tang X,
Chen J and Wang J: miR-126-3p and miR-451a correlate with
clinicopathological features of lung adenocarcinoma: The underlying
molecular mechanisms. Oncol Rep. 36:909–917. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu XJ, Zhao ZF, Kang XJ, Wang HJ, Zhao J
and Pu XM: MicroRNA-126-3p suppresses cell proliferation by
targeting PIK3R2 in Kaposi's sarcoma cells. Oncotarget.
7:36614–36621. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ji H, Chen M, Greening DW, He W, Rai A,
Zhang W and Simpson RJ: Deep sequencing of RNA from three different
extracellular vesicle (EV) subtypes released from the human LIM1863
colon cancer cell line uncovers distinct miRNA-enrichment
signatures. PLoS One. 9:e1103142014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Suresh R, Sethi S, Ali S, Giorgadze T and
Sarkar FH: Differential expression of microRNAs in papillary
thyroid carcinoma and their role in racial disparity. J Cancer Sci
Ther. 7:145–154. 2015.PubMed/NCBI
|
|
50
|
Pullen TJ, da Silva Xavier G, Kelsey G and
Rutter GA: miR-29a and miR-29b contribute to pancreatic
beta-cell-specific silencing of monocarboxylate transporter 1
(Mct1). Mol Cell Biol. 31:3182–3194. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sulpizio S, Franceschini N, Piattelli A,
Di Sebastiano P, Innocenti P and Selvaggi F: Cathepsins and
pancreatic cancer: The 2012 update. Pancreatology. 12:395–401.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou X, Huang Z, Xu L, Zhu M, Zhang L,
Zhang H, Wang X, Li H, Zhu W, Shu Y and Liu P: A panel of 13-miRNA
signature as a potential biomarker for predicting survival in
pancreatic cancer. Oncotarget. 7:69616–69624. 2016.PubMed/NCBI
|