Introduction
Renal cell carcinoma (RCC) is the most common type
of kidney tumor and the most lethal urologic tumor (1). Despite increased early detection of
RCC and more frequent surgery, the prognosis remains poor for
locally advanced and metastatic cases of RCC (2,3).
Therefore, understanding the underlying molecular mechanisms of RCC
progression is required, for the development of a novel therapy
against advanced RCC.
MicroRNAs (miRNAs/miRs) are a class of small, single
stranded, non-coding RNA molecules of 19–25 nucleotides that bind
to the 3′ untranslated regions (UTR) of specific target mRNAs,
leading to direct mRNA degradation or translational repression
(4). miRNAs may regulate multiple
target genes and pathways simultaneously and are involved in
regulation of various biological processes, including cell growth,
migration, invasion apoptosis, metabolism and cellular
differentiation (5,6). Accumulating evidence has suggested
that miRNAs may function as tumor suppressors or oncogenes in
various types of cancer by directly targeting tumor suppressor
genes or oncogenes (7,8). Recent studies demonstrated that a
number of miRNAs serve a crucial role in modulating RCC progression
(9–11).
miR-212, located at chromosome 17p13.3 (12), has been demonstrated to be
deregulated in various types of human cancer, including pancreatic
cancer (13), lung cancer
(14), prostate cancer (15), cervical cancer (16), glioblastoma (17), ovarian cancer (18) and hepatocellular carcinoma
(19). However, the clinical
significance of miR-212 and the associated molecular signaling
pathways involved in the progression of RCC remain poorly
understood. Therefore, the aim of the present study was to
investigate the role and underlying molecular mechanism of miR-212
in RCC. Results revealed that miR-212 was downregulated in RCC
tissues and cell lines, and that miR-212 functioned as a tumor
suppressor in RCC by suppressing cell proliferation, migration and
invasion, and inducing cell apoptosis. A direct target, forkhead
box protein A1 (FOXA1), was confirmed, which mediated the effects
of miR-212. These results may aid the development of an effective
therapeutic strategy for RCC.
Materials and methods
Clinical samples
A total of 48 RCC samples and adjacent non-tumor
tissues (ANT; >3 cm from the margin of resection) were collected
from patients including 20 males and 28 females, who underwent
resection of their primary RCC in the Department of General Surgery
at The Affiliated Hospital of Changchun University of Chinese
Medicine (Changchun, China) between April 2012 and December 2014.
Following surgical resection, all samples were immediately stored
in liquid nitrogen until RNA or protein extraction. The
clinicopathological data are listed in Table I. No patients received chemotherapy
or radiotherapy prior to surgery. All samples were obtained with
informed consent from each patient and the study was approved by
the Medicine Ethics Committee of Changchun University of Chinese
Medicine.
 | Table I.Correlation between
clinicopathological features and miR-212 expression in RCC
tissues. |
Table I.
Correlation between
clinicopathological features and miR-212 expression in RCC
tissues.
|
|
| miR-212
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | No. of cases | Low, n (%) | High, n (%) | P-value |
|---|
| Age |
|
|
| 0.714 |
| <55
years old | 23 | 12 (52.2) | 11 (47.8) |
|
| ≥55 years
old | 25 | 14 (56.0) | 11 (44.0) |
|
| Gender |
|
|
| 0.623 |
| Male | 21 | 11 (52.4) | 10 (47.6) |
|
|
Female | 27 | 15 (55.6) | 12 (44.4) |
|
| TNM stage |
|
|
| <0.01 |
|
T1-T2 | 34 | 13 (38.2) | 21 (71.8) |
|
|
T3-T4 | 14 | 13 (92.9) | 1 (7.1) |
|
| Tumor size |
|
|
| <0.01 |
| <5
cm | 31 | 12 (38.7) | 19 (61.2) |
|
| ≥5
cm | 17 | 14 (82.4) | 3
(17.6) |
|
| Lymph node
metastasis |
|
|
| <0.01 |
| No | 35 | 13 (37.1) | 24 (62.9) |
|
|
Yes | 13 | 13 (100) | 0
(0) |
|
Cell lines and transfection
A total of three clear cell RCC cell lines (786-O,
ACHN and Caki-1) (20,21), a papillary RCC cell line Caki-2
(22) and a human renal proximal
tubule epithelial cell line (HK-2) were purchased from the Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China), and were cultured in Dulbecco's modified Eagle medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal calf serum (FCS; Invitrogen; Thermo
Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 IU/ml
streptomycin at 37°C in an atmosphere containing 5%
CO2.
An miR-212 mimic (5′-CCGGCACUGACCUCUGACAAU-3′) and
corresponding negative control (Ctrl; 5′-GUCCTUGCUCGAGCGAGGUGA-3′)
mimic were purchased from GeneCopoeia, Inc. (Rockville, MD, USA).
Plasmids carrying human FOXA1 were purchased from OriGene
Technologies, Inc. (Rockville, MD, USA; cat. no. SC108256). Cells
were seeded in a six-well plate at a density of 1×103
cells/well and transfected with miR-212 (100 nM), miR-Ctrl (100 nM)
or FOXA1 overexpression plasmid (100 ng) using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol, and transfected cells were cultured for
1–3 days before the cell proliferation, migration, invasion and
apoptosis assays.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cultured cells and
frozen fresh tissues using TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.). To quantify miR-212, cDNA was synthesized using
the TaqMan miRNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.), and quantified using the TaqMan Human MicroRNA
Assay kit (Thermo Fisher Scientific, Inc.) and the ABI 7900
Sequence Detection system (Life Technologies; Thermo Fisher
Scientific, Inc.). Primers for miR-212 and U6 (GeneCopoeia, Inc.)
were used as follows: miR-212 forwards, 5′-CGCTAACAGTCTCCAGTC-3′
and reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forwards,
5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′. U6 was used as an internal control. U6
small nuclear RNA was used as an internal control. For detection of
FOXA1, total RNA was reverse transcribed to cDNA using the Primer
Script RT reagent kit (Takara Biotechnology Co., Ltd., Dalian,
China), and quantified on the ABI 7900HT Fast Real-Time PCR system
using the SYBR Green PCR Master mix (Takara Biotechnology Co.,
Ltd.) using primers specific for FOXA1 and GAPDH, as described
previously (23). GAPDH was used
as an internal control. The following PCR conditions were used:
Denaturation at 94°C for 3 min, followed by 40 cycles of
amplification (denaturation at 94°C for 15 sec, annealing at 60°C
for 30 sec and extension at 72°C for 45 sec) The comparative
2−∆∆Cq method was used for relative quantification
(24).
Cell proliferation
Transfected cells were seeded in 24-well plates
(3,000 cells/well), then cell proliferation was determined at
different time points (24, 48 and 72 h) by incubating cells with
0.5 mg/ml MTT reagent (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) for 1 h, and dissolved in dimethyl sulfoxide
(Sigma-Aldrich; Merck KGaA) for 10 min at room temperature. The
absorbance of samples was measured using a wavelength of 490 nm
with a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Cell migration and invasion assay
The migratory and invasive capability of RCC cells
was determined using Transwell chambers of diameter 6.5 mm with an
8-µm membrane (Corning Incorporated, Corning, NY, USA). A total of
3×104 transfected cells suspended in serum-free medium
were added to the upper chamber without Matrigel (for migration) or
coated with Matrigel (for invasion), and DMEM containing 10% FCS
was used as an attractant in the lower chamber. After 24 h (for
migration) or 48 h (for invasion), cells remaining in the upper
chamber were removed with a sterile swab, whereas migratory or
invasive cells in the bottom chamber were fixed with 70% ethanol
for 30 min at room temperature (20–25°C) and stained with 0.2%
crystal violet (Sigma-Aldrich; Merck KGaA) for 10 min at room
temperature (20–25°C). Cell numbers were counted in five randomly
selected microscopic fields under a X71 inverted light microscope
(Olympus Corporation, Tokyo, Japan) at ×200 magnification.
Cell apoptosis assay
Cell apoptosis was determined using an
Annexin-V-FLUOS Staining kit (Roche Applied Science, Penzberg,
Germany) under a fluorescence activated cell sorting Calibur
instrument (BD Biosciences, Franklin Lakes, NJ, USA), following the
manufacturer's protocol. The percentage apoptotic cells was
calculated using Cell Quest software (version 3.4; BD
Biosciences).
Luciferase reporter assay
Prediction of miR-212 targets was performed using
three publicly available algorithms: TargetScan (www.targetscan.org), miRanda (www.microrna.org) and PicTar (www.pictar.org). The 3′-UTR sequence of FOXA1
predicted to bind to miR-212 or a mutated sequence within the
predicted target sites was synthesized and inserted into the pGL3
control vector (Promega Corporation, Madison, WI, USA) at the
XbaI and FseI sites, and were referred to as
wild-type (Wt) FOXA1-3UTR or mutant (Mut) FOXA1-3′UTR,
respectively. For the reporter assay, Caki-1 cells were seeded into
24-well plates at density of 2×105 for 24 h, and then
cotransfected with the Wt/Mut FOXA1 3′UTR and miR-212/miR-Ctrl, and
the pRL-TK plasmid (Promega Corporation), which was used for
internal normalization, using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Following 48 h transfection, the cells were harvested,
and a Renilla luciferase activity was determined using the
Dual-Luciferase® Reporter assay system (Promega
Corporation), following the manufacturer's protocol.
Western blotting
Total protein was extracted from cultured cells
using a Total Protein Extraction kit (KeyGen Biotech Co., Ltd.,
Nanjing, China) and was quantified using a bicinchoninic acid
protein assay kit (Boster Biological Technology, Pleasanton, CA,
USA). A total of 30 µg protein/lane was separated using SDS-PAGE on
10% gels (Bio-Rad Laboratories, Inc.) and then transferred onto
nitrocellulose membranes (Bio-Rad Laboratories, Inc.). The membrane
was blocked with 5% non-fat milk (Sigma-Aldrich; Merck KGaA) for 2
h at room temperature, and then incubated with the following
antibodies: Mouse monoclonal anti-human FOXA1 (1:1,000; cat no:
sc-101058; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and
mouse monoclonal anti-human GAPDH (1:5,000; cat no. sc-365062;
Santa Cruz Biotechnology, Inc.) antibodies were incubated overnight
at 4°C. The blots were incubated with polyclonal goat anti-mouse
horseradish peroxidase-conjugated immunogloblin G (1:5,000; cat.
no. sc-2005; Santa Cruz Biotechnology, Inc.) for 2 h at room
temperature (20–25°C) and visualized using the enhanced
chemiluminescence system (Thermo Fisher Scientific, Inc.). GAPDH
was used to normalize the expression levels of the target
protein.
Statistical analysis
Results are expressed as the mean ± standard
deviation from ≥3 independent experiments. Statistical analysis was
performed using SPSS software (version 17; SPSS, Inc., Chicago, IL,
USA) and GraphPad Prism version 5.0 software (GraphPad Software,
Inc., La Jolla, CA, USA). An independent t-test was used to compare
the differences between two groups. One-way analysis of variance
followed by the Bonferroni post-hoc test was performed to compare
the differences between three or more groups. Correlations between
miR-212 expression and FOXA1 were determined by Spearman's rank
correlation coefficient. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-212 expression is downregulated in
RCC cell lines and tissues
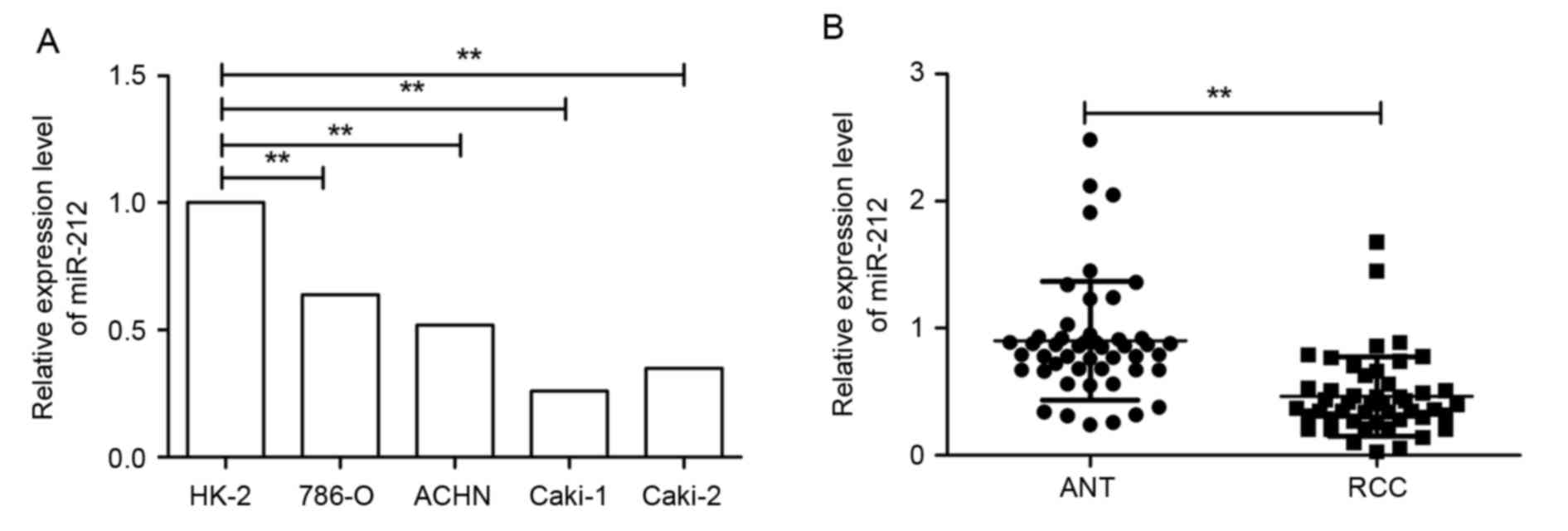
To determine the expression status of miR-212 in
RCC, the expression levels of miR-212 in four RCC cell lines was
analyzed by RT-qPCR. Reduced expression levels of miR-212 were
observed in all four RCC cell lines compared with the human renal
proximal tubule epithelial cell line (P<0.01; Fig. 1A). The expression of miR-212 in 48
pairs of HCC tissues and ANT was then measured. The expression of
miR-212 in RCC tissues was significantly decreased compared with
that in matched ANT (P<0.01; Fig.
1B). All RCC samples were divided into miR-212 low-expression
group (n=26) and high-expression group (n=22) according to the
cut-off value, which was defined as the mean value (0.446) in all
RCC samples. miR-212 expression was significantly associated with
tumor size, tumor, nodes, metastasis (TNM) stage and lymph node
metastasis (all P<0.01); however, miR-212 expression was not
significantly associated with gender and age (Table I). These results suggested that
miR-212 may serve a key role in the development and progression of
RCC.
miR-212 inhibits RCC cell
proliferation, migration and invasion, and induces apoptosis
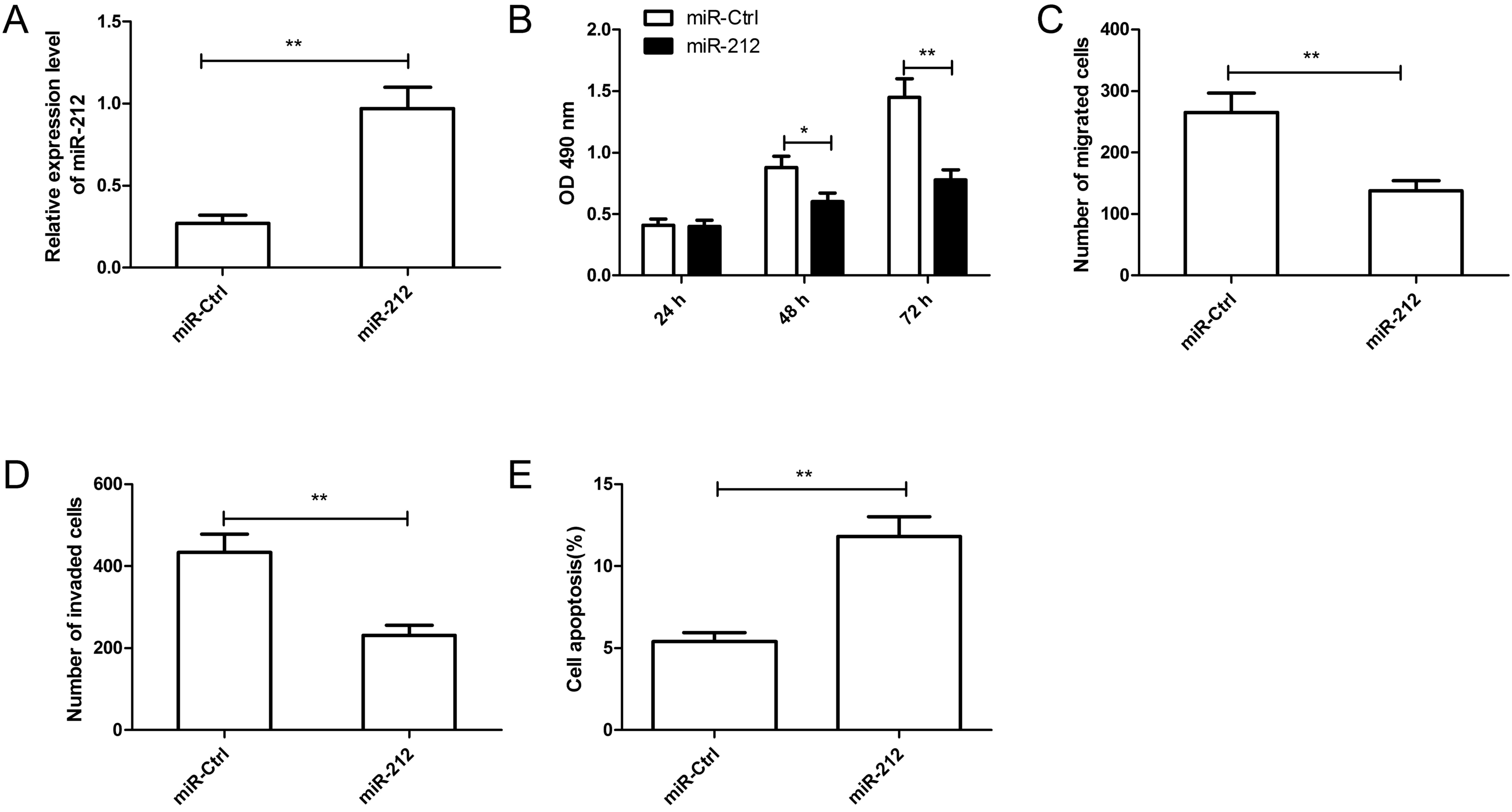
To further investigate the biological function of
miR-212 in RCC, miR-212 mimic or miR-Ctrl was transfected into
Caki-1 cells (Fig. 2A). As
measured by RT-qPCR, the miR-212 mimic significantly increased the
level of miR-212 in Caki-1 cells compared with the miR-Ctrl
(P<0.01; Fig. 2A). The MTT
assay demonstrated that overexpression of miR-212 significantly
inhibited cell proliferation of Caki-1 cells after 48 and 72 h
compared with the miR-Ctrl (Fig.
2B). A Transwell chamber was then used to investigate the roles
of miR-212 in cell migration and invasion in Caki-1 cells. Results
revealed that overexpression of miR-212 significantly inhibited the
migratory and invasive capabilities in Caki-1 cells compared with
the miR-Ctrl (Fig. 2C and D). In
addition, compared with the miR-Ctrl, overexpression of miR-212
significantly induced apoptosis in Caki-1 cells (Fig. 2E). These results suggested that
miR-212 may serve a suppressive role in RCC.
FOXA1 is a functional target of
miR-212 in RCC
To investigate the underlying molecular mechanism by
which miR-212 inhibits RCC tumor growth, analysis of bioinformatic
databases (TargetScan, PicTar and miRanda) were used to predict
putative miR-212 targets. Target prediction analysis revealed that
the 3′-UTR of FOXA1 mRNA contained the complementary sequence of
miR-212 at position 164–171 (Fig.
3A). To further assess whether FOXA1 is a direct target of
miR-212, a luciferase reporter assay was performed. The results
demonstrated that miR-212 significantly inhibited the luciferase
activity of FOXA1 containing the Wt 3′-UTR, but no alteration in
FOXA1 activity was observed with the Mut 3′-UTR, compared with
Caki-1 cells transfected with the miR-Ctrl (Fig. 3B). In addition, RT-qPCR and western
blotting results confirmed that overexpression of miR-212 resulted
in downregulation of FOXA1 expression at the mRNA (Fig. 3C) and protein (Fig. 3D) level in Caki-1 cells.
FOXA1 is inversely correlated with
miR-212 in RCC tissues
FOXA1 mRNA expression in 48 pairs of RCC tissue
samples and ANT was investigated by RT-qPCR. FOXA1 mRNA expression
was increased in RCC specimens compared with ANT (Fig. 4A). Using Spearman's rank
correlation coefficient analysis, an inverse correlation between
miR-212 and FOXA1 mRNA expression levels was confirmed in 48 RCC
tissues (r=−0.667; P<0.0001; Fig. 4B).
FOXA1 overexpression reverses the
inhibitory effects of miR-212 in RCC cells
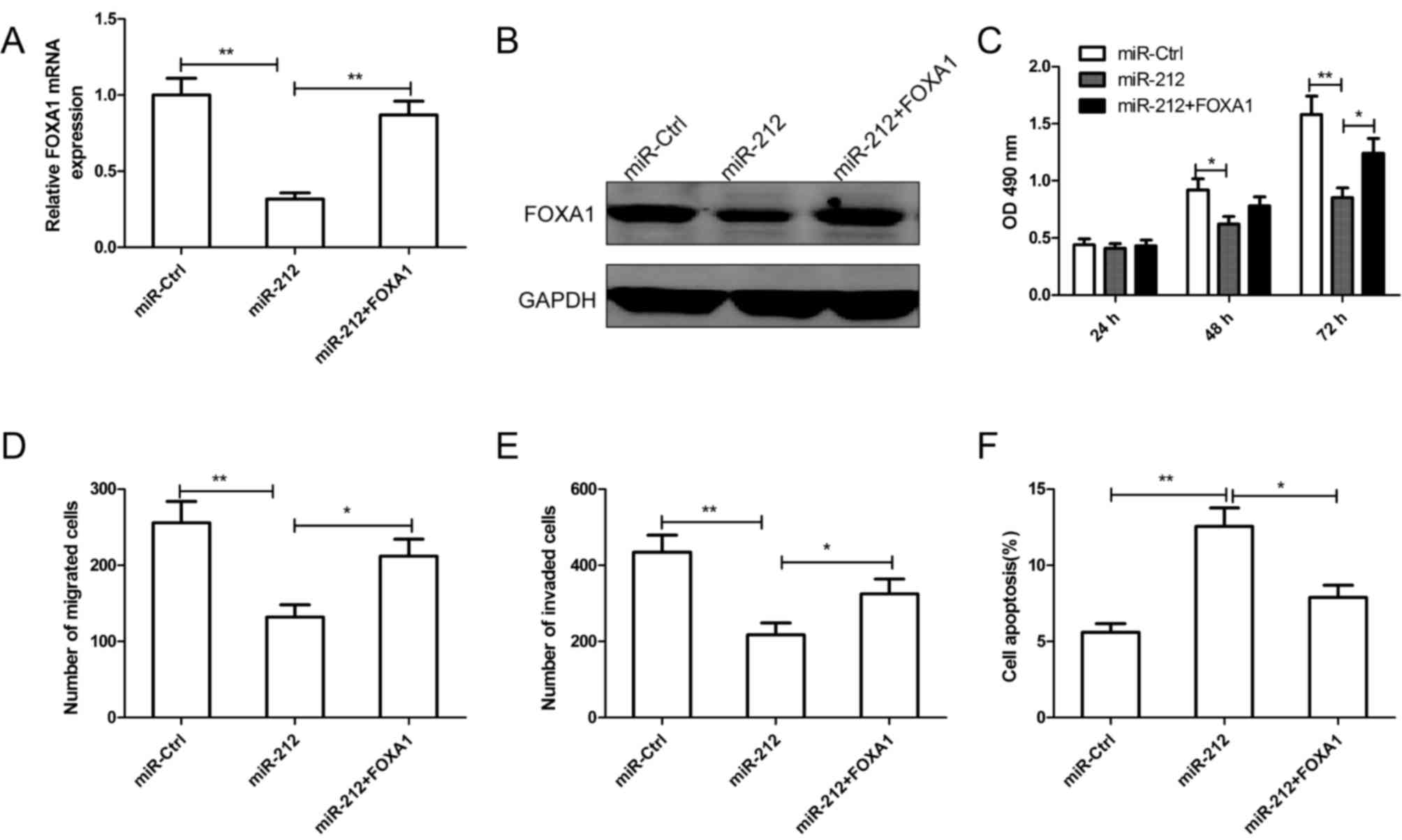
To determine whether FOXA1 mediates miR-212 in cell
proliferation, migration and invasion, a rescue experiment was
performed. Caki-1 cells were cotransfected with miR-212 mimic and
FOXA1 overexpression plasmid, in parallel with controls (miR-Ctrl).
The expression of FOXA1 was confirmed by RT-qPCR (Fig. 5A) and western blotting (Fig. 5B). In addition, it was revealed
that overexpression of FOXA1 restored cell proliferation (Fig. 5C), migration (Fig. 5D) and invasion (Fig. 5E) in Caki-1 cells that were
inhibited by miR-212. The increased apoptotic rate induced by
miR-212 in Caki-1 cells was also reversed by FOXA1 overexpression
(Fig. 5F).
Discussion
Accumulating evidence has identified a number of
miRNAs with aberrant expression in RCC tissues or cell lines, which
are involved in the initiation and progression of RCC as a tumor
suppressor or oncogene (9–11). Therefore, investigating the
function of miRNAs specifically involved in RCC development and
progression may contribute to the understanding RCC carcinogenesis,
and provide novel diagnostics and therapeutic targets for this
disease. In the present study, the results revealed that miR-212
expression was downregulated in RCC cells lines and tissues.
Furthermore, reduced expression of miR-212 was associated with poor
prognostic features of RCC. Overexpression of miR-212 suppressed
the cell proliferation, migration and invasion, and induced cell
apoptosis. These results suggested that miR-212 may be a novel
potential therapeutic strategy for RCC.
miR-212 has been reported to be downregulated and
act as a tumor suppressor in several types of cancers, including
lung cancer (14), cervical cancer
(16), glioblastoma (17), ovarian cancer (18), hepatocellular carcinoma (19) and gastric carcinoma (25). However, several other studies have
suggested that miR-212 exhibits oncogenic roles in pancreatic
cancer (13), prostate cancer
(15) and colorectal cancer
(26). This suggests that the
biological functions of miR-212 are cancer type specific, partly
resulting from the different cellular contexts of various types of
cancer. In the present study, the expression and biological
function of miR-212 in RCC was investigated. miR-212 expression in
RCC tissues was significantly downregulated as compared with that
in adjacent non-tumor tissues. In addition, results demonstrated
that reduced expression of miR-212 was associated with TNM stage
and lymph node metastasis. miR-212 significantly suppressed RCC
proliferation, migration and invasion, and induced cell apoptosis.
These results suggested that miR-212 has a possible tumor
suppressor role in RCC.
To further investigate the underlying molecular
mechanisms by which miR-212 exerts its anti-tumor effect on RCC
cells, identification of its downstream functional targets is
necessary. Using three public databases (TargetScan, PicTar, and
miRanda), it was predicted that FOXA1 was a direct target of
miR-212. FOXA1, a member of the FOXA gene family, has been reported
to serve an important regulatory role in proliferation, apoptosis
and the cell cycle (27). FOXA1
has been demonstrated to be upregulated in various types of cancer,
including RCC (28), breast cancer
(29), hepatocellular carcinoma
(18,19), prostate cancer (30) and gastric cancer (31). In addition, FOXA1 may promote
cancer cell proliferation and inhibit apoptosis partly by
upregulating Yes-associated protein expression, suggesting its
oncogenic role in various types of cancer (27,31,32).
FOXA1 has been reported to be a target of miR-212 in hepatocellular
carcinoma (19,23); however, the interaction between
miR-212 and FOXA1 has not been experimentally validated in RCC. In
the present study, it was confirmed that FOXA1 was a direct
downstream target of miR-212 as evidenced by the observation that
ectopic expression of miR-212 reduced luciferase activity of the
FOXA1 promoter and miR-212 overexpression downregulated FOXA1
expression. An inverse correlation between the expression of
miR-212 and FOXA1 mRNA expression was observed in RCC tissues.
Overexpression of FOXA1 expression partly abrogated the functional
effect of miR-212 on RCC cell proliferation, migration, invasion
and apoptosis. These data may suggest that miR-212 partly exerts it
antitumor role in RCC by targeting FOXA1.
In conclusion, the present study revealed that
miR-212 is downregulated in RCC cell lines and tissues. Low
expression of miR-212 was prominently associated with large tumor
size, advanced TNM stage and lymph node metastasis. It was also
demonstrated that miR-212 overexpression significantly inhibited
cell proliferation, migration and invasion, and induced cell
apoptosis in RCC cells by suppressing FOXA1 expression. These
results suggested that miR-212 may potentially act as a clinical
biomarker and a therapeutic target for RCC.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pantuck AJ, Zisman A and Belldegrun AS:
The changing natural history of renal cell carcinoma. J Urol.
166:1611–1623. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Ann Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garzon R and Marcucci G: Potential of
microRNAs for cancer diagnostics, prognostication and therapy. Curr
Opin Oncol. 24:655–659. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu L, Li H, Chen L, Ma X, Gao Y, Li X,
Zhang Y, Fan Y and Zhang X: MicroRNAs as prognostic molecular
signatures in renal cell carcinoma: A systematic review and
meta-analysis. Oncotarget. 6:32545–32560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang S, Zhang D, Yi C, Wang Y, Wang H and
Wang J: MicroRNA-22 functions as a tumor suppressor by targeting
SIRT1 in renal cell carcinoma. Oncol Rep. 35:559–567. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jingushi K, Ueda Y, Kitae K, Hase H, Egawa
H, Ohshio I, Kawakami R, Kashiwagi Y, Tsukada Y, Kobayashi T, et
al: miR-629 Targets TRIM33 to Promote TGFβ/Smad Signaling and
Metastatic Phenotypes in ccRCC. Mol Cancer Res. 13:565–574. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ucar A, Vafaizadeh V, Jarry H, Fiedler J,
Klemmt PA, Thum T, Groner B and Chowdhury K: miR-212 and miR-132
are required for epithelial stromal interactions necessary for
mouse mammary gland development. Nat Genet. 42:1101–1108. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma C, Nong K, Wu B, Dong B, Bai Y, Zhu H,
Wang W, Huang X, Yuan Z and Ai K: miR-212 promotes pancreatic
cancer cell growth and invasion by targeting the hedgehog signaling
pathway receptor patched-1. J Exp Clin Cancer Res. 33:542014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Incoronato M, Urso L, Portela A, Laukkanen
MO, Soini Y, Quintavalle C, Keller S, Esteller M and Condorelli G:
Epigenetic regulation of miR-212 expression in lung cancer. PLoS
One. 6:e277222011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Y, Jia D, Kim H, Elmageed Abd ZY,
Datta A, Davis R, Srivastav S, Moroz K, Crawford BE, Moparty K, et
al: Dysregulation of miR-212 promotes castration resistance through
hnRNPH1-Mediated regulation of AR and AR-V7: Implications for
racial disparity of prostate cancer. Clin Cancer Res. 22:1744–1756.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao JL, Zhang L, Guo X, Wang JH, Zhou W,
Liu M, Li X and Tang H: miR-212/132 downregulates SMAD2 expression
to suppress the G1/S phase transition of the cell cycle and the
epithelial to mesenchymal transition in cervical cancer cells.
IUBMB Life. 67:380–394. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu H, Li C, Shen C, Yin F, Wang K, Liu Y,
Zheng B, Zhang W, Hou X, Chen X, et al: MiR-212-3p inhibits
glioblastoma cell proliferation by targeting SGK3. J Neurooncol.
122:431–439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei LQ, Liang HT, Qin DC, Jin HF, Zhao Y
and She MC: MiR-212 exerts suppressive effect on SKOV3 ovarian
cancer cells through targeting HBEGF. Tumour Biol. 35:12427–12434.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dou C, Wang Y, Li C, Liu Z, Jia Y, Li Q,
Yang W, Yao Y, Liu Q and Tu K: MicroRNA-212 suppresses tumor growth
of human hepatocellular carcinoma by targeting FOXA1. Oncotarget.
6:13216–13228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang JL, Liao Y, Qiu MX, Li J and An Y:
Long non-coding RNA CCAT2 promotes cell proliferation and invasion
through regulating Wnt/β-catenin signaling pathway in clear cell
renal cell carcinoma. Tumour Biol. 39:10104283177113142017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu J, Cui L, Xu A, Yin X, Li F and Gao J:
MEIS1 inhibits clear cell renal cell carcinoma cells proliferation
and in vitro invasion or migration. BMC Cancer. 17:1762017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Furge KA, Chen J, Koeman J, Swiatek P,
Dykema K, Lucin K, Kahnoski R, Yang XJ and Teh BT: Detection of DNA
copy number changes and oncogenic signaling abnormalities from gene
expression data reveals MYC activation in high-grade papillary
renal cell carcinoma. Cancer Res. 67:3171–3176. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tu H, Wei G, Cai Q, Chen X, Sun Z, Cheng
C, Zhang L, Feng Y, Zhou H, Zhou B and Zeng T: MicroRNA-212
inhibits hepatocellular carcinoma cell proliferation and induces
apoptosis by targeting FOXA1. OncoTargets Ther. 8:2227–2235.
2015.
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiping Z, Ming F, Lixiang W, Xiuming L,
Yuqun S, Han Y, Zhifang L, Yundong S, Shili L, Chunyan C and Jihui
J: MicroRNA-212 inhibits proliferation of gastric cancer by
directly repressing retinoblastoma binding protein 2. J Cell
Biochem. 114:2666–2672. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meng X, Wu J, Pan C, Wang H, Ying X, Zhou
Y, Yu H, Zuo Y, Pan Z, Liu RY and Huang W: Genetic and epigenetic
down-regulation of microRNA-212 promotes colorectal tumor
metastasis via dysregulation of MnSOD. Gastroenterology.
145(426–436): e1–e6. 2013.
|
|
27
|
Bernardo GM and Keri RA: FOXA1: A
transcription factor with parallel functions in development and
cancer. Biosci Rep. 32:113–130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Neely BA, Wilkins CE, Marlow LA,
Malyarenko D, Kim Y, Ignatchenko A, Sasinowska H, Sasinowski M,
Nyalwidhe JO, Kislinger T, et al: Proteotranscriptomic analysis
reveals stage specific changes in the molecular landscape of
clear-cell renal cell carcinoma. PLoS One. 11:e01540742016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shou J, Lai Y, Xu J and Huang J:
Prognostic value of FOXA1 in breast cancer: A systematic review and
meta-analysis. Breast. 27:35–43. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang YA and Yu J: Current perspectives on
FOXA1 regulation of androgen receptor signaling and prostate
cancer. Genes Dis. 2:144–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ren H, Zhang P, Tang Y, Wu M and Zhang W:
Forkhead box protein A1 is a prognostic predictor and promotes
tumor growth of gastric cancer. OncoTargets Ther. 8:3029–3039.
2015.
|
|
32
|
Augello MA, Hickey TE and Knudsen KE:
FOXA1: Master of steroid receptor function in cancer. EMBO J.
30:3885–3894. 2011. View Article : Google Scholar : PubMed/NCBI
|