Introduction
Oral lichen planus (OLP) is a chronic inflammatory
disorder in which T-cell mediated autoimmunity is implicated
(1). OLP affects 0.5–2% of the
population of China and disease onset occurs between 30 and 60
years of age (2). Although the
etiology of OLP remains unclear, genetic predisposition,
autoimmunity and viral infection are reported to be associated with
OLP (3).
During inflammation, osteopontin (OPN) is primarily
expressed by immunocytes, including natural killer cells, activated
T cells and macrophages (4). CD44
is a receptor for OPN and is associated with the mediation of cell
chemotaxis and attachment (5). It
has been demonstrated that OPN is overexpressed in various types of
autoimmune diseases (6–8). Additionally, there are reports that
OPN is overexpressed in the serum and oral epithelia of patients
with OLP, which indicates a potential role for OPN in the
pathogenesis of OLP (9,10).
MicroRNAs (miRNA) are a group of endogenous, short
non-coding RNAs, which regulate gene expression through targeting
the 3′untranslated region (UTR) of mRNA. miRNAs have been
identified in various organisms and many of them are evolutionary
conserved. Additionally, it is estimated that >50% of all human
protein-coding genes are potentially regulated by miRNAs (11). It has been previously reported that
the miRNA profile was altered in the oral mucosa of patients with
OLP (12). However, the effect of
altered miRNA levels in OLP requires further investigation.
In the present study, 11 miRNAs that may target CD44
directly were selected. The expression levels of the selected
miRNAs in oral mucosa samples from 33 patients and 33 controls were
detected. In addition, the expression of the CD44 protein was
examined by western blot analysis. The results of the present study
demonstrated that two miRNAs were significantly reduced in OLP
samples compared with controls and the expression of CD44 was
upregulated in OLP samples. Subsequently, correlation analysis was
performed to assess the correlation between the expression of
miRNAs and CD44 in vitro and in patients with OLP.
Materials and methods
Participants and sample
collection
Oral mucosa samples were collected during incisional
biopsy procedures following surgical removal from 33 patients (age,
23–37 years; mean age, 38.7 years; 18 female; 15 male), who were
recruited from Jining First People's Hospital (Jining, China)
between February 2013 and November 2014. Following collection, one
half of the specimens were examined at the pathology department of
Jining Hospital (Jining, China) to verify the diagnosis of OLP. The
other half were directly frozen in liquid nitrogen and were stored
at −80°C until further analysis. The control group was composed of
tissue samples from elective oral surgery whereby a
histopathological examination excluded any diseases of the oral
mucosa. The study was approved by the ethics board of Jining
hospital of Stomatology (Jining, China). All participants were
informed by a patient information sheet about the intention of the
study and written informed consent according to the Helsinki
convention was obtained from all participants.
Cell culture
HeLa human cervical carcinoma cells (National
Infrastructure of Cell Line Resource, Beijing, China) were
maintained in Dulbecco's modified Eagle's medium (HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal calf serum (FCS; HyClone; GE Healthcare Life Sciences) and 50
U/ml penicillin and 0.1 mg/ml streptomycin (HyClone; GE Healthcare
Life Sciences) at 37°C in a humidified atmosphere containing 5%
CO2. Raji Burkitt lymphoma B-cell lines and Jurkat
T-lymphocytes were obtained from the National Infrastructure of
Cell Line Resources, and were cultured in RPMI 1640 medium
(HyClone; GE Healthcare Life Sciences) supplemented with 10%
heat-inactivated FCS (Hyclone; GE Healthcare Life Sciences), 100
U/ml penicillin, 0.1 mg/ml streptomycin, 2 mM L-glutamine, and 1 mM
sodium pyruvate at 37°C in a 5% CO2 atmosphere.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Prior to RT-qPCR, TargetScan (www.targetscan.org) was used to predict miRNAs that
may target the 3′UTR of CD44, and 11 miRNAs were selected for
RT-qPCR analysis. Total RNA was extracted from tissue samples by
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. The expression of miRNAs was detected by TaqMan miRNA
RT-qPCR. Single-stranded cDNA was synthesized by using TaqMan
MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.) at 16°C for 30 min, followed by 42°C for
30 min and 85°C for 5 min. cDNA was subsequently amplified by using
TaqMan Universal PCR Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) together with miRNA-specific TaqMan MGB probes
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer
sequences for OPN were: OPN forward, 5′-CATATGATGGCCGAGGTGATAG-3′
and OPN reverse 5′-AGGTGATGTCCTCGTCTGTA-3′. The primer sequences
for CD44 were: CD44 forward 5′-GATGTCACAGGTGGAAGAAGAG-3′ and CD44
reverse 5′-TTCCTTCGTGTGTGGGTAATG-3′. The catalog numbers for the
miRNA probes were: MiR-135a, 000460; miR-135b, 002261; miR-143-3p,
002249; miR-199a, 002304; miR-204, 000508; miR-211, 477502_mat;
miR-214, 000517; miR-216a, 002220; miR-216b, 002326; miR-320a,
002277; and miR-489, 466,550_mat. The thermocycling conditions were
as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15
sec and 60°C for 1 min (ABI7500; Applied Biosystems; Thermo Fisher
Scientific, Inc.). Results were normalized using the ΔΔCq method
and U6 snRNA was used as internal control (13). Each sample in each group was
measured in triplicate and the experiment was repeated at least
three times. The negative control was reacted with all PCR reagents
except the reverse transcriptase.
Dual luciferase assay
A segment of 722 bp CD44 3′UTR containing 2
predicted miR-214 target regions was cloned into pmirGLO vector
between XbaI and XhoI sites, downstream of the
firefly luciferase coding region (Promega Corporation, Madison, WI,
USA) to generate luciferase reporter vectors. Mutant CD44 3′UTR was
directly amplified using primers with 3 mutated nucleotides, and
then cloned into the pmirGLO vector to generate the mutant reporter
vector. For luciferase reporter assays, HeLa cells were seeded in
48-well plates (5×104 cells/well). miRNA mimics or
inhibitors, or short nucleotide oligo controls, and luciferase
reporter vectors, were co-transfected into HeLa cells by
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 6 h. At 2 days post-transfection,
cells were harvested and assayed with the
Dual-Luciferase® Reporter Assay System (Promega
Corporation). Each treatment was performed in triplicate in 3
independent experiments. The results were expressed as relative
luciferase activity (Firefly luciferase/Renilla luciferase).
The sequence of the 2′-O-methoxyethyl (OMe) modified double strand
miR-214 mimic is 5′-ACAGCAGGCACAGACAGGCAGU-3′ and the sequence of
the 2′-OMe modified single strand miR-214 inhibitor is
5′-ACUGCCUGUCUGUGCCUGCUGU-3′. Scrambled 2′-OMe-modified RNA
(5′-AAGGCAAGCUGACCCUGAAGU-3′) was used as a negative control.
Western blot analysis
Protein was extracted from cells and tissue samples
using radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher
Scientific, Inc.), following the manufacturer's protocol. Cells
were washed with cold PBS twice and incubated with cold RIPA buffer
on ice for 5 min. Samples were centrifuged at ~14,000 × g at 4°C
for 15 min to pellet the cell debris and the supernatant was
transferred to a new tube. For tissue samples, the tissue was
dissected with clean tools on ice and ice-cold RIPA buffer was
added rapidly to the tube, followed by homogenizing with an
electric homogenizer. Samples were centrifuged for 20 min at 15,000
× g at 4°C and the supernatant was transferred to a new tube. The
protein concentration was determined using a Bicinchoninic Acid
Protein Assay kit (Thermo Fisher Scientific, Inc.). Protein
extracts were boiled in SDS/β-mercaptoethanol sample buffer
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 10 µg protein
from tissue or 20 µg from cells were loaded into each lane of 8%
polyacrylamide gels. The proteins were separated by
electrophoresis, and the proteins in the gels were blotted onto
polyvinylidene fluoride membranes (GE Healthcare Life Sciences,
Chalfont, UK) by electrophoretic transfer. Membranes were
subsequently incubated with rabbit anti-CD44 polyclonal antibody
(cat. no. ab51037, 1:1,000 dilution; Abcam, Cambridge, MA, USA),
rabbit anti-osteopontin polyclonal antibody (cat. no. ab8448,
1:1,000 dilution; Abcam) or mouse anti-β-actin monoclonal antibody
(cat. no. sc-47778, 1:5,000 dilution; Santa Cruz Biotechnology
Inc., Dallas, TX, USA) for 1 h at 37°C. The specific
protein-antibody complex was detected by using horseradish
peroxidase-conjugated goat anti-rabbit (cat. no. ab6721, 1:5,000
dilution; Abcam) or rabbit anti-mouse (cat. no. ab6728, 1:5000
dilution; Abcam) antibody by incubation at 37°C for 2 h. Detection
by chemiluminescence was performed using the Pierce™ ECL Western
Blotting Substrate (Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. The β-actin signal was used as a
loading control. Densitometric analysis was performed using
Quantity One software version 4.6.2 (Bio-Rad Laboratories, Inc.
Hercules, CA, USA.) Results are exhibited as the mean ± standard
deviation of 2 replicates.
Histological analysis
The cases were classified histopathologically
following the diagnostic criteria by van der Waal et al
(14). All cases and controls
underwent a hematoxylin and eosin standard histological procedure
(15). Results were analyzed by
χ2 analysis.
Statistical analysis
Data were analyzed by using SPSS Statistical Package
version 15 (SPSS, Inc., Chicago, IL, USA). The correlation between
miRNAs level and relative CD44 expression were analyzed by Spearman
correlation analysis. Independent two group analyses were conducted
using unpaired Student's t-test. Associations between the
expression of miR-214, OPN or CD44 and histopathological features
were analyzed by χ2−analysis. P<0.05 was considered
to indicate a statistically significant difference.
Results
OPN and CD44 mRNA and protein levels
increase in patients with OLP
As OPN and CD44 have important roles during the
pathogenesis of OLP, the present study initially detected the
expression of OPN and CD44 in mucosa samples of patients with OLP
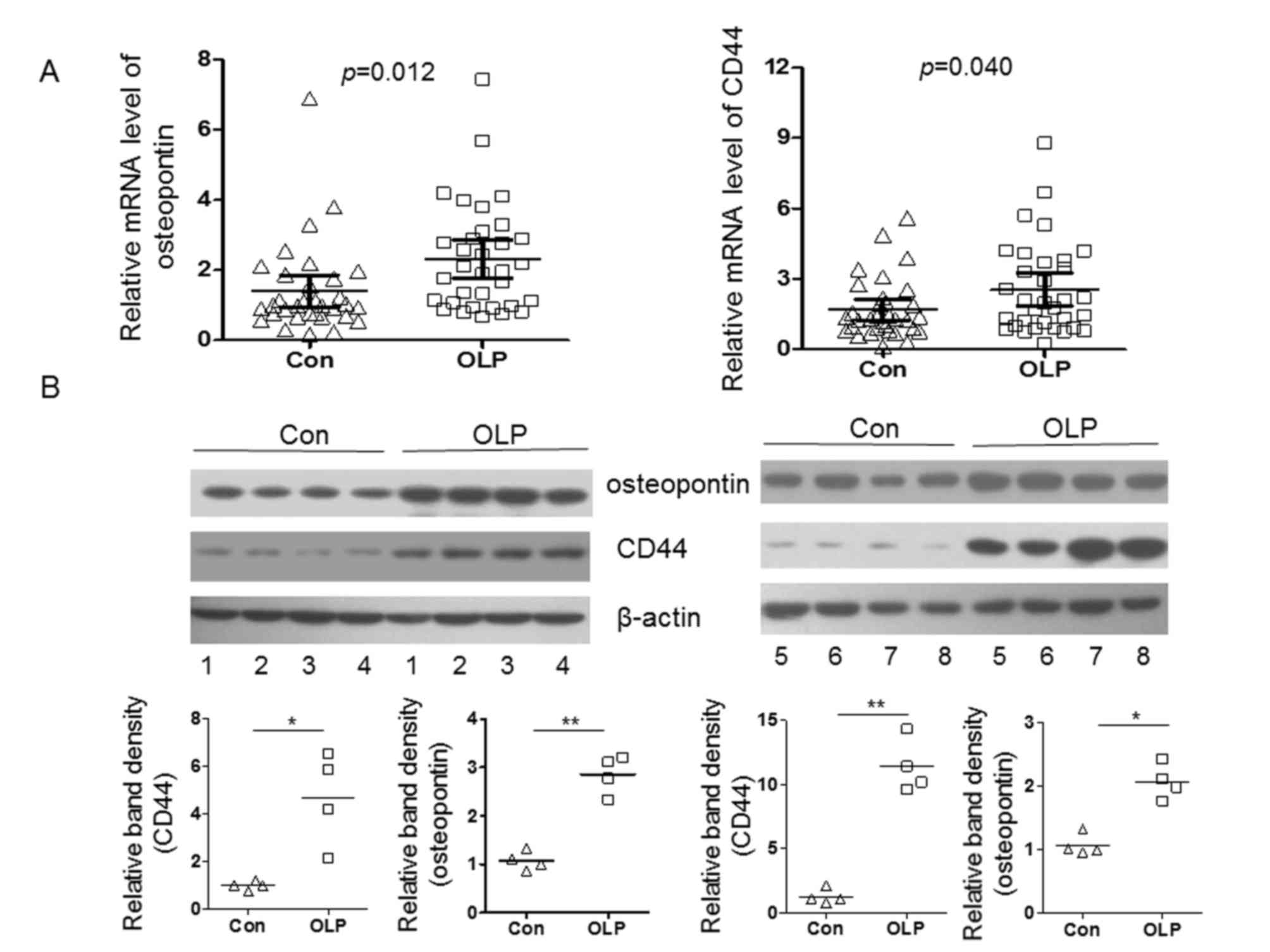
and healthy controls. As presented in Fig. 1A, the mRNA expression levels of OPN
and CD44 were significantly upregulated in patients with OLP,
compared with controls (P<0.05). Protein levels of OPN and CD44
were also examined by western blot analysis. As demonstrated in
Fig. 1B, protein expression of OPN
(P<0.05) and CD44 (P<0.01) was markedly increased in patients
with OLP compared with controls. However, the protein expression of
CD44 increased to a greater degree compared with increases in CD44
mRNA expression, indicating that post-transcriptional regulatory
factors may contribute to the increased expression of CD44.
miR-214, miR-216a and miR-216b
expression decreases in patients with OLP
As an important negative regulator of expression,
miRNAs regulate the expression of target genes by directly
targeting 3′UTRs through post-transcriptional methods. To identify
the miRNAs that repress the expression of CD44 and may be
associated with the pathogenesis of OLP, miRNAs that target the
CD44 3′UTR were predicted by using the following online
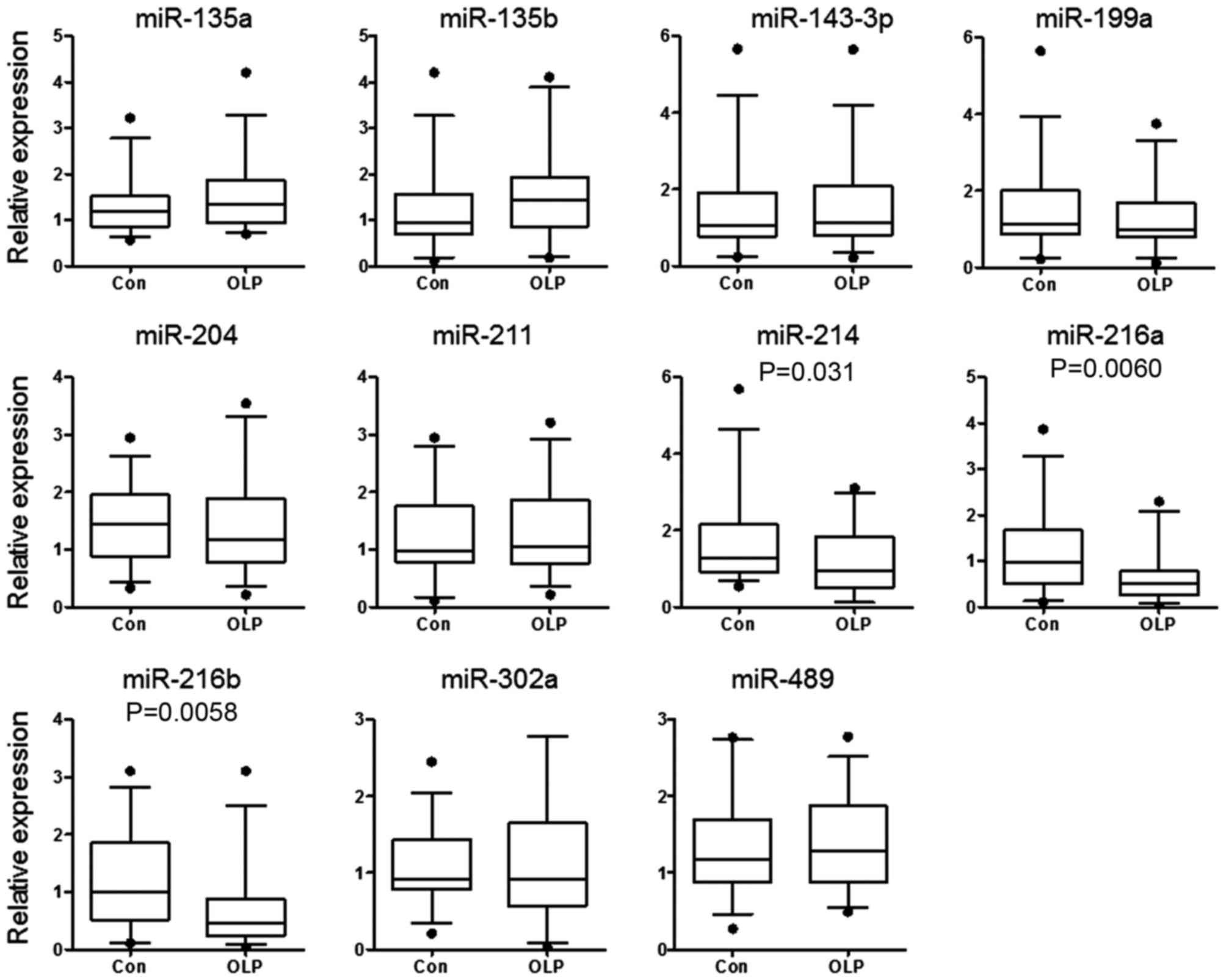
bioinformatics tool: TargetScan (www.targetscan.org). A total of 11 miRNAs were
selected, with target sites conserved among mammals. The expression
levels of the selected candidate miRNAs were detected in the tissue
samples of patients with OLP and controls by RT-qPCR. As presented
in Fig. 2, the expression of
miR-214, miR-216a and miR-216b was reduced significantly in
patients with OLP compared with controls (P=0.031, P=0.0060 and
P=0.0058, respectively).
CD44 exhibits a negative correlation
with miR-214
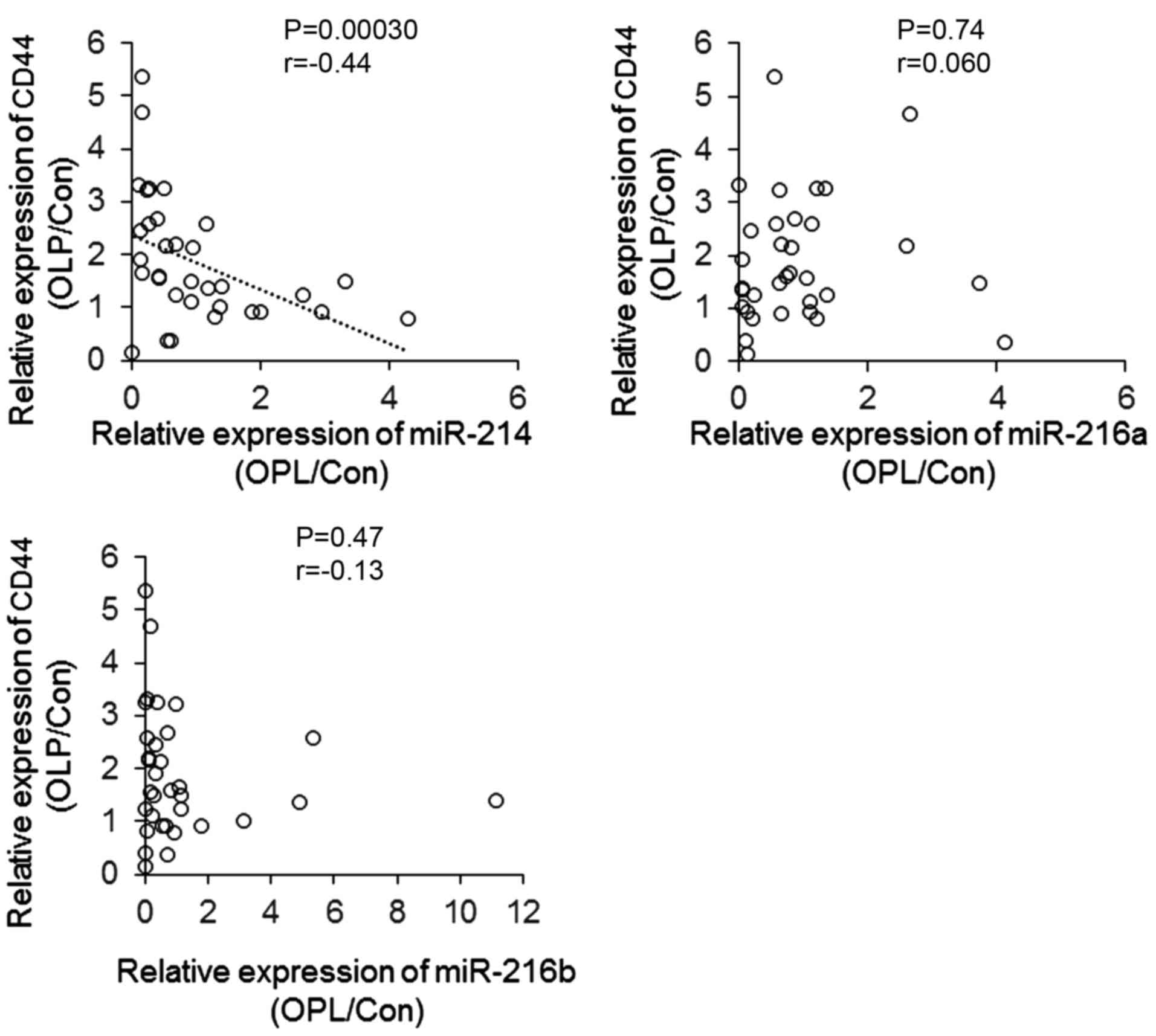
To investigate the association between the
expression of CD44 and the 3 miRNAs that exhibited reduced
expression in patients with OLP, the CD44 band density results
presented in Fig. 1B were used for
further analysis, to assess the correlation between the expression
of CD44 and miRNAs. As presented in Fig. 3, the expression of miR-214 was
negatively correlated with the protein level of CD44 (P=0.00030;
r=−0.44), indicating that reduced miR-214 may contribute to the
increased expression of CD44 observed in patients with OLP compared
with controls.
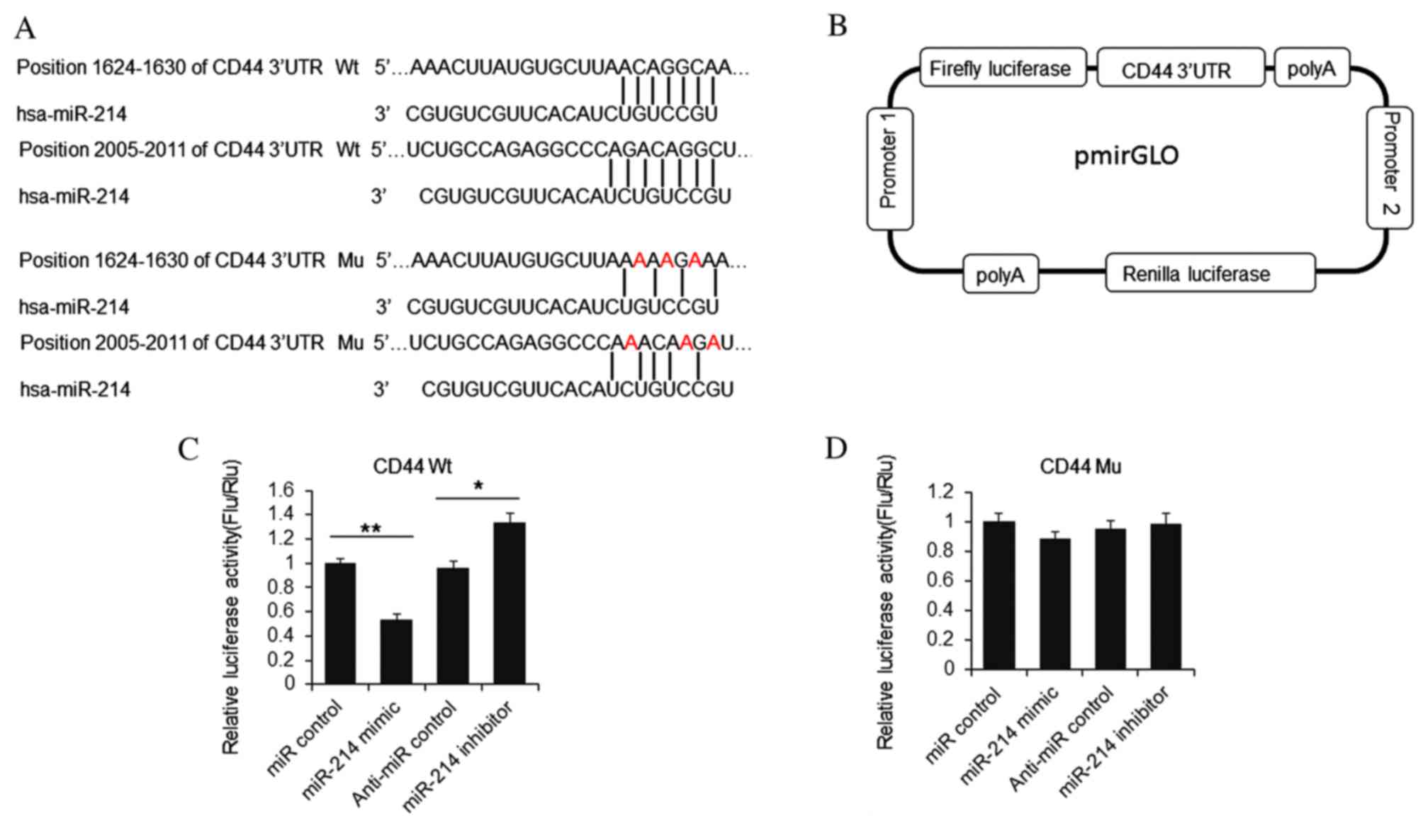
miR-214 suppresses luciferase activity
in cell lines
To confirm whether CD44 is a direct target of
miR-214, a luciferase activity reporter vector was constructed,
attaching the 3′UTR of CD44 to the end of the firefly luciferase
coding region (Fig. 4A and B).
HeLa cells were transfected with miR-214 mimic or inhibitor and
reporter vector. At 48 h post-transfection, cells were lysed and
the luciferase activities were detected. As presented in Fig. 4C, firefly luciferase activity was
reduced significantly by miR-214 mimic, compared with miR control
group, and increased by miR-214 inhibitor addition compared with
anti-miR control group. Furthermore, when 6 nucleotides were
mutated, the luciferase activity was not repressed by miR-214
(Fig. 4D). The results indicated
that miR-214 repressed luciferase activity by targeting the CD44
3′UTR.
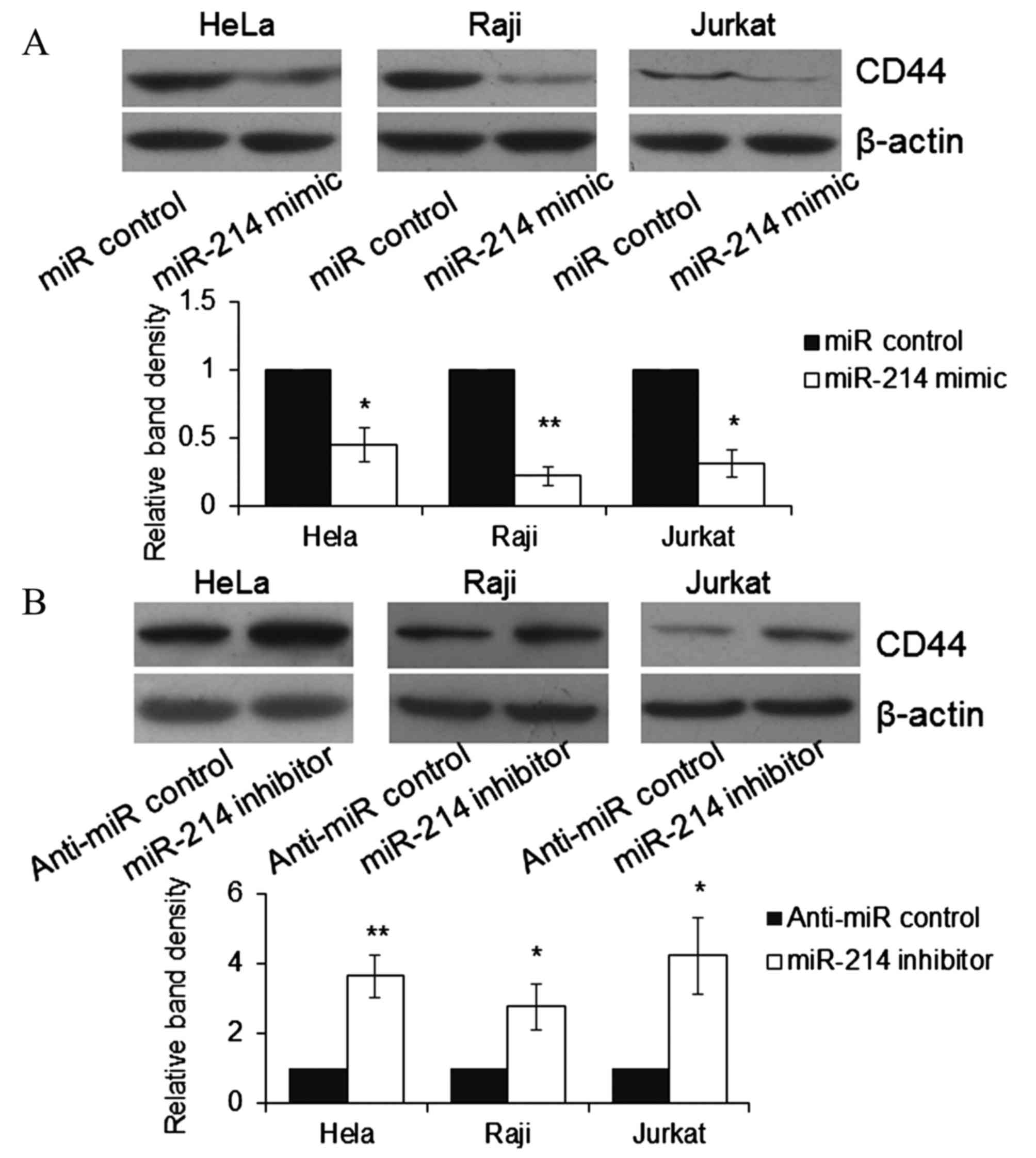
Expression of CD44 in HeLa, Raji and
Jurkat cell lines varies with transfection with miR-214 mimic or
inhibitor
To further understand whether endogenous CD44 was
repressed by miR-214, HeLa, Raji and Jurkat cells were transfected
with miR-214 mimic or inhibitor. At 48 h post-transfection, cells
were lysed and the expression of CD44 was detected by western blot
analysis. As demonstrated in Fig.
5, the expression of endogenous CD44 was significantly reduced
by miR-214 mimic compared with miR control group (P<0.05) and
significantly increased by miR-214 inhibitor compared with anti-miR
control group (P<0.05) in HeLa, Raji and Jurkat cells. The
results indicate that CD44 is a direct target of miR-214.
Histological analysis
The histopathological features of patients with
downregulated miR-214 were analyzed and it was observed that those
with downregulated miR-214 had a significantly greater probability
of exhibiting parakeratosis, lengthened epithelial crests and deep
inflammatory infiltrates (Table
I). Furthermore, overexpression of CD44 was associated with
basal degeneration and deep inflammatory infiltrate (Table I). Overexpressed OPN was associated
with basal degeneration, superficial inflammatory infiltrate and
deep inflammatory infiltrate (Table
I).
 | Table I.Association between the major
histopathological features and the expression of miR-214, CD44 and
OPN. |
Table I.
Association between the major
histopathological features and the expression of miR-214, CD44 and
OPN.
|
| miR-214
downregulation | CD44
overexpression | OPN
overexpression |
|---|
|
|
|
|
|
|---|
| Histopathological
feature | No (%) | Yes (%) | P-value | No (%) | Yes (%) | P-value | No (%) | Yes (%) | P-value |
|---|
| Parakeratosis | 2 (18.2) | 14 (63.6) | 0.014 | 3 (33.3) | 13 (54.2) | >0.05 | 4 (50) | 12 (48) | >0.05 |
| Orthokeratosis | 9 (81.2) | 8 (36.4) | 0.014 | 6 (66.7) | 11 (45.8) |
| 4 (50) | 13 (52) | >0.05 |
| Hyperkeratosis | 5 (45.5) | 9 (40.9) | >0.05 | 4 (44.4) | 10 (41.7) | >0.05 | 3 (37.5) | 11 (44) | >0.05 |
| atrophy | 3 (27.3) | 7 (31.8) | >0.05 | 2 (22.2) | 8 (33.3) | >0.05 | 3 (37.5) | 7 (28) | >0.05 |
| Lengthened epithelial
crest | 1 (9.1) | 12 (54.5) | 0.012 | 2 (22.2) | 15 (62.5) | 0.020 | 2 (25) | 11 (44) | >0.05 |
| Basal
degeneration | 9 (81.8) | 19 (86.4) | >0.05 | 6 (66.7) | 22 (91.7) | 0.00050 | 4 (50) | 24 (96) | 0.0016 |
| Dyskeratosis | 7 (63.6) | 15 (68.2) | >0.05 | 5 (55.6) | 17 (70.8) | >0.05 | 5 (62.5) | 17 (68) | >0.05 |
| Inflammatory
infiltrate | 5 (45.5) | 17 (77.3) | >0.05 | 4 (44.4) | 18 (75) | 0.037 | 3 (37.5) | 19 (76) | 0.0074 |
| Lymphocytes | 3 (27.3) | 15 (68.2) | >0.05 | 4 (44.4) | 14 (58.3) | >0.05 | 4 (50) | 14 (56) | >0.05 |
| Plasmocytes | 2 (18.2) | 8 (36.4) | >0.05 | 3 (33.3) | 7 (29.2) | >0.05 | 2 (25) | 8 (32) | >0.05 |
| Band-like
inflammatory infiltrate | 4 (36.4) | 14 (63.6) | >0.05 | 3 (33.3) | 15 (62.5) | >0.05 | 2 (25) | 16 (64) | >0.05 |
| Perivascular
inflammatory infiltrate | 1 (9.1) | 3 (13.6) | >0.05 | 1 (11.1) | 3 (12.5) | >0.05 | 0 (0) | 4 (16) | >0.05 |
| Superficial
inflammatory infiltrate | 3 (27.3) | 13 (59.1) | >0.05 | 2 (22.2) | 14 (58.3) | >0.05 | 1 (12.5) | 15 (60) | 0.019 |
| Deep inflammatory
infiltrate | 3 (27.3) | 15 (68.2) | 0.026 | 1 (11.1) | 17 (70.8) | 0.0022 | 2 (25) | 16 (64) | 0.019 |
Discussion
CD44 is a transmembrane glycoprotein that acts as a
receptor for a wide variety of extracellular matrix ligands,
including hyaluronan, collagen, fibronectin, laminin and OPN. CD44
and receptor tyrosine kinases have been previously demonstrated to
promote cell survival in a co-operative manner (16). CD44 is one of the characterized
receptors of OPN that mediate the chemotaxis and attachment of
immune cells. In OLP, abundant T lymphocytes have been previously
demonstrated to infiltrate the oral mucosa, in which the activated
T cells trigger apoptosis of oral epithelial cells. Overexpression
of OPN and CD44 was previously demonstrated to be present in the
mucosa of patients with OLP, and it has been confirmed that OPN
suppresses the apoptosis of activated CD8+ T cells via
CD44 (17,18). The present study initially
confirmed the overexpression of OPN and CD44 in the mucosa of
patients with OLP, compared with controls, by RT-qPCR and western
blot analysis. As the upregulation of CD44 protein expression in
OLP samples compared with controls, was greater than the observed
increase in CD44 mRNA expression, and the fact that gene expression
is regulated transcriptionally and post-transcriptionally, it was
hypothesized that miRNAs may participate in the regulation of CD44
expression. By analyzing the association between CD44 protein level
and relative miRNA expression, a strong negative correlation
between the levels of miR-214 and CD44 in the mucosa samples of
patients with OLP was demonstrated. Subsequently, the present study
indicated that miR-214 represses endogenous CD44 expression by
targeting the 3′UTR of CD44 in HeLa, Raji and Jurkat cells. The
present study identified that reduced miR-214 is associated with
OLP and miR-214 may be a potential candidate for drug
development.
The present study demonstrated a direct association
between miR-214 and CD44. However, the effect of reduced miR-214
expression on the survival of T cells requires further
investigation. Furthermore, miR-214 has been previously identified
as a tumor suppressor. Reports indicate that reduced miR-214
expression has been implicated in patients with ovarian, esophageal
and pancreatic cancer (19–21).
Furthermore, overexpression of miR-214 increased the drug
sensitivity of cancer cells (22,23).
Therefore, reduced miR-214 expression may increase the risk of the
carcinogenesis. The experiments in the present study were performed
using biopsy samples and the identification of which specific cell
types were involved in the observed alterations in expression was
not possible. Therefore, further experiments, including laser
capture microdissection, are required to determine which cell types
have the most important role during the pathogenesis of OLP. In
conclusion, the present study demonstrated a direct correlation
between the expression of miR-214 and CD44 in mucosa samples of
patients with OLP. In addition, the results confirmed CD44 as a
direct target of miR-214. The current study identified that reduced
miR-214 is associated with OLP and may be a candidate molecule for
drug development.
References
|
1
|
Khan A, Farah CS, Savage NW, Walsh LJ,
Harbrow DJ and Sugerman PB: Th1 cytokines in oral lichen planus. J
Oral Pathol Med. 32:77–83. 2003. View Article : Google Scholar
|
|
2
|
Gupta S and Jawanda MK: Oral Lichen
Planus: An update on etiology, pathogenesis, clinical presentation,
diagnosis and management. Indian J Dermatol. 60:222–229. 2015.
View Article : Google Scholar :
|
|
3
|
Dudhia BB, Dudhia SB, Patel PS and Jani
YV: Oral lichen planus to oral lichenoid lesions: Evolution or
revolution. J Oral Maxillofac Pathol. 19:364–370. 2015. View Article : Google Scholar :
|
|
4
|
Ashkar S, Weber GF, Panoutsakopoulou V,
Sanchirico ME, Jansson M, Zawaideh S, Rittling SR, Denhardt DT,
Glimcher MJ and Cantor H: Eta-1 (osteopontin): An early component
of type-1 (cell-mediated) immunity. Science. 287:860–864. 2000.
View Article : Google Scholar
|
|
5
|
Weber GF, Ashkar S, Glimcher MJ and Cantor
H: Receptor-ligand interaction between CD44 and osteopontin
(Eta-1). Science. 271:509–512. 1996. View Article : Google Scholar
|
|
6
|
Chabas D, Baranzini SE, Mitchell D,
Bernard CC, Rittling SR, Denhardt DT, Sobel RA, Lock C, Karpuj M,
Pedotti R, et al: The influence of the proinflammatory cytokine,
osteopontin, on autoimmune demyelinating disease. Science.
294:1731–1735. 2001. View Article : Google Scholar
|
|
7
|
Sakata M, Tsuruha JI, Masuko-Hongo K,
Nakamura H, Matsui T, Sudo A, Nishioka K and Kato T: Autoantibodies
to osteopontin in patients with osteoarthritis and rheumatoid
arthritis. J Rheumatol. 28:1492–1495. 2001.
|
|
8
|
Chiocchetti A, Indelicato M, Bensi T,
Mesturini R, Giordano M, Sametti S, Castelli L, Bottarel F,
Mazzarino MC, Garbarini L, et al: High levels of osteopontin
associated with polymorphisms in its gene are a risk factor for
development of autoimmunity/lymphoproliferation. Blood.
103:1376–1382. 2004. View Article : Google Scholar
|
|
9
|
Zhou ZT, Wei BJ and Shi P: Osteopontin
expression in oral lichen planus. J Oral Pathol Med. 37:94–98.
2008. View Article : Google Scholar
|
|
10
|
Santarelli A, Mascitti M, Rubini C,
Bambini F, Zizzi A, Offidani A, Ganzetti G, Laino L, Cicciù M and
Lo Muzio L: Active inflammatory biomarkers in oral lichen planus.
Int J Immunopathol Pharmacol. 28:562–568. 2015. View Article : Google Scholar
|
|
11
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Rese. 19:92–105. 2009. View Article : Google Scholar
|
|
12
|
Gassling V, Hampe J, Açil Y, Braesen JH,
Wiltfang J and Häsler R: Disease-associated miRNA-mRNA networks in
oral lichen planus. PLoS One. 8:e630152013. View Article : Google Scholar :
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
14
|
van der Waal I, Schepman KP and van der
Meij EH: A modified classification and staging system for oral
leukoplakia. Oral Oncol. 36:264–266. 2000. View Article : Google Scholar
|
|
15
|
Fischer AH, Jacobson KA, Rose J and Zeller
R: Hematoxylin and eosin staining of tissue and cell sections. CSH
Protoc. 2008:pdb prot4986. 2008.
|
|
16
|
Chanmee T, Ontong P, Kimata K and Itano N:
Key roles of Hyaluronan and its CD44 receptor in the stemness and
survival of cancer stem cells. Front Oncol. 5:1802015. View Article : Google Scholar :
|
|
17
|
Chaiyarit P, Thongprasom K, Satayut S,
Dhanuthai K, Piboonratanakit P, Phothipakdee P, Subarnbhesaj A,
Limlertmongkol S and Chaimusig M: Alteration of the expression of
CD44 [corrected] isoforms in oral epithelia and saliva from
patients with oral lichen planus. J Clin Immunol. 28:26–34. 2008.
View Article : Google Scholar
|
|
18
|
Liu GX, Sun JT, Yang MX, Qi XM, Shao QQ,
Xie Q, Qu X, Wei FC and Sun SZ: OPN promotes survival of activated
T cells by up-regulating CD44 in patients with oral lichen planus.
Clin Immunol. 138:291–298. 2011. View Article : Google Scholar
|
|
19
|
Kuninty PR, Bojmar L, Tjomsland V, Larsson
M, Storm G, Östman A, Sandström P and Prakash J: MicroRNA-199a and
−214 as potential therapeutic targets in pancreatic stellate cells
in pancreatic tumor. Oncotarget. 7:16396–16408. 2016. View Article : Google Scholar :
|
|
20
|
Liu Y, Zhou H, Ma L, Hou Y, Pan J, Sun C,
Yang Y and Zhang J: MiR-214 suppressed ovarian cancer and
negatively regulated semaphorin 4D. Tumour Biol. 37:8239–8248.
2016. View Article : Google Scholar
|
|
21
|
Xu Y and Lu S: Regulation of
β-catenin-mediated esophageal cancer growth and invasion by
miR-214. Am J Transl Res. 7:2316–2325. 2015.
|
|
22
|
Yu X, Luo A, Liu Y, Wang S, Li Y, Shi W,
Liu Z and Qu X: MiR-214 increases the sensitivity of breast cancer
cells to tamoxifen and fulvestrant through inhibition of autophagy.
Mol Cancer. 14:2082015. View Article : Google Scholar :
|
|
23
|
Phatak P, Byrnes KA, Mansour D, Liu L, Cao
S, Li R, Rao JN, Turner DJ, Wang JY and Donahue JM: Overexpression
of miR-214-3p in esophageal squamous cancer cells enhances
sensitivity to cisplatin by targeting survivin directly and
indirectly through CUG-BP1. Oncogene. 35:2087–2097. 2016.
View Article : Google Scholar
|